Abstract
Background: A variety of next-generation sequencing methods have been used to investigate the genomic landscape of primary lymphoma patient samples, including whole-genome, whole-exome, and RNA sequencing. In diffuse large B cell lymphoma (DLBCL), sequencing studies identified numerous genomic alterations (GAs) of potential clinical relevance, but the distribution and frequency of GAs have not been precisely determined. Discrepancies in existing data likely arise from variability in types of specimens examined, sequencing technologies employed, and depth of coverage utilized to identify GAs. In this study, we performed comprehensive DNA/RNA sequencing of genes known to be important across the spectrum of hematologic malignancies in order to determine the nature and prevalence of GAs with potential diagnostic, prognostic, or therapeutic implications in a cohort of 112 well-annotated clinical DLBCL cases.
Methods: We performed hybridization capture of 405 cancer-related genes and 31 genes commonly rearranged in cancer (FoundationOne Heme) and 265 frequently rearranged genes for RNA-seq applied to ³50ng of DNA and sequenced to high, uniform coverage. Genomic alterations, including short variants (small indels and base substitutions), rearrangements, and copy number alterations, were determined. All captured libraries were sequenced to high depth (Illumina HiSeq), averaging >500x for DNA and >20,000,000 total pairs for RNA, to enable the sensitive and specific detection of GAs. Significant non-synonomous variants were identified as mutations from the COSMIC database, amplifications of established oncogenes, or homozygous deletions and/or clear loss-of-function mutations of known tumor suppressors.
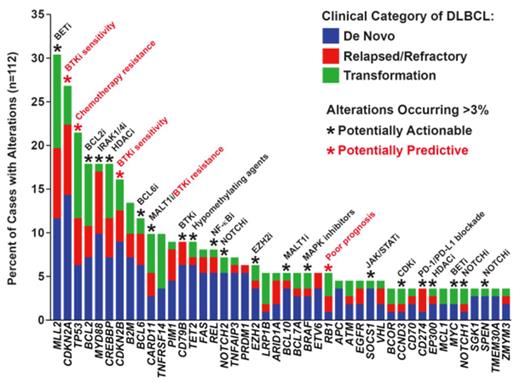
Results: GAs with diagnostic, prognostic, or therapeutic relevance were identified in 96% of cases, with a median of 5 (range 0 to 12) alterations per sample. Figure 1 shows the frequencies of different GAs. De novo DLBCL tended to exhibit fewer GAs (median 4) than relapsed/refractory (median 6) or transformed disease (median 6) (p=0.179; Wilcoxon rank sum). Compared with reported frequencies ranging from 5 to 44%, we detected alterations in CREBBP/EP300 in 21% of cases, with CREBBP mutations preferentially found in germinal-center B cell-like (GCB) compared to non-GCB DLBCL (26% vs. 8%; p=0.02; Pearson's chi-squared). Although previously described only in Burkitt lymphoma, we identified mutations in ID3 (L70P, P98R, Q100P) and TCF3 (N551K) in GCB-phenotype DLBCL cases with aggressive pathologic features and no MYC expression by IHC. Additional novel findings included alterations of genes involved in cellular metabolism in 18% of cases, including one IDH2 R172M mutation and two cases with SDHA L649fs* truncating mutations. We also identified several mutations more commonly found in solid tumors and leukemias, including BRAF V600E and KRAS G13D. With respect to genomic rearrangements, combined DNA/RNA capture and sequencing detected translocations/fusions in MYC, BCL2, and BCL6, which were concordant with cytogenetics/FISH analysis. We also identified rearrangements involving TBXL1XR1-P63, NOTCH2, SOCS1, and ETV6. Copy number alterations were detected in 26 different genes, including amplifications of CD274/PDCD1LG2 (PD-L1/PD-L2) (n=4) that were restricted to non-GCB cases. Alterations in TP53 were more frequently observed in transformed (42%) and relapsed/refractory (26%) compared to de novo DLBCL (12%; p=0.007; Pearson's chi-squared). TP53 mutations predicted for lack of response to chemotherapy, with chemo-refractory disease occurring in 48% of TP53-mutant patients compared to 10% of patients with intact TP53 (p=0.0004; Fisher's exact). Truncating mutations or deletions of RB1, albeit rare (5% of cases), were the most significant negative prognostic factor and were associated with therapeutic resistance and poor overall survival.
Conclusions: Our results demonstrate the utility of comprehensive combined DNA/RNA next-generation sequencing as a promising method to identify clinically relevant GAs in clinical lymphoma specimens. This streamlined approach has the potential to combine multiple molecular and cytogenetic tests into a single platform. Future efforts will be directed at incorporating this approach both retrospectively and prospectively into clinical trials to identify predictive biomarkers to guide therapeutic decisions.
Intlekofer:Foundation Medicine, Inc: Consultancy. Levine:Foundation Medicine, Inc: Consultancy. Zelenetz:Foundation Medicine, Inc: Consultancy. Dogan:Foundation Medicine, Inc: Consultancy. Palomba:Foundation Medicine, Inc: Consultancy. van den Brink:Foundation Medicine, Inc: Consultancy. Brennan:Foundation Medicine, Inc: Employment. Young:Foundation Medicine, Inc: Employment. He:Foundation Medicine: Employment. Nahas:Foundation Medicine, Inc: Employment. Yelensky:Foundation Medicine, Inc: Employment. Otto:Foundation Medicine, Inc: Employment. Lipson:Foundation Medicine, Inc: Employment. Stephens:Foundation Medicine, Inc: Employment. Miller:Foundation Medicine, Inc: Employment. Younes:Foundation Medicine, Inc: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal