Abstract
Background: MYC mRNA overexpression has been described in acute myeloid leukemia (AML), but no studies have assessed MYC protein expression in AML where its clinical significance is unknown. In this study our aim is to assess MYC protein expression across all AML subtypes and explore its prognostic value in in a subset of patients (pts) with particularly poor prognosis, namely those with AML with myelodysplastic syndrome (MDS)-related changes (AML-MRC) and therapy-related AML (t-AML). Characterized by unfavorable cytogenetics, the prognosis of patients with AML-MRC and t-AML is often dismal, even in patients who undergo allogeneic stem cell transplant. There is a need for further molecular characterization of AML-MRC and t-AML to identify prognostic markers to optimally risk-stratify to better guide treatment decisions in these pts.
Objectives: In the current study we sought to: (1) assess MYC protein expression by immunohistochemistry (IHC) performed on bone marrow (BM) specimens of pts with all types of AML, including acute promyelocytic leukemia (APL), and myelodysplastic syndromes (MDS), and (2) explore the prognostic significance of MYC protein expression in AML-MRC and t-AML (2008 WHO criteria).
Methods: MYC protein expression was assessed by IHC on BM obtained from pts with AML, APL, and MDS, and from patients without hematologic neoplasms (normal controls). MYC expression was considered positive if > 5% blasts in BM showed nuclear reactivity. For cases classified as t-AML and AML-MRC by WHO 2008, MYC expression was correlated with molecular, cytogenetic, and clinical outcome data. X-tile software was used to identify the optimal cutoff point to dichotomize pts by MYC protein expression.
Results: We evaluated BM MYC expression in 306 pts during 2006-2013 with newly diagnosed AML (n=246) or APL (n=11), previously treated AML (induction failure or relapse) (n=30), MDS (n=19), AML-MRC (n=68), t-AML (n=10), and normal BM (n=11). The median age was 61 yrs (13-88) with 54% men. Normal BM showed negligible MYC expression, ≤2% positive nuclei. The median MYC expression varied across diseases: newly diagnosed AML 25% (range 0-38%), APL 50% (range 19-75%), previously treated AML 20% (range 0-80), MDS 5% (range 0-30%) (differences between groups, p=0.0001).
High MYC expression correlated with high LDH (rs = 0.285, p<0.0001), WBC (rs = 0.225, p<0.0001) and BM blasts (rs 0.417, p<0.0001). Among newly diagnosed AML pts 20 had normal MYC expression ≤ 2%. MYC rearrangement by FISH was not detected in tested normal karyotype AML cases with variable MYC expression levels. MYC rearrangement by FISH was not detected in tested normal karyotype AML cases with variable MYC expression levels.
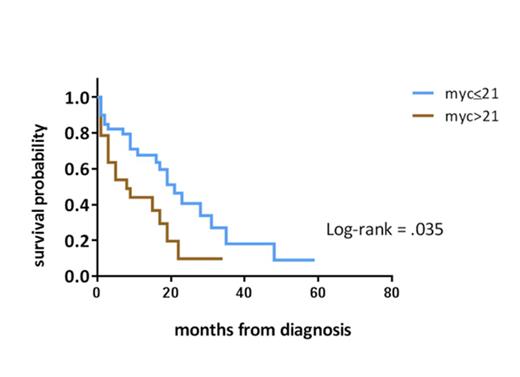
The clinical significance of MYC expression was examined in 78 pts with previously untreated AML-MRC and t-AML (59% men, median age 64 years). Pts with MYC expression above a determined cutoff of 21% had a significantly worse median overall survival (OS) of 8 months compared to 17 months in those with MYC ≤ 21% (p=0.035).
Disease free survival (DFS) was also inferior in pts with higher MYC expression. Median DFS was 3 months in pts with MYC >21% and 8 months for pts with MYC ≤ 21% (p=0.064). The rate of CR was 63% in pts with MYC≤ 21% and 37% in those >21%, however this difference was not statistically significant.
In pts with newly diagnosed AML-MRC and t-AML, high MYC expression did not correlate with markers of proliferation (high LDH, WBC and BM blasts). Furthermore, MYC levels were similar across karyotypes: trisomy 8 (where MYC resides), complex, and core binding factors.
Conclusion: MYC protein expression is a strong predictor of survival and an important biomarker of prognosis in patients with t-AML and AML-MRC. On BM IHC, pts with >21% MYC-positive blasts have an inferior OS, DFS, and remission rate than pts with ≤ 21% MYC-positive blasts. Increased MYC protein expression is independent of rearrangement status by FISH.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal