Key Points
The threonine kinase TAK1 is a survival-related gene that is strongly upregulated in AML CD34+ cells vs NBM CD34+ cells.
Genetic and pharmacologic inhibition of TAK1-induced cell death in an NF-κB–dependent manner in AML cells in vitro and in vivo.
Abstract
Development and maintenance of leukemia can be partially attributed to alterations in (anti)-apoptotic gene expression. Genome-wide transcriptome analyses revealed that 89 apoptosis-associated genes were differentially expressed between patient acute myeloid leukemia (AML) CD34+ cells and normal bone marrow (NBM) CD34+ cells. Among these, transforming growth factor-β activated kinase 1 (TAK1) was strongly upregulated in AML CD34+ cells. Genetic downmodulation or pharmacologic inhibition of TAK1 activity strongly impaired primary AML cell survival and cobblestone formation in stromal cocultures. TAK1 inhibition was mainly due to blockade of the nuclear factor κB (NF-κB) pathway, as TAK1 inhibition resulted in reduced levels of P-IκBα and p65 activity. Overexpression of a constitutive active variant of NF-κB partially rescued TAK1-depleted cells from apoptosis. Importantly, NBM CD34+ cells were less sensitive to TAK1 inhibition compared with AML CD34+ cells. Knockdown of TAK1 also severely impaired leukemia development in vivo and prolonged overall survival in a humanized xenograft mouse model. In conclusion, our results indicate that TAK1 is frequently overexpressed in AML CD34+ cells, and that TAK1 inhibition efficiently targets leukemic stem/progenitor cells in an NF-κB–dependent manner.
Introduction
Due to a high incidence of relapse, the survival rate of acute myeloid leukemia (AML) patients is still <30%, despite intensive treatment with chemotherapy. It is assumed that a small population of quiescent leukemic stem cells (LSCs) with self-renewal properties persists within the bone marrow microenvironment. These LSCs are responsible for relapse of the disease posttreatment,1,2 suggesting that current therapies change the more rapidly dividing leukemic blasts, whereas the LSCs generally survive.
Leukemogenesis of stem cells is a process in which various cellular programs can be affected, including those that regulate apoptosis and differentiation.3 Modification of programmed cell death may be important not only for leukemic transformation, but also could contribute to tumor maintenance and chemoresistance. Dysregulation of a number of cell survival pathways, such as BCL2, p53, and nuclear factor κB (NF-κB), have been associated with aberrations in apoptotic responses of AML cells.4 In contrast to normal bone marrow (NBM) CD34+ cells, constitutive activation of NF-κB has been observed in AML CD34+ cells.5-8 In accordance with this activation, NF-κB-associated pathways are related to tumor formation and maintenance.9 Pharmacologic inhibition of NF-κB by proteasome inhibitors has been reported to induce cell death in the AML CD34+CD38- subfraction, both in vitro and in vivo.7,8
Transforming growth factor-β activated kinase 1 (TAK1)/MAP3K7 is a kinase upstream of NF-κB, which can be activated by a variety of cytokines including tumor necrosis factor (TNF)α, transforming growth factor-β, and IL-1. Subsequently, phosphorylation of TAK1 leads to downstream activation of several pathways, including the NF-κB, c-Jun N-terminal kinase (JNK), extracellular signal-regulated kinase (ERK), and p38 pathways.
Throughout embryonic development, TAK1 is required for angiogenesis and regulates the survival of endothelial cells and normal hematopoietic stem cells (HSCs).10,11 A strong reduction of HSCs in TAK−/− mice has been observed that can partially be rescued by the knockout of tumor necrosis factor receptor 1 and tumor necrosis factor receptor 2, suggesting that apoptosis upon TAK1 inhibition is largely mediated by TNFα signaling.12,13 In various cancer models, including colorectal cancer, skin tumors and mantle cell lymphoma, TAK1 has been shown to be essential for the survival of cancer cells, where it might affect tumor metastasis in a WNT-dependent manner.14-18 Furthermore, reduction of TAK1 activity specifically induced cell death within all these different tumors, both in vitro and in vivo. Studies in patients with esophageal squamous cell carcinomas and clear cell renal cell carcinoma have also demonstrated that high TAK1 expression is associated with unfavorable prognosis.19,20
We performed a detailed analysis of the apoptotic programs within the LSC-enriched CD34+ fraction of AML patients in comparison with normal CD34+ stem/progenitor cells. We discovered that TAK1 expression is elevated in a large subset of AML CD34+ cells. Pharmacologic or genetic inhibition of TAK1-induced cell death in AML CD34+ cells in an NF-κB-dependent manner and significantly prolonged survival in vivo. These results indicate that targeting of TAK1 could be a potential new strategy in the treatment of AML.
Methods
Micro-array analysis
Gene expression profiling apoptosis-related genes was based on previous studies using Illumina HumanHT-12 Expression BeadChips.21,22 These studies included 100 samples and were divided as follows: 62 AML CD34+ (4 in duplicate) and 38 NBM CD34+ samples. All samples were corrected for background using Illumina GenomeStudio and then jointly forced to positive values, normalized and transformed using the R packages Bioconductor and Lumi.23,24 Probes with a detection P > .01 in all samples, as provided by GenomeStudio, were deleted. Log2 transformation and quantile normalization were applied. As a measure of quality control, we performed a principal component analysis on the correlation matrix of all 100 samples.25 The first component was removed from the data.26 To ensure reliability and reproducibility of the results, we used multivariate permutations to determine the significance of our results.
A gene list composed of 386 apoptosis-related genes (in total, 650 probes) was constructed. This list was largely based on gene ontology terms 0097191 (extrinsic apoptotic signaling pathway) and 0097193 (intrinsic apoptotic signaling pathway) combined with human apoptotic proteins present in the UniProtKB database. Differential expression was considered significant at P < .0001. Average linkage hierarchical clustering with the centered correlation distance metric was performed using Cluster 3.0 and TreeView software (http://www.eisenlab.org/eisen/). Microarray data have been deposited in GEO, accession number GSE30029.
Cell culture
The human promyelocytic leukemia cell line HL-60 and the human monocytic cell line MOLM13 were cultured in RPMI 1640 supplemented with 10% fetal bovine serum. The human monocytic leukemia cell line Oci-AML3 was cultured in RPMI 1640 supplemented with 20% fetal bovine serum. The human erythroleukemic cell line TF-1 was cultured in RPMI 1640 supplemented with 10% fetal calf serum and 10 ng/mL granulocyte macrophage–CSF (Genetics Institute, Cambridge, MA).
Primary AML cells, NBM, and long-term cultures on stroma
Informed consent was obtained to use the AML blasts in accordance with the Declaration of Helsinki; the protocol was approved by the Medical Ethics Committee of the University Medical Center Groningen (UMCG).
AML mononuclear cells were isolated by density gradient centrifugation. CD34+ cells were selected by MiniMacs (Miltenyi Biotec, Amsterdam, The Netherlands). Thereafter, cells were expanded on MS5 stromal cells in long-term culture medium (αMEM supplemented with heat-inactivated 12.5% fetal calf serum (Sigma, Zwijndrecht, The Netherlands), penicillin, streptomycin, 2 mM glutamine, 57.2 μM β-mercaptoethanol (Sigma), 1 μM hydrocortisone (Sigma) with IL-3 (Gist-Brocades, Delft, The Netherlands), granulocyte colony-stimulating factor (Rhone-Poulenc Rorer, Amstelveen, The Netherlands), and thrombopoietin (Kirin, Tokyo, Japan) (20 ng/mL each), as previously described.27,28 Cultures were kept at 37°C and 5% CO2. Cultures were demipopulated weekly for analysis.
After achieving informed consent, bone marrow aspirates were obtained from patients who underwent a total hip replacement. The protocol for bone marrow collection was approved by the Institutional Review Board of the UMCG. All participants had normal general health, normal peripheral blood counts, and they did not suffer from a hematologic disorder.
Flow cytometry analysis
Antibodies were obtained from Beckton Dickinson and Biolegend (Alphen a/d Rijn, The Netherlands). Cells were incubated with antibodies at 4°C for 30 minutes. All fluorescence-activated cell sorter (FACS) analyses were performed on a FACScalibur (Becton Dickinson) and data were analyzed using FlowJo 7.6.1. Cells were sorted on a MoFLo-XDP or Astrios (DakoCytomation, Carpinteria, CA).
Lentiviral and retroviral transductions
ExtGLuc was retrieved from the rPLSI1180-ExtGLuc-IRES-hr-green fluorescent protein (GFP) vector (kindly provided by the Brentjens Laboratory29 ) by EcoRI/StuI (blunt), and ligated into EcoRI/SalI (blunt) sites of the third generation lentivector CD711B_1_pCDH_MSCV. Lentiviral particles were produced by transient transfection in 293T cells using pVSV, pREV, and pMDL-PRRE helper plasmids.
The lentivirus short hairpin RNA vectors targeting human TAK1 were obtained from Open Biosystems (Thermo Scientific, Clone ID: TRCN0000001554, TRCN0000001555, TRCN0000001556, and TRCN0000001558) targeting the following sequences: (1) AAACAATCCAAGAATCACTGC, (2) TATTAGGATGGTTCACACGGG, (3) ATTCCATCACAAGACACACTG, and (4) ATAGTATCATTTGTGGCAGGA. These hairpins were cloned into the pLKO.1 lentiviral vector containing GFP (kindly provided by Dr. J. Larsson) or mCherry. The pLKO.1 GFP vector containing a scrambled (SCR) short hairpin was used as control vector. Lentiviral particles were produced by transient transfection of 293T cells with the lentiviral expression vectors, and stable transduction of AML cell lines or CD34+ AML cells was performed, which have both been extensively described.28 The retroviral construct pCMV IKKβ S177E S181E has been previously described.30 Retroviral particles were made by transient transfection of PG13, and OCI-M3 cells were transduced with these particles. Transduction efficiency was measured by FACS analysis. Knockdown was investigated by quantitative reverse-transcription polymerase chain reaction and western blot analysis.
NF-κB assay
NF-κB activity was measured by enzyme-linked immunosorbent assay (ELISA). ELISA was performed using the TRANS-AM NF-κB p65 Transcription Factor Assay Kit (Active Motif, North America, Carlsbad, CA) by following the manufacturer’s recommendations and as previously described.31
NF-κB luciferase reporter assay
Migration assay
Migration assay of MOLM13 cells was performed in a transwell system (Corning Costar, Cambridge, UK) with an 8 μm pore size. Two days after transduction, cells were resuspended in 100 μL medium and added to the upper chamber; 600 uL of medium with and without 100 ng/mL stromal derived factor-1 was added to the lower chamber. Cells were incubated for 4 hours at 37°C and migrated and nonmigrated cells were counted.
Animal experiments
Eight- to 10-week-old female NSG (NOD.Cg-Prkdcscid ll2rgtm1Wjl/SzJ) were purchased from the Central Animal Facility breeding facility within the UMCG. Mouse experiments were performed in accordance with national and institutional guidelines, and all experiments were approved by the Institutional Animal Care and Use Committee of the University of Groningen. Prior to transplantations, mice were sublethally irradiated with a dose of 1.0 Gy. After irradiation, mice received Neomycin (3.5 g/L in drinking water), and soft food daily for 2 weeks. Mice were lateral tail vein injected with 200 000 MOLM13 luciferase GFP+ transduced with SCR or shTAK1 mCherry lentivirus.
Bioluminescence imaging was performed 17 days after injection of the cells using the IVIS Spectrum, whereby 100 μg of coelenterazine was intravenously injected into the retro-orbital plexus and the mice were imaged for 30 seconds. Bioluminescence was quantified using Living Image 2.50. Mice were sacrificed upon detection of clinical signs of disease.
The supplemental Methods are available online at the Blood Web site.
Results
Transcriptome profiling identified differences in apoptotic signaling in AML CD34+ cells compared with NBM CD34+ cells
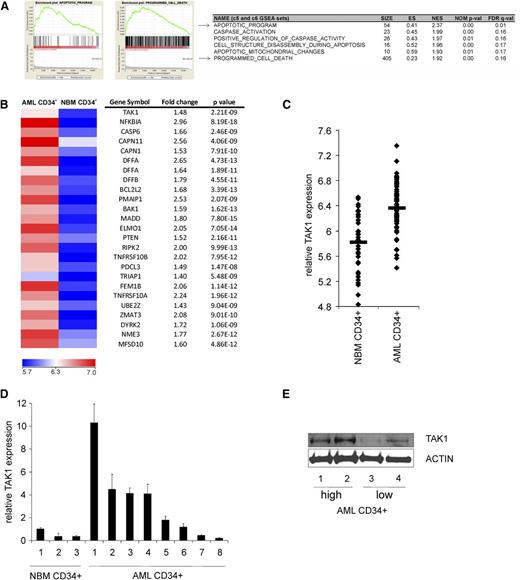
To evaluate the apoptotic programming in LSC-enriched AML CD34+ cells, transcriptome analyses was performed on a previously established gene expression data set of a large cohort of patient AML CD34+ cells (n = 62) and compared with NBM CD34+ cells (n = 38).21,22 Gene set enrichment analysis was performed, which revealed multiple apoptosis-associated sets to be highly enriched in AML CD34+ cells. In the top 30 significantly enriched gene sets, 6 sets were related to apoptosis, suggesting a major change in apoptotic programming in AML CD34+ cells compared with normal CD34+ cells (Figure 1A). Next, we determined the apoptosis-associated genes that were differentially expressed between AML and normal CD34+ cells by performing transcriptome analysis on 386 apoptosis-associated genes (supplemental Table 2). In total, 89 genes were differentially expressed between AML and normal CD34+ cells (P < 1e−6). Supervised cluster analysis using these 89 genes revealed that AML CD34+ cells and NBM CD34+ cells indeed clustered into 2 groups (supplemental Figure 1). Interestingly, the expression of various antiapoptotic and proapoptotic genes was increased in AML CD34+ cells compared with NBM CD34+ (supplemental Table 3). A heat map of the 25 most significantly upregulated genes is shown in Figure 1B, where the survival-related gene TAK1 was significantly higher expressed in AML CD34+ cells compared with NBM CD34+ cells. TAK1 was previously identified as one of the components of the LSC-related gene profile, which were significantly higher expressed in the AML LSC compared with the non-LSC fraction.33 Gene array data for TAK1 expression in all individual samples is shown in Figure 1C, showing an increased expression of TAK1 in a large subset of AML CD34+ cells. Elevated expression of TAK1 was independently confirmed by quantitative polymerase chain reaction (Figure 1D) and western blotting (Figure 1E). These findings indicate that TAK1 is overexpressed in AML CD34+ cells.
TAK1 expression is increased in AML CD34+ cell vs NBM CD34+ cells. (A) Gene set enrichment analysis profiles generated from the AML CD34+ vs NBM CD34+ gene list.22 (B) There were 25 most significantly upregulated apoptosis-associated genes in AML CD34+ cells vs NBM CD34+ cells. (C) Relative expression of TAK1 in all individual AML CD34+ and NBM CD34+ samples analyzed by gene profiling. (D) Relative expression of TAK1 AML CD34+ and NBM CD34+ cells by using quantitative polymerase chain reaction. (E) Relative protein levels of TAK1 in 2 AML CD34+ cells expressing high RNA levels of TAK1 and 2 AML CD34+ cells expressing low RNA levels of TAK1. ES, enrichment score; FDR, false discovery rate; GSEA, gene set enrichment analysis; NES, normalized enrichment score; NOM, nominal P value.
TAK1 expression is increased in AML CD34+ cell vs NBM CD34+ cells. (A) Gene set enrichment analysis profiles generated from the AML CD34+ vs NBM CD34+ gene list.22 (B) There were 25 most significantly upregulated apoptosis-associated genes in AML CD34+ cells vs NBM CD34+ cells. (C) Relative expression of TAK1 in all individual AML CD34+ and NBM CD34+ samples analyzed by gene profiling. (D) Relative expression of TAK1 AML CD34+ and NBM CD34+ cells by using quantitative polymerase chain reaction. (E) Relative protein levels of TAK1 in 2 AML CD34+ cells expressing high RNA levels of TAK1 and 2 AML CD34+ cells expressing low RNA levels of TAK1. ES, enrichment score; FDR, false discovery rate; GSEA, gene set enrichment analysis; NES, normalized enrichment score; NOM, nominal P value.
Inhibition of TAK1-impaired expansion of AML cell lines by increased apoptosis
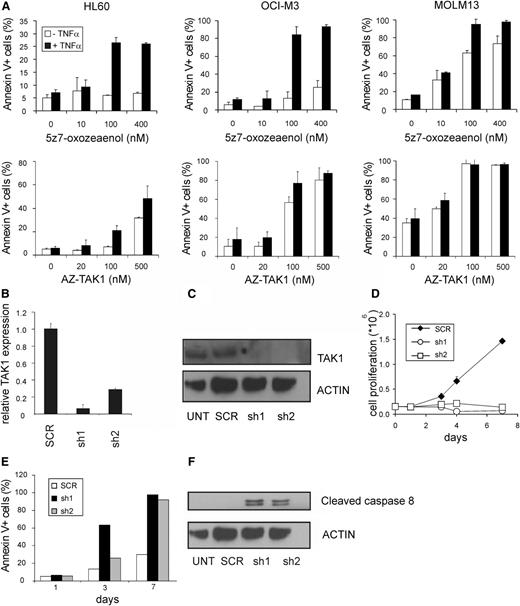
To study the functional consequences of TAK1 expression in AML cells, AML cell lines were exposed to the TAK1 inhibitors 5z-7-oxozeaenol and AZ-TAK1, which block the phosphorylation, and thereby the activity of TAK1.34 Whereas 5z-7-oxozeaenol is a resorcyclic acid lactone of fungal origin, AZ-TAK1 has recently been discovered in a small molecule-based lead identification to find compounds that specifically inhibit TAK1 phosphorylation. Both compounds inhibit the kinase activity at a nanomolar range (8 nM) by binding into the adenosine triphosphate-binding pocket.14,34,35 In the present study, we tested the functionality of these inhibitors on the AML cell lines HL60, OCI-M3, and MOLM13. Low concentrations of 5z-7-oxozeaenol and AZ-TAK1 resulted in increased apoptosis in most of the cell lines (Figure 2A) and could be further promoted when 5z-7-oxozeaenol was combined with TNFα, as previously indicated.36 With the AZ-TAK1 inhibitor, comparable effects were observed with or without TNFα addition; therefore, the additional experiments with AZ-TAK1 were performed without TNFα. To confirm the specificity of the effects, we constructed 2 independent pLKO.1 GFP short hairpins targeting TAK1. Efficient downmodulation of TAK1 in HL60 cells was confirmed at the RNA and protein level (Figure 2B-C). TAK1 depletion resulted in a growth disadvantage of AML cell lines (Figure 2D). This growth disadvantage was due to enhanced apoptosis as indicated by an increase of annexin V positive cells over time and the presence of cleaved caspase 8 (Figure 2E-F). Taken together, these results indicate that TAK1 is critical for the cell survival of AML cell lines.
TAK1 inhibition induces apoptosis in AML cells. (A) HL60, OCI-M3, and MOLM13 cells were incubated with various concentrations of TAK1 inhibitors 5z-7-oxozeaenol and AZ-TAK1, alone, or in combination with TNFα, and apoptosis was quantified by using annexin V staining. (B) Relative expression of TAK1 in HL60 cells using 2 independent hairpins targeting TAK1. (C) Relative TAK1 protein expression of short hairpin TAK1 (shTAK1) or scrambled transduced HL60 cells. (D) Cell growth of MOLM13 cells transduced with TAK1 hairpins or scrambled hairpin. (E) Annexin V staining of the shTAK1 or scrambled transduced MOLM13 cells. (F) Cleaved caspase-8 protein expression of shTAK1 and scrambled transduced HL60 cells at day 1 after transduction. UNT, untransduced.
TAK1 inhibition induces apoptosis in AML cells. (A) HL60, OCI-M3, and MOLM13 cells were incubated with various concentrations of TAK1 inhibitors 5z-7-oxozeaenol and AZ-TAK1, alone, or in combination with TNFα, and apoptosis was quantified by using annexin V staining. (B) Relative expression of TAK1 in HL60 cells using 2 independent hairpins targeting TAK1. (C) Relative TAK1 protein expression of short hairpin TAK1 (shTAK1) or scrambled transduced HL60 cells. (D) Cell growth of MOLM13 cells transduced with TAK1 hairpins or scrambled hairpin. (E) Annexin V staining of the shTAK1 or scrambled transduced MOLM13 cells. (F) Cleaved caspase-8 protein expression of shTAK1 and scrambled transduced HL60 cells at day 1 after transduction. UNT, untransduced.
Cell death induced by TAK1 inhibition was clearly associated with inhibition of the NF-κB pathway
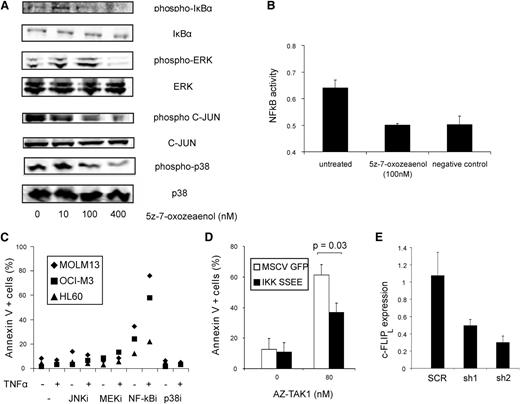
To unravel the downstream signaling pathways responsible for the TAK1-mediated cell death, various known pathways downstream of TAK1 were studied. Upon inhibition of TAK1 by 5z-7-oxozeaenol, phosphorylation of JNK, p38, ERK, and IκBα was inhibited (Figure 3A). Downstream activation of NF-κB was completely abolished, as demonstrated by a p65 activity assay (Figure 3B). Furthermore, a decrease in IL8 messenger RNA levels, which transcription is NF-κB dependent, was observed upon addition of 5z-7-oxozeaenol or transduction with TAK1 hairpins (supplemental Figure 2A-B). To determine which downstream pathways were mainly responsible for TAK1-mediated cell death, AML cell lines were treated with either the JNK inhibitor SP600125, the mitogen-activated protein kinase (MEK)/ERK inhibitor U0126, the NF-κB inhibitor BMS-345541, and p38 inhibitor SB203580, alone, or in combination with TNFα. Addition of the NF-κB inhibitor BMS-345541 induced apoptosis in MOLM13, OCI-M3, and HL60 cells, which increased significantly in combination with TNFα (2.4-fold; P = .02) (Figure 3C). Interestingly, the sensitivity of the 3 AML cell lines to the NF-κB inhibitor was comparable to the sensitivity to both TAK1 inhibitors. In contrast, inhibition of the p38, MEK/ERK, and JNK signaling pathways, either alone, or in combination with TNFα, caused limited cell death (<14%) (Figure 3C), although the p38 inhibitor, MEK/ERK inhibitor, and the JNK inhibitor were effectively inhibiting their target (supplemental Figure 2C). To show that TAK1-mediated survival was mainly due to decreased NF-κB activation, we transduced the OCI-M3 cells with IKKβ S177E, S181E (IKK SSEE) mutant, which mimics the active state of p65.37 Constitutive activation of NF-κB by these constructs was detected by performing an NF-κB luciferase reporter assay. This assay confirmed that expression of IKK SSEE constitutively triggered NF-κB activation in these cells (supplemental Figure 3). The cell death induced by the TAK1 inhibitor AZ-TAK1 was significantly reduced (P = .03) upon overexpression of IKK SSEE OCI-AML3 cells, indicating that NF-κB is an important mediator of the prosurvival pathway downstream of TAK1 (Figure 3D).
Apoptosis induced by TAK1 inhibition is mainly dependent on NF-κB pathway. (A) OCI-M3 cells were treated with various concentrations of 5z-7-oxozeaenol, and western blot analysis was performed on phospho-IκBα, total IκBα, phospho-ERK, total ERK, phospho-c-JUN, total C-JUN, phospo-p38, and total p38. (B) NF-κB activity of OCI-M3 cells treated with 100 nM 5z-7-oxozeaenol. NF-κB activity was measured by p65 DNA-binding enzyme-linked immunosorbent assay. The negative control is the background (no nuclear extract). (C) MOLM13, OCI-M3, and HL60 cells were treated with 10 μM JNK inhibitor SP600125, 5 μM MEK/ERK inhibitor U0126, 5 μM NF-κB inhibitor BMS-345541, or 1 μM p38 inhibitor SB203580, alone, or in combination with TNFα. After 24 hours of incubation, apoptosis was quantified by annexin V staining. (D) OCI-M3 cells were transduced with control MIGR1 or IKK SSEE vector and incubated with 80 nM AZ-TAK1. After 24 hours, annexin V+ cells were measured. (E) Relative cFLIPL levels of SCR HL60 and shTAK1 cells were quantified by quantitative polymerase chain reaction. MSCV, murine stem cell virus.
Apoptosis induced by TAK1 inhibition is mainly dependent on NF-κB pathway. (A) OCI-M3 cells were treated with various concentrations of 5z-7-oxozeaenol, and western blot analysis was performed on phospho-IκBα, total IκBα, phospho-ERK, total ERK, phospho-c-JUN, total C-JUN, phospo-p38, and total p38. (B) NF-κB activity of OCI-M3 cells treated with 100 nM 5z-7-oxozeaenol. NF-κB activity was measured by p65 DNA-binding enzyme-linked immunosorbent assay. The negative control is the background (no nuclear extract). (C) MOLM13, OCI-M3, and HL60 cells were treated with 10 μM JNK inhibitor SP600125, 5 μM MEK/ERK inhibitor U0126, 5 μM NF-κB inhibitor BMS-345541, or 1 μM p38 inhibitor SB203580, alone, or in combination with TNFα. After 24 hours of incubation, apoptosis was quantified by annexin V staining. (D) OCI-M3 cells were transduced with control MIGR1 or IKK SSEE vector and incubated with 80 nM AZ-TAK1. After 24 hours, annexin V+ cells were measured. (E) Relative cFLIPL levels of SCR HL60 and shTAK1 cells were quantified by quantitative polymerase chain reaction. MSCV, murine stem cell virus.
The long isoform of cellular FLICE-inhibitory protein (c-FLIPl) is one of the anti-apoptotic gene targets of NF-κB and has also been shown to overcome the cell death-inducing effect of TAK1 in MEFs.38 Therefore, to determine whether a correlation exists between TAK1 and cFLIPL, RNA levels of c-FLIPL were measured in shTAK1-transduced HL60 cells. Downmodulation of TAK1 resulted in reduced gene expression levels of cFLIPL (relative expression level is 40% ± 14%; P < .05) (Figure 3E), indicating that c-FLIPL is one of the pro-survival signals provided by the TAK1–NF-κB axis.
Whereas NF-κB is commonly constitutively activated in AML,5,7 we verified whether AML CD34+ cells expressing high levels of TAK1 have also increased NF-κB activity by determining IL8 levels. We observed that AML CD34+ cells indeed express increased levels of IL8 (supplemental Figure 2D). Nevertheless, TAK1 levels were not linearly correlated with IL8 levels (supplemental Figure 2E-F), and a number of AMLs that expressed low levels of TAK1 also expressed high levels of IL8. This is possibly due to enhanced NF-κB activity that is observed in a wide spectrum of leukemia subtypes, which may reflect abnormalities or mutations of activators of NF-κB that activate NF-κB independent of TAK1, such as RAS.5
Impaired long-term growth of AML CD34+ cells on bone marrow stroma after inhibition of TAK1
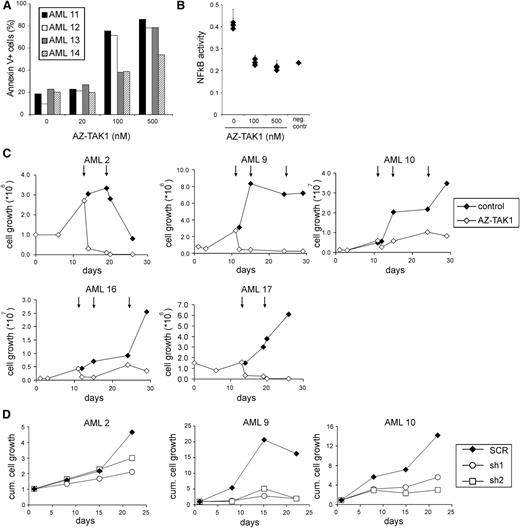
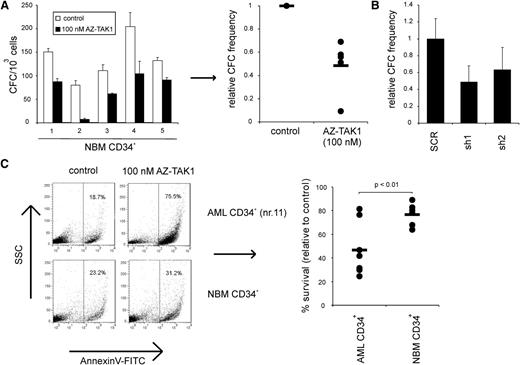
To assess whether TAK1 inhibition also affects the survival of primary AML CD34+ cells, we investigated the effectiveness of the TAK1 inhibitor AZ-TAK1 on primary AML CD34+ cells. After 24 hours of incubation with AZ-TAK1 of AML CD34+ cells, we observed a concentration-dependent effect on cell death (Figure 4A), which equally affected the AML CD34+CD38- and CD34+CD38− cell fraction (data not shown). In line with the reduction of NF-κB activity in AML cell lines upon TAK1 inhibition, a decrease in NF-κB activity was also seen in AML CD34+ patient cells after addition of 100 nM and 500 nM AZ-TAK1 (n = 3) (Figure 4B). More importantly, long-term growth of primary AML CD34+ cells (n = 5) on MS5 stromal layer was reduced upon addition of 100 nM AZ-TAK1 (Figure 4C). Moreover, addition of 500 nM AZ-TAK1 completely abrogated the outgrowth of AML CD34+ cells and no cobblestone formation could be observed under the MS5 stromal layer (supplemental Figure 4).
Targeting of TAK1 impairs expansion of primary AML CD34+ cells. (A) Primary AML CD34+ cells were incubated with various concentrations of AZ-TAK1 for 24 hours and cell death was quantified by annexin V staining. (B) NF-κB activity of primary AML CD34+ cells treated with 100 nM AZ-TAK1. NF-κB activity was measured by p65 DNA binding enzyme-linked immunosorbent assay. (C) Growth of primary AML CD34+ cells treated with 100 nM AZ-TAK1. AZ-TAK1 was added after initial growth was observed and added at the indicated time points (↓). (D) Growth of primary AML CD34+ cells transduced with hairpins targeting TAK1 or control hairpins.
Targeting of TAK1 impairs expansion of primary AML CD34+ cells. (A) Primary AML CD34+ cells were incubated with various concentrations of AZ-TAK1 for 24 hours and cell death was quantified by annexin V staining. (B) NF-κB activity of primary AML CD34+ cells treated with 100 nM AZ-TAK1. NF-κB activity was measured by p65 DNA binding enzyme-linked immunosorbent assay. (C) Growth of primary AML CD34+ cells treated with 100 nM AZ-TAK1. AZ-TAK1 was added after initial growth was observed and added at the indicated time points (↓). (D) Growth of primary AML CD34+ cells transduced with hairpins targeting TAK1 or control hairpins.
To verify the specificity of the effects, AML CD34+ cells (n = 3) were transduced with both TAK1 hairpins, and after 2 days the GFP+ cells were cultured on MS5 stromal layer. A significant reduction in growth of TAK1-hairpin transduced AML cells was observed in time, whereas scrambled transduced cells were able to expand on the stromal cells (Figure 4D). Also, a reduction of cobblestone frequency was observed in the shTAK1 AML cell cultures. Loss of suspension and adhesion cells in these long-term assays indicated that the primitive leukemic progenitor and stem cells were targeted in vitro.
NBM cells less affected upon TAK1 inhibition
To evaluate the effect of TAK1 inhibition on the survival of normal hematopoietic cells, NBM CD34+ cells (n = 5) were incubated with 100 nM AZ-TAK1 for 24 hours and were thereafter plated in methylcellulose. TAK1 inhibition resulted in a twofold decrease in colony forming cell (CFC) numbers (Figure 5A). In line with this data, shTAK1 transduced cord blood CD34+ cells also resulted in a twofold reduction in CFC numbers when plated in methylcellulose (Figure 5B).
Difference in sensitivity to TAK1 inhibition between NBM CD34+ cells and AML CD34+ cells. (A) NBM CD34+ cells (n = 5) were incubated with 100 nM AZ-TAK1 for 24 hours and after 2 weeks CFCs were determined. (B) Cord blood CD34+ cells were transduced with hairpins targeting TAK1; after 2 weeks, CFCs were determined. (C) AML CD34+ cells (n = 8) and NBM CD34+ cells (n = 6) were incubated with 100 nM AZ-TAK1; after 24 hours cell death was quantified by annexin V staining.
Difference in sensitivity to TAK1 inhibition between NBM CD34+ cells and AML CD34+ cells. (A) NBM CD34+ cells (n = 5) were incubated with 100 nM AZ-TAK1 for 24 hours and after 2 weeks CFCs were determined. (B) Cord blood CD34+ cells were transduced with hairpins targeting TAK1; after 2 weeks, CFCs were determined. (C) AML CD34+ cells (n = 8) and NBM CD34+ cells (n = 6) were incubated with 100 nM AZ-TAK1; after 24 hours cell death was quantified by annexin V staining.
Interestingly, upon treatment with 100 nM AZ-TAK1 for 24 hours, a small increase in annexin V+ cells was observed in NBM CD34+ cells (relative survival is 77% ± 9%; n = 6) upon TAK1 inhibition. However, the suppressive effect was significantly less pronounced (P < .01) compared with AML CD34+ cells (n = 8) treated for 24 hours with 100 nM AZ-TAK1 (Figure 5C). The observed variability in sensitivity between the various AML CD34+ cells was not related to expression levels of TAK1.
TAK1 inhibition resulted in impaired leukemia development and prolonged survival in an MOLM13 xenograft model
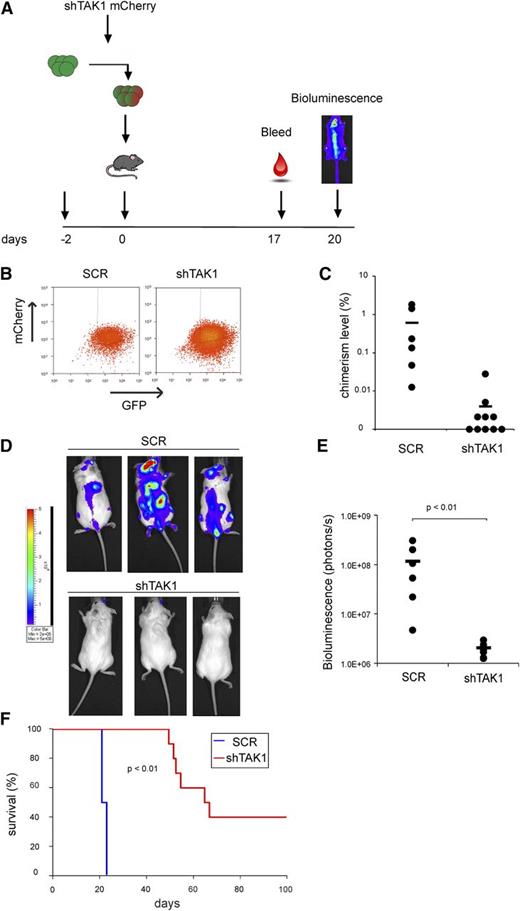
Because the bone marrow microenvironment may provide important signals that attenuate the effects seen upon TAK1 inhibition, we verified the effect of TAK1 inhibition on AML cells in vivo. MOLM13 Gaussia luciferase GFP-positive cells were transduced with lentiviral TAK1 mCherry hairpin or control constructs (Figure 6A). Two days after transduction, cells were sorted and intravenously injected into NSG mice (Figure 6B). The effectiveness of knockdown of TAK1 mCherry hairpins was verified by quantitative polymerase chain reaction (supplemental Figure 5A). Two weeks after injection, chimerism levels in peripheral blood were significantly lower in shTAK1 mice compared with SCR mice (Figure 6C). Moreover, luciferase activity was detectable in all SCR mice within the bone marrow, liver, and lungs, whereas shTAK1 mice only had minimal bioluminescence activity (Figure 6D-E). Kaplan Meier curves indicated that SCR mice survived for only 3 weeks (Figure 6F), with leukemia penetrance in the bone marrow, blood, spleen, and liver (supplemental Figure 5C). In contrast, TAK1 knockdown resulted in a significantly improved survival (P < .01) (Figure 6F). Efficiency of TAK1 knockdown on the day of sacrifice of some of the shTAK1 mice was lower than on the day of injection, suggesting that leukemia developed due to improper knockdown in these cells (supplemental Figure 5D). In the 40% of the shTAK1 mice that did survive throughout the experiment, no leukemic cells could be detected in the blood, bone marrow, liver, and spleen at the time these mice were sacrificed. An impaired homing of the transduced cells is unlikely to be responsible for the observed findings because MOLM 13 cells transduced with shTAK1 had a comparable CXCR4 expression level as the control cells (supplemental Figure 6A). In addition, no difference in migration to stromal derived factor-1 was observed between both cell populations (supplemental Figure 6B). These data indicate that targeting TAK1 strongly impairs leukemia development in vivo and results in a significantly improved survival.
TAK1 inhibition results in impaired leukemia development and increased survival in a MOLM13 xenograft model. (A) Schematic representation of mouse experiment. (B) Sorting strategy of MOLM13 luciferase GFP-positive cells transduced with either control mCherry vector or shTAK1 mCherry hairpin. (C) Chimerism levels in peripheral blood after 2 weeks of injection of MOLM13 luciferase GFP/SCR mCherry cells or MOLM13 luciferase GFP/shTAK1 mCherry cells. (D) Bioluminescence pictures of SCR mice and shTAK1 mice. (E) Quantification of bioluminescence assay of SCR mice (n = 6) and shTAK1 mice (n = 10). (F) Survival curve of SCR mice and shTAK1 mice.
TAK1 inhibition results in impaired leukemia development and increased survival in a MOLM13 xenograft model. (A) Schematic representation of mouse experiment. (B) Sorting strategy of MOLM13 luciferase GFP-positive cells transduced with either control mCherry vector or shTAK1 mCherry hairpin. (C) Chimerism levels in peripheral blood after 2 weeks of injection of MOLM13 luciferase GFP/SCR mCherry cells or MOLM13 luciferase GFP/shTAK1 mCherry cells. (D) Bioluminescence pictures of SCR mice and shTAK1 mice. (E) Quantification of bioluminescence assay of SCR mice (n = 6) and shTAK1 mice (n = 10). (F) Survival curve of SCR mice and shTAK1 mice.
Discussion
Evasion of apoptosis is one of the hallmarks during the development of cancer.39 Overexpression of multiple antiapoptotic genes of the mitochondrial related BCL-2 family has been related to the development and/or maintenance of AML and is prognostically significant for the treatment outcome of this group of patients.40-47 By comparing the apoptotic gene profile of AML CD34+ cells vs normal CD34+ cells, we observed a high expression of several antiapoptotic genes and, surprisingly, also some proapoptotic genes (supplemental Table 2). This has been previously observed in an independent cohort of AML and chronic myeloid leukemia patients.42,48 It is assumed that the increased expression of antiapoptotic genes, leading to oncogene addiction, is counterbalanced by elevated expression of proapoptotic genes, thus creating a “primed to cell death” status.49 In our present study, we observed increased expression of TAK1 in a large panel of AML CD34+ cells at RNA and protein level. Notably, TAK1 expression was recently shown to be part of a gene expression signature that defines AML stem cells.33 These findings led us hypothesize that TAK1 might play an important role in AML stem cell maintenance. The results of the present study indicate that TAK1 is highly relevant for the in vitro and in vivo survival of AML cells. TAK1 inhibition by genetic or pharmacologic inhibition strongly triggered cell death in AML CD34+ cells.
We observed that the prosurvival function of TAK1 was largely dependent on NF-κB activity, and less dependent on MEK, p38, and JNK activity, although all these pathways are triggered by TAK1 activation. NF-κB inhibition not only phenocopied the effects observed with the TAK1 inhibitors, but cell death upon TAK1 inhibition also could be partially rescued by overexpression of NF-κB. It has been demonstrated that NF-κB activity is elevated in patient AML blasts and AML CD34+ cells compared with normal CD34+ cells, in part due to the autocrine and paracrine production of growth factors.5,7,50 So far it appears that the enhanced NF-κB expression in AML is a more general result of the malignant transformation and is not related to specific AML subtypes or genetic abnormalities. It was recently demonstrated that NF-κB is activated in several leukemia mouse model systems, including MLL-ENL and MOZ-TIF2, and in the double hit model of BCR-ABL with NUP98-HOXA9. Moreover, in these studies the constitutive activation of NF-κB affected the primitive LSC fraction by expanding the numbers of leukemic initiating cells.51,52
An important mediator for the constitutive activation of NF-κB is the autocrine production of TNFα by leukemic cells.53 Apparently, the transforming events triggered by various mutated genes favor pathways with a prosurvival signature, which are subsequently highly relevant for tumor maintenance. An alternative pathway for NF-κB activation may be the TNF receptor associated factor 6 (TRAF6) and interleukin receptor associated kinase-1 (IRAK1), which are interacting proteins and mediators of toll-like receptors and IL-1 receptors. Activation of toll-like receptors or IL-1 receptors results in phosphorylation of IRAK1, leading to binding and activation of TRAF6. Thereafter, TAK1 will be phosphorylated, resulting in the subsequent activation of NF-κB. Recently, it was shown that IRAK1 is overexpressed and highly activated in high-risk MDS and AML.54 Inhibition of IRAK1 exhibited an impaired expansion and apoptosis and the cotreatment with BCL2 inhibitors eliminated the MDS clones.
Multiple transcriptome data sets of AML and normal CD34+ cells, including ours, indicate that besides TAK1, TRAF6 and IL1RAP are also significantly overexpressed in AML CD34+ cells,22,55-57 suggesting an ongoing activation of these pathways, subsequently leading to prosurvival signals. The higher expression of TAK1 in AML CD34+ cells compared with normal CD34+ cells suggests that leukemic cells are more dependent on this pathway than normal CD34+ cells for their survival, indicating a potential therapeutic window for drug targeting. The in vitro results of the TAK1 inhibitor are in line with these data, showing a difference in susceptibility for leukemic vs normal cells. Apparently, this type of NF-κB inhibition is more effective than NF-κB inhibition by proteasome inhibition.31,58
However, it cannot be excluded that other pathways besides NF-κB are affected because NF-κB overexpression could only partially rescue the phenotype. In addition, knockout of the components of the NF-κB complex, p65/RelA and p52/RelB, results in a significant decline in HSCs frequency,59,60 but the phenotypes appear to be less severe in comparison with the TAK1 knockout mice.11 These results are in line with the findings that different pathways converge at TAK1 and that various pathways, including NF-κB, are activated downstream of TAK1. In summary, our data demonstrate that TAK1 is a critical component in LSC maintenance and may be an innovative target for patient treatment.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Aneta Schaap-Oziemlak for generation of the extGLuc vector, Bibi van Keeken for her input in this project, and Roelof Jan van der Lei, Henk Moes, and Geert Mesander for their help with cell sorting.
This work was supported by an Ubbo Emmius grant from the University of Groningen.
Authorship
Contributions: M.C.J.B., W.J.Q., J.J.S., and E.V. conceived and designed the experiments; M.C.J.B., H.S., J.J., A.Z.B.-V., and J.J.S. performed the experiments; M.C.J.B., J.J.S., and E.V. analyzed the data; and M.C.J.B., J.J.S. and E.V. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jan Jacob Schuringa, Hanzeplein 1, 9713 GZ, Groningen, The Netherlands; e-mail: j.j.schuringa@umcg.nl; and Edo Vellenga, Hanzeplein 1, 9713 GZ, Groningen, The Netherlands; e-mail: e.vellenga@umcg.nl.
References
Author notes
J.J.S. and E.V. contributed equally as the last authors.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal