Key Points
MLL-AF6 leads to aberrant activation of RAS and its downstream targets.
RAS targeting is a novel potential therapeutic strategy in AML patients carrying t(6;11).
Abstract
A rare location, t(6;11)(q27;q23) (MLL-AF6), is associated with poor outcome in childhood acute myeloid leukemia (AML). The described mechanism by which MLL-AF6, through constitutive self-association and in cooperation with DOT-1L, activates aberrant gene expression does not explain the biological differences existing between t(6;11)-rearranged and other MLL-positive patients nor their different clinical outcome. Here, we show that AF6 is expressed in the cytoplasm of healthy bone marrow cells and controls rat sarcoma viral oncogene (RAS)-guanosine triphosphate (GTP) levels. By contrast, in MLL-AF6-rearranged cells, AF6 is found localized in the nucleus, leading to aberrant activation of RAS and of its downstream targets. Silencing MLL-AF6, we restored AF6 localization in the cytoplasm, thus mediating significant reduction of RAS-GTP levels and of cell clonogenic potential. The rescue of RAS-GTP levels after MLL-AF6 and AF6 co-silencing confirmed that MLL-AF6 oncoprotein potentiates the activity of the RAS pathway through retention of AF6 within the nucleus. Exposure of MLL-AF6-rearranged AML blasts to tipifarnib, a RAS inhibitor, leads to cell autophagy and apoptosis, thus supporting RAS targeting as a novel potential therapeutic strategy in patients carrying t(6;11). Altogether, these data point to a novel role of the MLL-AF6 chimera and show that its gene partner, AF6, is crucial in AML development.
Introduction
The mixed lineage leukemia (MLL) protein is a histone H3 lysine 4-specific methyltransferase, commonly associated with transcriptional activation.1 MLL is essential for both embryonic development and normal hematopoiesis, mainly through transcriptional regulation of the homeobox (HOX) gene.2 Chromosome translocations involving MLL locus are 1 of the major genetic lesions leading to acute leukemia. MLL translocations are detected in up to 80% of infant acute leukemia and in approximately 10% to 15% of childhood acute myeloid leukemia (AML).3,4 Aberrant proteins resulting from translocations, duplications, or amplifications of the MLL gene cause alteration of the differentiation program with severe effects on leukemogenesis.5,6 To date, more than 60 fusion partners of MLL have been described, which result in AML, acute lymphoid, and biphenotypic or chemotherapy-related leukemias.7,8 The underlying mechanisms for MLL-mediated leukemogenesis have been extensively studied; however, they still remain elusive for many of the described translocations. MLL-rearranged AML is, in fact, a heterogeneous disease, which depends on the MLL partner gene for its biological and clinical features, such as gene expression and genomic imbalances.4 Among diverse fusion genes, the 1 that has been consistently associated with the worst outcome both in adult and pediatric AML is MLL-AF6.9
The t(6;11)(q27;q23) translocation is not rare in childhood AML3,4 and has been demonstrated to impart a worse prognosis with respect to other forms of MLL-rearranged AML. AF6 is a cytoplasmic protein with 2 distinctive features: 1 single PDZ and 2 rat sarcoma viral oncogene (RAS)-association (RA) domains. The PDZ domain drives AF6 to specialized sites on the membrane, where it can interact with many molecules10-15 ; RA domains are homologous to RA domains of RAS effectors.12,16,17 Altogether, these characteristics enable AF6, either alone or when fused to MLL, to modulate multiple signal transduction pathways in vivo, especially those involving RAS, Notch, and Wnt.16 In the MLL-AF6 chimera, AF6 protein maintains its functional domains, both PDZ and RA, showing no homology, either for sequence or function, to the product of any other MLL partner gene.18,19
To define the role of MLL-fusion genes, several researches were conducted mostly on MLL-AF9 and MLL-AF10 chimeras, revealing that a functional hallmark of MLL fusion-chimera is a block of hematopoietic differentiation.20-23 The proposed tumorigenic mechanism of MLL-AF6 is based on the acquired aberrant transcriptional capability. In fact, the transcriptional activation of MLL is described to be dependent on MLL-AF6 homodimerization mediated by AF6, which acts as a scaffold protein that permits the interaction with target genes.24 Furthermore, Amstrong’s group25 recently showed that MLL-AF6 requires DOT1L histone-methyltransferase activity to maintain its gene-expression program, which is considered to be its main oncogenic force.25
In this study, we demonstrate that MLL-AF6 affects AF6 localization to be able to aberrantly activate RAS and its downstream signaling to empower the tumorigenic potential of myeloid cells. In particular, we show that MLL-AF6 sequesters AF6 in the nucleus, leading to increased levels of RAS-GTP in the cytoplasm. Silencing of the chimera re-localized the AF6 protein back into the cytoplasm, thus leading to reduction of both RAS levels and activity. These results imply that RAS may play a crucial oncogenic role in AML, prompting us to perform further experiments aimed at disrupting its function. Chemical inhibition of RAS signaling affected the proliferation of t(6;11)-rearranged cells to the same extent as that observed after silencing the chimera. Therefore, we explored the effects of a new targeted treatment, namely a farnesyltransferase inhibitor, tipifarnib, and demonstrated its efficacy in primary cultures from patients with t(6;11)-rearranged AML.
Materials and methods
Cell lines, primary cell cultures, and patient samples
MLL-AF6-rearranged ML2 and SHI-1 cell lines, MLL-AF9-rearranged NOMO1 and THP1 cell lines (DSMZ), and mononucleated cells obtained from whole bone marrow collected from pediatric healthy donors, and newly diagnosed t(6;11)MLL-AF6 and t(9;11)MLL-AF9-rearranged AML blasts were cultured in RPMI1640 (Invitrogen-Life Technologies, Monza, Italy) as previously described.26 Diagnosis of leukemia was established according to standard criteria based on immunohistochemical, immunophenotyping, and cytogenetic studies, as detailed in the AIEOP-2001/02 AML treatment protocol.27 In compliance with the Helsinki Declaration, informed consent was obtained from the parents of the patients.
Sequencing
Bone marrow samples of t(6;11) pediatric AML were analyzed for mutations affecting p53 (exon 6-exon 8); N-RAS, and K-RAS mutation were searched in the hotspot region of exon 1 and 2 at codon G12, G13, and Q61 by Sanger sequencing.
Immunofluorescence microscopy
Cytospins were incubated overnight at 4°C with 1:500 anti-AF6 (BD Biosciences, Milan, Italy) and anti-RAS (Cell Signaling Technology, Danvers, MA) antibodies. Slides were incubated with secondary antibodies conjugated to Alexa dyes (Invitrogen-Life Technologies). Cells were counterstained with 4ʹ,6-diamidino-2-phenylindole (DAPI; 1:10c000; Sigma-Aldrich, St. Louis, MO) to label nuclei. Staining was visualized by epifluorescence (video-confocal, Vico; Nikon, Tokyo, Japan).
Western blot
Total proteins lysates (20 μg) were isolated as previously described,26 and were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Immunoblots were hybridized with anti-C-terminal portion of AF6 (BD Biosciences); anti-β-actin, anti-c-RAF, anti-MEK1/2 (Sigma-Aldrich); anti-HDAC1 (Santa Cruz Biotechnology, Dallas, TX); anti-RAS, anti-phospho-c-RAF (Ser338), anti-phospho-MEK1/2 (Ser217/221), anti-total and phospho- extracellular signal-regulated kinase (ERK)1/2 (Thr202/Tyr204), anti-LC3 (Novus Biologicals, Littelton, CO), and anti-p62 (Abnova, Taipei City, Taiwan). Enhanced chemiluminescence western blot detection reagents and films (GE Healthcare, Cleveland, OH) were used. Densitometric analyses for protein quantification were carried out using the ImageJ 1.38x software (http://rsbweb.nih.gov/ij/index.html). The value of each band was normalized to the value of either β-actin or total-RAS protein.
RPPA
RAS activation assay
A total of 20 × 106 healthy bone marrow (HBM) or transiently silenced ML2 and SHI-1 cells were lysed, and 500 μg of protein extract were used for the RAS activation kit (ENZO Life Sciences, Lausen, Switzerland) according to the manufacturer’s instructions. Positive control samples were obtained by treating the lysates with guanosine-5′-O-(3-thio) triphosphate (GTP-γS) at a final concentration of 0.1 mM to activate endogenous RAS.
Immunoprecipitation
ML2, SHI-1, and HBM cells were immunoprecipitated with 8 μg of anti-RAS or anti-AF6 antibody, as previously described.26 The immunoprecipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
RNA interference
Two MLL-AF6-specific small-interfering (si)RNAs (siMAF6; QIAGEN GmbH, Hilden, Germany) were designed on the fusion breakpoint to selectively silence the chimera. To silence AF6, siAF6 (FlexiTube siRNA Hs_MLLT4; QIAGEN GmbH) was transfected into HBM cells. Double silencing with both siMLL-AF6 and siAF6 was also performed for rescue experiments. AllStars Negative Control siRNA (scramble RNA [scRNA]; QIAGEN GmbH) was used as control in each experiment. Cell transfections were performed using the Nucleofector systems (Amaxa Biosystems, Lonza Sales Ltd., Basel, Switzerland) according to the manufacturer’s instructions.
RNA isolation and SYBR green qRT-PCR assays
Total RNA was extracted with Trizol reagent (Invitrogen-Life Technologies). RNA (1 μg) was reverse-transcribed using the SuperScript II system (Invitrogen-Life Technologies) and random hexamers, according to the manufacturer’s instructions. Quantitative real-time polymerase chain reaction (qRT-PCR) reactions were performed in triplicate on the Applied Biosystems 7900 HT (Applied Biosystems-Life Technologies). The comparative threshold cycle (Ct) method of relative quantification was applied to interpret the results.30
Luciferase assay
A mixture of pFOS WT-GL3 (Addgene, #11983) 31 plasmid, Renilla plasmid (REN), siMAF6 or siMAF6 and siAF6 were used to co-transfect cell lines, whereas a mixture of pFOS WT-GL3, REN, and scRNA was used as a control. The qRT-PCR was used to monitor gene silencing. Protein lysates were analyzed for RAS activity by measuring LUC and REN levels using the Dual Luciferase Assay System (Promega Corporation, Madison, WI). LUC activity was normalized to REN activity.
Soft agar colony assay
After MLL-AF6 silencing, a total of 2 × 103 ML2 and SHI-1 cells were seeded onto a minimum methylcellulose semisolid dish (StemCell Technologies, Vancouver, Canada) and incubated at 37°C. Fourteen days after transduction, colonies were counted by light microscopy after incorporation of 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT).
Apoptosis analysis
Transiently transfected cells were collected, double-stained with Annexin-V/propidium iodide (Immunostep-Valter Occhiena, Turin, Italy) and analyzed using Cytomics FC500 (Beckman Coulter, Brea, CA). Relative apoptosis was calculated and expressed as the percentage of Annexin-V-positive/propidium iodide–positive cells. Cell lines or cultures of primary AML blasts harboring either t(6;11) or t(9;11) were seeded at 106 cells per well and treated for 24 hours with a concentration of tipifarnib ranging from 0.1 μM to 10 μM.
Microarray analysis
RNA was extracted from bone marrow of 11 MLL-AF6-rearranged patients, as well as from a series of 11 HBM. RNA quality was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). The GeneChip Human Genome U133 Plus 2.0 array was used and analyzed as previously described.32 The Student t test was used for supervised analysis between t(6;11)-rearranged patients and HBM samples (GSE 19577). To control for false discovery rate, multiplicity corrections were used; probes with adjusted P values <.01 were declared significant using Partek Genomic Suite Software. DAVID Functional Annotation Bioinformatics Microarray Analysis was used to identify molecular networks among differentially expressed genes.
Reagents and treatments
ML2 and SHI-1 cell lines were treated with PD98059 (40 μM, Calbiochem, Merck Group, Darmstadt, Germany), a MEK inhibitor. ML2, SHI-1, harboring the t(6;11), and NOMO-1 and THP-1 cell lines harboring t(9;11), as well as primary AML cells from patients with the same 2 translocations were treated with tipifarnib (0.1-100 μM, Aurogene Srl, Rome, Italy). The MTT test was used to assess cell proliferation.
Data analysis
Statistical analysis was performed using Prism 4.02 (Graph Pad Software, San Diego, CA). Experiments were performed in duplicate or triplicate, and results were presented as mean ± standard error of the mean of replicate experiments. Statistical significance was evaluated by the unpaired Student t test. Differences were considered to be statistically significant at P values <.05 and were indicated with an asterisk.
Results
MLL-AF6 modifies AF6 localization, maintaining high RAS-GTP levels
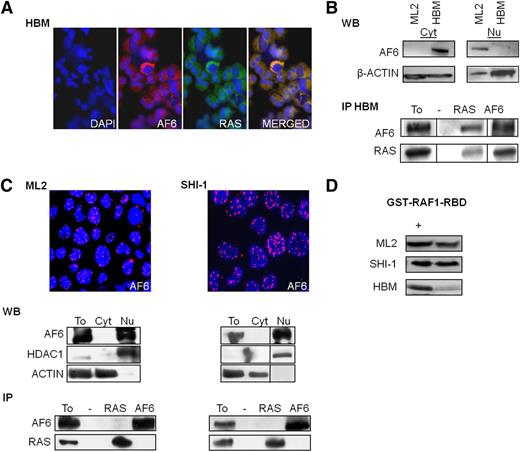
There are alternative AF6 isoforms with described specific subcellular localization. In particular, AF6 has been found ubiquitously expressed in different cell types, having a cytoplasmic localization. A shorter splice variant of AF6 with nuclear localization was reported exclusively in cells of the central nervous system.33 To define the distribution of AF6 in normal hematopoietic cells, HBM cells were immunostained with an AF6-specific antibody. In these cells, AF6 was expressed within the cytoplasm (Figure 1A). Western blot analysis confirmed its localization; in fact, only the cytoplasmic (Cyt) protein fraction showed AF6 expression. Furthermore, in HBM, AF6 was found to co-localize with RAS; these data were confirmed by coimmunoprecipitation experiments, as previously described in other systems (Figure 1B).14,16,31 ML2 and SHI-1, 2 leukemia cell lines carrying the t(6;11)(q27;q23)MLL-AF6 translocation, were also investigated for AF6 localization. Immunofluorescence showed a nuclear punctate localization for AF6 in both t(6;11)-rearranged cell lines. Nuclear subcellular position of AF6 protein was then confirmed by western blot analysis. Immunoprecipitation assay demonstrated no interaction between AF6 and RAS in t(6;11)-rearranged cell lines due to the nuclear localization of AF6 (Figure 1C). We subsequently measured RAS activity in HBM and in leukemia cell lines by glutathione S-transferase (GST)-pulldown of the active form of RAS. ML2 and SHI-1 cells had an elevated amount of RAS in its active GTP-bound status, whereas the amount of active RAS was low in HBM cells, thus supporting the hypothesis that AF6 may act as repressor of RAS activity in normal hematopoietic cells (Figure 1D).
MLL-AF6 modifies AF6 localization from cytosol to nuclear. (A) AF6 colocalizes with RAS (merged) in cytosol of healthy bone marrow (HBM) cells: (left) immunofluorescence of HBM primary cells stained with DAPI and respective antibodies against AF6 and RAS (×20 zoom). (B) (Top) Western blot analysis (WB) of AF6 and RAS expression in cytoplasmic (Cyt) and nuclear (Nu) cell extracts. (Bottom) coimmunoprecipitation (IP) of AF6 and RAS in HBM cells. Total lysates (To) were used as positive controls; negative controls (−). (C) (Top) Nuclear localization of AF6 in ML2 and SHI-1 cell lines by immunofluorescence (AF6 red, nuclei stained with DAPI in blue, ×20 zoom). (Middle) WB of AF6 and RAS expression in total (To), cytoplasmic (Cyt) and nuclear (Nu) cell extracts; anti-HDAC1 and anti-ACTIN were used as endogenous controls for nuclear and cytoplasmic proteins, respectively. (Bottom) coimmunoprecipitations (IP) between RAS and AF6 showed no interaction between the 2 proteins in neither of t(6;11) leukemic cell lines. (D) Active RAS-GTP levels in ML2, SHI-1, and HBM cells; positive control (+). Vertical lines have been inserted to indicate repositioned gel lanes.
MLL-AF6 modifies AF6 localization from cytosol to nuclear. (A) AF6 colocalizes with RAS (merged) in cytosol of healthy bone marrow (HBM) cells: (left) immunofluorescence of HBM primary cells stained with DAPI and respective antibodies against AF6 and RAS (×20 zoom). (B) (Top) Western blot analysis (WB) of AF6 and RAS expression in cytoplasmic (Cyt) and nuclear (Nu) cell extracts. (Bottom) coimmunoprecipitation (IP) of AF6 and RAS in HBM cells. Total lysates (To) were used as positive controls; negative controls (−). (C) (Top) Nuclear localization of AF6 in ML2 and SHI-1 cell lines by immunofluorescence (AF6 red, nuclei stained with DAPI in blue, ×20 zoom). (Middle) WB of AF6 and RAS expression in total (To), cytoplasmic (Cyt) and nuclear (Nu) cell extracts; anti-HDAC1 and anti-ACTIN were used as endogenous controls for nuclear and cytoplasmic proteins, respectively. (Bottom) coimmunoprecipitations (IP) between RAS and AF6 showed no interaction between the 2 proteins in neither of t(6;11) leukemic cell lines. (D) Active RAS-GTP levels in ML2, SHI-1, and HBM cells; positive control (+). Vertical lines have been inserted to indicate repositioned gel lanes.
Silencing of MLL-AF6 in t(6;11)-rearranged AML cell lines restores AF6 localization in the cytoplasm and reduces RAS hyperactivation
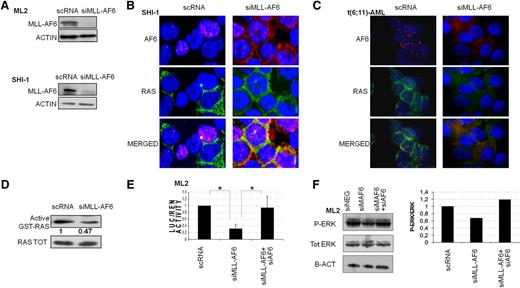
Silencing of MLL-AF6 in ML2 and SHI-1 decreased its messenger RNA expression as compared with cell lines transfected with a nonsilencing scramble siRNA (scRNA). In particular, ML2 showed an average reduction of 46% and 38% at 24 and 48 hours, respectively, whereas SHI-1 showed an average reduction of 53% and 19% at the same time points (by qRT-PCR, data not shown). Chimera protein levels were decreased, as shown by western blot experiments (Figure 2A). AF6 and RAS localization was investigated after silencing. We demonstrated that AF6 was in the cytoplasm in the MLL-AF6-silenced cell lines (Figure 2B), as well as in a primary sample of a t(6;11)-rearranged patient (Figure 2C), whereas it was retained in nuclear foci in scRNA-transfected cells. We also showed a restored colocalization of AF6 and RAS in the cytoplasm (Figure 2B-C), as observed in HBM. We measured the active GTP-bound status of RAS and found that it was decreased in cells silenced for MLL-AF6 compared with scRNA (0.47 vs 1, respectively) (Figure 2D), suggesting that the chimera maintained AF6 within the nucleus, preventing its control over RAS activation. To confirm our hypothesis, we set up a luciferase assay system in which t(6;11)-rearranged cell lines were cotransfected together with siRNA for MLL-AF6 and a luciferase reporter of RAS activity. Results showed that luciferase activity was reduced after MLL-AF6 silencing and AF6 relocalization in the cytoplasm, supporting RAS activity impairment (n = 3; P = .008) (Figure 2E) and the correlation between these events. We then confirmed that cosilencing both MLL-AF6 and AF6 increased the luciferase activity because of RAS rescued expression (n = 3; P = .05) (Figure 2E). This phenomenon was also reinforced by measuring the main RAS target, ERK, which showed a decrease in phosphorylated ERK (p-ERK) form in MLL-AF6-silenced ML2, over the total ERK protein which remained at the same levels. By contrast, the ratio p-ERK/ERK increased again after MLL-AF6 and AF6 cosilencing (the ratio is represented in the histogram). In line with our hypothesis, the chimera silencing promoted the relocalization of AF6 into the cytoplasm, thus restoring its control over RAS activity. In the same system, by reducing AF6 levels, we rescued RAS activity.
Silencing of MLL-AF6 in t(6;11)(q27;q23) rearranged cells restores AF6 in the cytoplasm. (A) Western blot (WB) analysis revealed decreased levels of MLL-AF6 after silencing (siMLL-AF6) compared with negative controls (scRNA) in both, ML2 and SHI-1. Anti-ACTIN was used as endogenous control. (B) The siMLL-AF6 cells restored AF6 protein in the cytoplasm. Immunofluorescence shows colocalization of AF6 (red) and RAS (green) in SHI-1 after MLL-AF6 silencing (merged signals, yellow, nuclei blue, ×60 zoom). In the negative control (scRNA), the punctuate pattern of AF6 nuclear localization is visible (red AF6, nuclei blue, ×60 zoom). (C) The siMLL-AF6 cells restored AF6 protein in the cytoplasm. Immunofluorescence shows colocalization of AF6 (red) and RAS (green) in primary t(6;11)-AML after MLL-AF6 silencing (merged signals yellow, nuclei blue, ×60 zoom). In the negative control (scRNA), the punctuate pattern of AF6 nuclear localization is visible (red AF6, nuclei blue, ×60 zoom). (D) Active RAS-GTP levels in ML2 cell line silenced for the chimera showed a decreased activity of RAS (0.47) compared with scRNA. (E) Luciferase (LUC) activity of ML2 transfected with a pFOS (FBJ murine osteosarcoma viral oncogene homolog) WT-GL3 plasmid and siMLL-AF6 show a reduction of LUC activity compared with scRNA. Introduction of both siRNA for MLL-AF6 and AF6 show a rescue of LUC activity in ML2. (F) WB of p-ERK1/2 and total ERK in ML2 silenced for MLL-AF6 and in double silencing of MLL-AF6 and AF6 compared with scRNA. (Right) Histogram represents the ratio between p-ERK and total ERK: a reduction of p-ERK is visible after MLL-AF6 silencing, and a rescue of p-ERK is documented when also AF6 was silenced. *P > .05.
Silencing of MLL-AF6 in t(6;11)(q27;q23) rearranged cells restores AF6 in the cytoplasm. (A) Western blot (WB) analysis revealed decreased levels of MLL-AF6 after silencing (siMLL-AF6) compared with negative controls (scRNA) in both, ML2 and SHI-1. Anti-ACTIN was used as endogenous control. (B) The siMLL-AF6 cells restored AF6 protein in the cytoplasm. Immunofluorescence shows colocalization of AF6 (red) and RAS (green) in SHI-1 after MLL-AF6 silencing (merged signals, yellow, nuclei blue, ×60 zoom). In the negative control (scRNA), the punctuate pattern of AF6 nuclear localization is visible (red AF6, nuclei blue, ×60 zoom). (C) The siMLL-AF6 cells restored AF6 protein in the cytoplasm. Immunofluorescence shows colocalization of AF6 (red) and RAS (green) in primary t(6;11)-AML after MLL-AF6 silencing (merged signals yellow, nuclei blue, ×60 zoom). In the negative control (scRNA), the punctuate pattern of AF6 nuclear localization is visible (red AF6, nuclei blue, ×60 zoom). (D) Active RAS-GTP levels in ML2 cell line silenced for the chimera showed a decreased activity of RAS (0.47) compared with scRNA. (E) Luciferase (LUC) activity of ML2 transfected with a pFOS (FBJ murine osteosarcoma viral oncogene homolog) WT-GL3 plasmid and siMLL-AF6 show a reduction of LUC activity compared with scRNA. Introduction of both siRNA for MLL-AF6 and AF6 show a rescue of LUC activity in ML2. (F) WB of p-ERK1/2 and total ERK in ML2 silenced for MLL-AF6 and in double silencing of MLL-AF6 and AF6 compared with scRNA. (Right) Histogram represents the ratio between p-ERK and total ERK: a reduction of p-ERK is visible after MLL-AF6 silencing, and a rescue of p-ERK is documented when also AF6 was silenced. *P > .05.
MLL-AF6 knockdown controls RAS-GTP levels influencing both RAS signaling pathway and cell proliferation
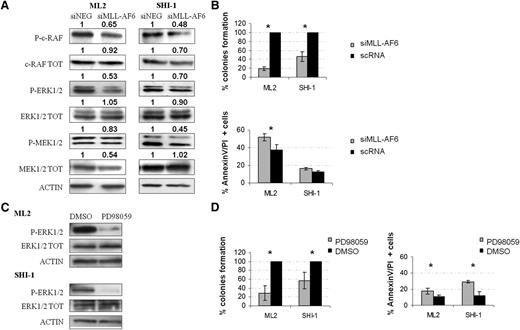
To demonstrate the relationship between MLL-AF6 and the RAS pathway, we analyzed the expression of its main downstream targets. Figure 3A shows that after MLL-AF6-silencing, expression of the phosphorylated active form of c-RAF, MEK1/2 and ERK1/2 is decreased in both ML2 and SHI-1. Densitometry was used to assess the extent of protein downregulation (results are detailed in Figure 3A). We confirmed the findings of western blot analyses by using the sandwich-based enzyme-linked immunosorbent assay technique because we obtained a 21% reduction of phospho-ERK1/2 levels (P = .02) in ML2 and SHI-1 cell lines after silencing of the chimera, whereas other analyzed proteins that are not targets of RAS signaling, such as phospho-p53, phospho-AKT, and phospho-JUN, were not affected by silencing (data not shown). Moreover, using an RPPA assay, we demonstrated that silencing of the chimera induces a reduction of p-ERK and p-MEK, as well as an increase of the expression levels of proteins involved in apoptosis, such as the cleaved caspase 7 and Poly (ADP ribose) polymerase (PARP) (supplemental Figure 1). We confirmed the oncogenic potential of the chimera also by measuring the clonogenic capacity after its silencing. A reduction of approximately 80% of colony number for ML2 and 55% for SHI-1 was found after silencing (Figure 3B) (n = 2; P < .05). Under these conditions, apoptosis was also increased compared with controls for both cell lines (ML2: 37.8% vs 51.9%; n = 3; P < .01 and SHI-1: 12.8% vs 16.0%; n = 3; P < .05). To ascertain the newly discovered role of RAS pathway activation in childhood AML cells carrying the t(6;11) translocation, we treated ML2 and SHI-1 cells with a chemical inhibitor of RAS signaling, PD980596. In both lines, we confirmed downregulation of p-ERK, the main downstream target of RAS (Figure 3C), as well as decreased clonogenicity up to 70% and 30% in ML2 and SHI-1 cell lines, respectively (n = 3; P < .05). Furthermore, PD980596 treatment alone increased apoptosis of t(6;11)-rearranged cell lines (ML2: 17.6% vs 10.9% after DMSO treatment; n = 3; P < .05 and SHI-1: 29.2% vs 12.1% of DMSO; n = 3; P < .05) (Figure 3D). Taken together, these results demonstrate that inhibition of the RAS pathway, either by restoring localization of AF6 in the cytoplasm after chimera silencing or through the use of a chemical compound, concurred to blunt proliferation of MLL-AF6-rearranged cells.
AF6 controls RAS-GTP levels and RAS downstream pathway. (A) WB analyses of the RAF/MEK/ERK pathway after silencing of MLL-AF6 in ML2 and SHI-1 cell lines: a decreased level of phosphorylation for c-RAF, MEK1/2, and ERK1/2 is visible compared with negative controls (scRNA); anti-ACTIN was used as control for total protein amount. (B) (Top) A decrease in colony formation (percentage with respect to scRNA) on semisolid medium. (Bottom) An increase in apoptosis (Annexin and propidium iodide-positive cells) in ML2 and SHI-1 cell lines after MLL-AF6 silencing (siMLL-AF6) compared with scRNA are observed (n = 3; P < .05). (C) Treatment of ML2 and SHI-1 cells with PD98059 (40 µM). WB analysis shows p-ERK1/2 reduction after treatment. Anti-ACTIN was used as a positive control of protein amount. (D) (Left) An increased apoptosis and (right) a decreased percentage of colony formation is measured after treatment with PD98059 compared with negative controls treated with dimethylsulfoxide (n = 2). *P < 0.05.
AF6 controls RAS-GTP levels and RAS downstream pathway. (A) WB analyses of the RAF/MEK/ERK pathway after silencing of MLL-AF6 in ML2 and SHI-1 cell lines: a decreased level of phosphorylation for c-RAF, MEK1/2, and ERK1/2 is visible compared with negative controls (scRNA); anti-ACTIN was used as control for total protein amount. (B) (Top) A decrease in colony formation (percentage with respect to scRNA) on semisolid medium. (Bottom) An increase in apoptosis (Annexin and propidium iodide-positive cells) in ML2 and SHI-1 cell lines after MLL-AF6 silencing (siMLL-AF6) compared with scRNA are observed (n = 3; P < .05). (C) Treatment of ML2 and SHI-1 cells with PD98059 (40 µM). WB analysis shows p-ERK1/2 reduction after treatment. Anti-ACTIN was used as a positive control of protein amount. (D) (Left) An increased apoptosis and (right) a decreased percentage of colony formation is measured after treatment with PD98059 compared with negative controls treated with dimethylsulfoxide (n = 2). *P < 0.05.
To further confirm the role of AF6 in controlling RAS activity in hematopoietic cells, we demonstrated increased phosphorylation of RAF/MEK/ERK proteins after AF6 silencing in HBM (30% reduction as determined by RT-PCR; n = 2; P < .05) (supplemental Figure 2), as previously described in other systems.34
Gene expression profile of t(6;11)-rearranged pediatric patients supports the transcriptional activity of MLL-AF6 on HOXA genes and RAS pathway
Gene expression analysis using Human Genome U133 Plus 2.0 was performed on samples of 11 AML t(6;11)-rearranged patients and of 11 HBM. Supervised analysis between these 2 groups identified 2463 differently expressed probe-sets (1747 genes; false discovery rate, <0.01). Among the differentially expressed genes, we found genes typically involved in MLL-rearranged leukemia, such as genes belonging to the MEIS1 and HOXA families (Figure 4A and supplemental Table 1). With differentially expressed genes, we performed pathway analysis using the DAVID software, finding that the mitogen-activated protein kinase (MAPK) pathway and genes related to apoptosis were the most important deregulated pathways in t(6;11)-rearranged cells. Remarkably, in samples with t(6;11) rearrangement, we noted the downregulation of a gene, RASA2, which encodes for a RAS-GAP known to negatively regulate RAS activity, as shown in the box plot (Figure 4B) (P < .001), and confirmed by qRT-PCR (Figure 4C) (P < .01). These results support the existence of an independent mechanism that enhances RAS activity in this type of leukemia, bringing to light that the RAS pathway sustains the leukemogenic properties of MLL-AF6-rearranged leukemia. Furthermore, supporting this finding, RASA2 expression is confirmed to be downregulated in t(6;11)-rearranged patients, as compared with MLL-other leukemia (Figure 4D) (P = .06 and P = .05).
Hoxa genes and RASA2 are differentially expressed between t(6;11) patients and HBM cells. (A) Hierarchical clustering analysis of 11 patients with t(6;11) (blue) and 11 healthy bone marrow (orange). (B) Box-plot of RASA2 probe sets in t(6;11)-patients (white) vs HBM (gray) samples generated using Partek Genomic Suite Software 6.6. Expression values are indicated in the boxes as the median of each group. Y-axis probe set expression values in log2 scale. (C) Histogram confirmed the decreased messenger RNA levels of RASA2 by qRT-PCR (ΔΔCt method) in 11 MLL-AF6 rearranged patients. Results are calibrated to the 11 HBM (RQ = 1). (D) Box-plot of RASA2 probe sets in t(6;11)-patients vs MLL-other leukemic samples generated using Partek Genomic Suite Software. Expression values are indicated in the boxes as the median of each group. Y-axis probe set expression values in log2 scale. *P < .05.
Hoxa genes and RASA2 are differentially expressed between t(6;11) patients and HBM cells. (A) Hierarchical clustering analysis of 11 patients with t(6;11) (blue) and 11 healthy bone marrow (orange). (B) Box-plot of RASA2 probe sets in t(6;11)-patients (white) vs HBM (gray) samples generated using Partek Genomic Suite Software 6.6. Expression values are indicated in the boxes as the median of each group. Y-axis probe set expression values in log2 scale. (C) Histogram confirmed the decreased messenger RNA levels of RASA2 by qRT-PCR (ΔΔCt method) in 11 MLL-AF6 rearranged patients. Results are calibrated to the 11 HBM (RQ = 1). (D) Box-plot of RASA2 probe sets in t(6;11)-patients vs MLL-other leukemic samples generated using Partek Genomic Suite Software. Expression values are indicated in the boxes as the median of each group. Y-axis probe set expression values in log2 scale. *P < .05.
Tipifarnib promotes cell death of t(6;11)-translocated blasts
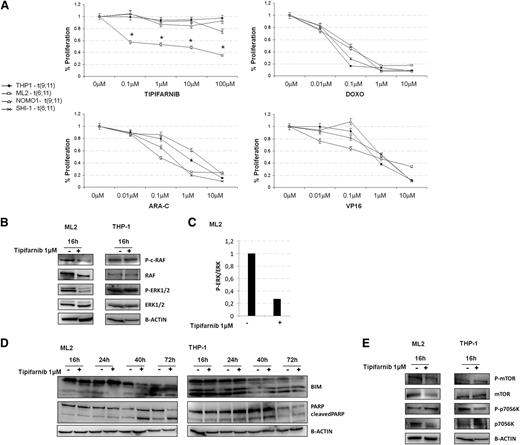
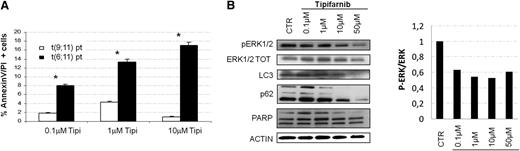
Because the prognosis of leukemia patients harboring t(6;11) is worse than that of patients with other MLL rearrangements,4 we speculated that our findings about the role of the RAS in this subtype of leukemia may lead to novel therapeutic opportunity by using tipifarnib. Tipifarnib is a drug not currently used in the treatment of pediatric AML, and it is currently adopted in clinical trials for different diseases35-38 for its ability to block the farnesyltransferase enzyme to inhibit targets among which there is Ras.39,40 We treated t(6;11)-translocated cell lines, ML2 and SHI-1, as well as cell lines with different rearrangements of MLL (ie, THP1 and NOMO1), both MLL-AF9 translocated, by comparing increasing concentrations of different cytotoxic drugs currently used for AML treatment, such as doxorubicin (Doxo), cytarabine (Ara-C), and etoposide (VP16) with tipifarnib. We observed that increasing concentrations of Doxo, Ara-C, and VP16 were able to reduce cell proliferation in all cell lines examined, and that ML2 were highly responsive to tipifarnib, whereas SHI-1 and the 2 other t(9;11)-rearranged cell lines were not (Figure 5A). This different behavior between the two t(6;11)-rearranged cell lines, ML2 and SHI-1, has been already investigated by Chen et al41 and found to be dependent on the presence of TP53 mutations, and associated with the documented greater multidrug resistance of this cell line. We performed western blot analysis to confirm RAS involvement, observing that its pathway, in particular p-ERK1/2, resulted to be decreased at 16-hours posttreatment with 1 µM tipifarnib (Figure 5B). We confirmed the downregulation of p-ERK1/2 by observing the upregulation of BCL2-like 11 (apoptosis facilitator) (BIM),39 and the apoptosis induction by PARP cleavage (Figure 5C-D). To check for off target effects, we analyzed the phosphorylation of mechanistic target of rapamycin (serine/threonine kinase) and of the p70S6K, denoting a decrease in the phosphorylated, as well as in both total proteins (Figure 5E). Consistent with our hypothesis, moreover, we cannot detect a perturbation of the RAS pathway in tipifarnib treated THP-1 cells. In view of these findings, we decided to treat with tipifarnib cultures of primary AML cells obtained from patients with or without the t(6;11) translocation. Tipifarnib increased cell mortality at 24 hours of treatment in cultured blasts of MLL-AF6-rearranged patients in comparison with cultures from patients with other aberrancies (Figure 6A) (n = 2; P < .05). Analyzing tipifarnib effects, we showed that low concentrations of tipifarnib (0.1 μM and 1 μM) increased autophagy, as shown by LC3 and p62 expression; by contrast, this drug at concentrations higher than 10 μM promotes apoptosis, as seen by PARP cleavage (Figure 6B). These results are consistent with previous reports demonstrating that RAS-induced tumorigenesis both in vitro and in vivo is always mediated by autophagy.42 ERK phosphorylation was shown to be inhibited at every concentrations of tipifarnib (Figure 6B), confirming the targeting of the RAS pathway.
Tipifarnib treatment provoked RAS inhibition and induced apoptosis of t(6;11)-rearranged cell line. (A) MLL-rearranged cell lines treated with increasing concentrations of chemotherapies (Doxo, Ara-C or VP16; 0.01-10 µM) showed a similar reduction in proliferation, whereas tipifarnib (0.1–100 µM) was specifically reducing ML2 cell proliferation. (B) WB analysis showed p-ERK1/2 reduced levels during increasing tipifarnib treatment in ML2. (C) Histogram represented the ratio between p-ERK and total ERK in ML2: a reduction of p-ERK was visible after tipifarnib treatment. (D) WB analysis showed BCL2-like 11 (apoptosis facilitator) (BIM) and PARP cleavage increased after tipifarnib treatment in ML2. (E) WB analysis showed mechanistic target of rapamycin (serine/threonine kinase) (mTOR) and p70S6K phosphorylated and total protein after tipifarnib treatment in ML2 and THP-1.
Tipifarnib treatment provoked RAS inhibition and induced apoptosis of t(6;11)-rearranged cell line. (A) MLL-rearranged cell lines treated with increasing concentrations of chemotherapies (Doxo, Ara-C or VP16; 0.01-10 µM) showed a similar reduction in proliferation, whereas tipifarnib (0.1–100 µM) was specifically reducing ML2 cell proliferation. (B) WB analysis showed p-ERK1/2 reduced levels during increasing tipifarnib treatment in ML2. (C) Histogram represented the ratio between p-ERK and total ERK in ML2: a reduction of p-ERK was visible after tipifarnib treatment. (D) WB analysis showed BCL2-like 11 (apoptosis facilitator) (BIM) and PARP cleavage increased after tipifarnib treatment in ML2. (E) WB analysis showed mechanistic target of rapamycin (serine/threonine kinase) (mTOR) and p70S6K phosphorylated and total protein after tipifarnib treatment in ML2 and THP-1.
Tipifarnib treatment provoked RAS inhibition and induces apoptosis of t(6;11)-rearranged primary cells. (A) The t(6;11) primary cell cultures treated with increasing concentration of tipifarnib showed an accentuated induction of apoptosis (percentage of Annexin V/propidium iodide-positive cells), especially with increasing drug concentrations with respect to primary AML cultures with different MLL-translocation. (B) (Left) WB analysis showed p-ERK1/2 reduced levels during increasingly tipifarnib treatment. LC3 and p62 documented autophagy induction at low tipifarnib doses, whereas PARP cleavage confirmed apoptosis when higher doses of tipifarnib was used. Anti-ACTIN was used as positive control for protein amount. (Right) Histogram represented the ratio between p-ERK and total ERK: a reduction of p-ERK was visible after tipifarnib treatment at any concentration.
Tipifarnib treatment provoked RAS inhibition and induces apoptosis of t(6;11)-rearranged primary cells. (A) The t(6;11) primary cell cultures treated with increasing concentration of tipifarnib showed an accentuated induction of apoptosis (percentage of Annexin V/propidium iodide-positive cells), especially with increasing drug concentrations with respect to primary AML cultures with different MLL-translocation. (B) (Left) WB analysis showed p-ERK1/2 reduced levels during increasingly tipifarnib treatment. LC3 and p62 documented autophagy induction at low tipifarnib doses, whereas PARP cleavage confirmed apoptosis when higher doses of tipifarnib was used. Anti-ACTIN was used as positive control for protein amount. (Right) Histogram represented the ratio between p-ERK and total ERK: a reduction of p-ERK was visible after tipifarnib treatment at any concentration.
Discussion
The MLL-AF6 fusion transcript has been found in a significant proportion of children who have AML, and it is associated with the worst prognosis among all variants of MLL-positive leukemia.2,4,6 MLL partner genes are broadly classified into 2 distinct groups based on their structural characteristics and cellular localization: gene codifying for the nuclear partner proteins with features of putative transcriptional regulators and genes codifying for cytoplasmic partners, associated with intracellular signaling.24 AF6 protein is the most frequent cytoplasmic partner in AML and has been previously found to ultimately orchestrate the aberrant transcription of MLL target genes as described by Cleary’s24 and Amstrong’s25 groups.
In this study, we identified a novel role for MLL-AF6 fusion protein in pediatric AML. We showed that AF6 and RAS colocalize and interact in the cytoplasm of healthy hematopoietic cells, whereas in t(6;11)-rearranged leukemia cells the 2 proteins have different localization, and hence the inhibitory effect of AF6 over RAS activation is lost.
Liedtke et al24 showed that AF6 exerts its function primarily as a scaffold protein for dimerization and activation of the transcriptional activity of MLL-AF6. These authors demonstrated the ability of the chimera to homodimerize and activate its oncogenic potential trough the AF6RA1 domain, which is capable of mediating self-association in vitro and is responsible of the self-association in the context of the MLL-AF6 fusion protein. Being documented as the ability of RA1 to self-associate, we infer that AF6 can also heterodimerize with MLL-AF6, leading to a previously unrecognized and parallel function of this chimera, able to delocalize the wild-type AF6 to the nucleus, thus enhancing the activation of RAS and its downstream pathway.24 Here, we showed that AF6 shuttling from cytoplasm to nucleus determines the level of the RAS-GTP active form, contributing to the tumorigenic effect exerted by MLL-AF6 as a transcription factor. Gene expression analysis, in fact, revealed that primary blasts of MLL-AF6-rearranged patients showed a gene expression profile typical of patients with other MLL-associated rearrangements, in which HOXA cluster is mainly overexpressed.24 However, we revealed that the MAPK pathway, acting downstream of RAS activation, is altered and that, among differentially expressed genes, RASA2, a known RAS inhibitor, is strongly downregulated in t(6;11)-rearranged cells, supporting the aberrant activation of RAS signaling. The hypothesis that RAS levels depend mainly on AF6 expression in hematopoietic cells has been here for the first time addressed and confirmed by rescue experiments. In t(6;11)-positive AML, AF6 is found within the nucleus, and after silencing of both MLL-AF6 and AF6, an evident increase of RAS activity was documented, with the concomitant rescue also of p-ERK levels when compared with silencing of MLL-AF6 alone.
Several studies previously demonstrated that uncontrolled RAS activation is 1 of the most common genetic alterations associated with development of several human cancers, including adult MLL-rearranged leukemias.45-48 This uncontrolled RAS activation is mainly due to known somatic activating mutations,49,50 or to the constitutive activation of several receptor tyrosine kinases, such as colony stimulating factor 1 and FLT3, or derives from the loss of function of tumor suppressor genes, such as NF1 and PTPN11.51 Even though the RAS pathway is overactivated in MLL-AF6-positive pediatric patients in our cohort, we never found mutations in the RAS gene, confirming our hypothesis of an active role of the chimera MLL-AF6 as the driving force of the observed aberrant RAS pathway activation.
Taken together, these results should encourage further studies aimed at evaluating novel treatment modalities in the t(6;11)(q27;q23) subgroup of childhood AML, considering that this subgroup still carries a dismal prognosis.4 To this purpose, we investigated the effect of the farnesyltransferase inhibitor tipifarnib,52 which hampers the attachment of the farnesyl moiety to the RAS protein, thereby repressing its activation. This drug is currently under evaluation in hematologic diseases with high rates of RAS mutation and activation.35-38,53,54 A phase 1 study showed that tipifarnib was well-tolerated by pediatric patients with resistant or refractory AML, although a poor clinical response was observed, partially due to the fact that RAS hyperactivation is not demonstrable in all AML variants. Support to the rationale of using tipifarnib is provided by the observation that apoptosis after drug exposure was enhanced in primary blasts carrying t(6;11), whereas blasts carrying other MLL rearrangement of childhood AML never showed relevant sensitivity to this drug. Notably, leukemia cells were sensitive to high concentrations of the drug, whereas low concentrations failed to induce apoptosis caused by a parallel activation of the autophagy process. This finding has been discussed in several works showing that many cancer cells with aberrant RAS activation have a high basal autophagy, and some depend on autophagy for normal growth.55 Autophagy is a dynamic process that prolongs survival for a short time under stress conditions56 ; therefore, blocking autophagy with conventional inhibitors,37 together with the use of tipifarnib, could be considered for t(6;11)-rearranged patients.
Because most patients with t(6;11) succumb within 1 year from diagnosis due to resistance to conventional cytotoxic therapy, RAS targeting promises to be a new valuable option for this subset of childhood AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Sabrina Gelain, Dr Alessandra Beghin, and Dr Samuela Francescato for the samples’ characterization, Dr Benedetta Accordi for the reverse phase protein array experiment, and Dr Ron Prywes for providing the pFOS WT-GL3 plasmid.
This work was supported by the CARIPARO, Istituto di Ricerca Pediatrica-Fondazione Città della Speranza and Università di Padova, and by the Special Grant “5 × 1000” #9962 from Associazione Italiana per la Ricerca sul Cancro, Milan, Italy to Franco Locatelli.
Authorship
Contribution: E.M. and E.B. performed experiments and interpreted results; E.M., E.B., G.B., and M.P. wrote the manuscript; C.T., V.B., and S.A. contributed to patients collection, immunofluorescence, and in vitro experiments; S.B. performed gene expression analysis; R.M. collected and interpreted clinical data of AML patients; F.L., G.B., and M.P. revised the manuscript; and G.B. and M.P. designed the study and interpreted results.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Martina Pigazzi, Istituto di Ricerca Pediatrica - Fondazione Città della Speranza ONLUS, Università degli Studi di Padova, Dipartimento della Salute della Donna e del Bambino, Clinica di Oncoematologia Pediatrica, Via Giustiniani, 3 – 35128 Padova, Italy; e-mail: martina.pigazzi@unipd.it.
References
Author notes
G.B. and M.P. contributed equally to this study.