Key Points
A new translocation t(2;11)(q22.1;q21) that involves CXCR4 and MAML2 has been described and characterized in CLL.
Abstract
Recent investigations of chromosomal aberrations in chronic lymphocytic leukemia (CLL) led to a better understanding of the molecular causes of CLL. Here we report a rearrangement between MAML2 (mastermind-like protein 2) and CXCR4 (specific receptor for CXC chemokine stromal cell-derived factor-1) in CLL cells of a patient with a t(2;11)(q22.1;q21) chromosomal translocation. The rearrangement between MAML2 and CXCR4, created by a t(2;11)(q22.1;q21) translocation, results in a new fusion gene in which a portion of CXCR4 is linked to the MAML2 gene. This fusion gene encodes for CXCR4/MAML2 protein chimera in which the N-terminal basic domain of MAML2 is replaced by the N-terminal domain of CXCR4.
Introduction
Chronic lymphocytic leukemia (CLL) is the most common human leukemia representing ∼30% of all leukemia cases.1 This disease occurs in 2 forms: aggressive and indolent; both forms are characterized by the clonal expansion of CD5-positive B lymphocytes.2 Genomic aberrations are detected in over 80% of CLL cases and numerous studies of lymphoid malignancies showed that chromosomal translocations often result in tumor-specific fusion oncogenes.3 The relevance of fusion oncogenes as therapeutic targets was first demonstrated when imatinib (a tyrosine kinase inhibitor) was found to inhibit the BCR-ABL tyrosine kinase fusion oncoprotein and to cause a complete remission in chronic myeloid leukemia.4,5 Here we describe a rearrangement between the MAML2 (mastermind-like protein 2) and CXCR4 (specific receptor for CXC chemokine stromal cell-derived factor-1) genes in CLL cells from a patient with a t(2;11)(q22.1;q21) chromosomal translocation. MAML2 belongs to the MAML protein family involved in the activation of the Notch signaling pathway including the HES1 transcription.6-8 Aberrant Notch activity is involved in the pathogenesis of human B-cell–derived malignancies, including CLL.9 MAML2 translocations have been found and characterized in different malignancies as mucoepidermoid carcinoma and in acute myeloid leukemia.10,11 C-X-C motif chemokine ligand 12 (CXCL12) and its receptor, CXCR4, are important components in the regulation of hematopoiesis and play an important role in retention of stem cells in the bone marrow.12,13 In this study, we identified the CXCR4 gene as a novel fusion partner of MAML2 and provided the evidence that the CXCR4/MAML2 fusion generates a transcribed gene and expressed fusion protein.
Study design
CLL sample
The study was carried out in accordance with institutional review board protocol approved by The Ohio State University. A CLL patient was enrolled in the CLL Research Consortium (CRC) and was diagnosed with CLL in 2008 at Rai stage 3. CLL samples were obtained from CLL patients enrolled in the CRC upon written informed consent. The study was conducted in accordance with the Declaration of Helsinki.
Cytogenetic analysis
G-banded chromosome analysis was performed on metaphase cells prepared by standard methods after 3-day culture in MarrowMax medium (Life Technologies) supplemented with 2 nmol/mL DSP30 CpG oligonucleotide (27 mer; TIB MOLBIOL) and 0.04 μg/mL interleukin 2 (Sigma).
Somatic cell hybrids
Somatic cell hybrids were created as described previously.14
Western blot analysis
Cell lines (LMTK−), hybrid cells, and cells from patients’ bone marrow were lysed in a lysis buffer (50mM Tris-HCl, 1mM EDTA, 20 g/L sodium dodecyl sulfate [SDS], 5mM dithiothreitol, 10mM phenylmethylsulfonyl fluoride). Equal amounts of protein lysates (30 µg each) were used to perform the western blot analysis as described previously.15 Anti-β-Actin (Sigma) was used as control. Anti-MAML2 was from Bethyl.
Southern blot
High-molecular-weight DNA was isolated by digestion with proteinase K, extraction with phenol/chloroform, and precipitation with ethanol. After digestion with EcoRI, DNA was electrophoresed using a 0.8% agarose gel, denaturated, and transferred onto nitrocellulose membrane (GE). Filters were hybridized overnight at 42°C with the 32P random priming labeled probe for MAML2 in Hybrisol Hybridization Solution (Sierological Corporation), washed 2 times for 15 minutes at 25°C in saline-sodium citrate 2 times, 0.1% SDS and 3 times at 55°C in saline-sodium citrate 0.2 times, 0.1% SDS, and exposed.
PCR
Polymerase chain reaction (PCR) was carried out using REDExtract-N-Amp PCR ReadyMix (Sigma) according to the manufacturer’s protocol. Commercial human DNA was used as control (Promega). All primers are described in supplemental Table 1 (available on the Blood Web site).
Real-time PCR
MAML2 messenger RNA (mRNA) expression was assayed by real time PCR (RT-PCR) using TaqMan Assay (assay ID HS00418423_m1) according to the manufacturer’s protocol and normalized using glyceraldehyde-3-phosphate dehydrogenase (assay ID HS 02758991_g1).
Results and discussion
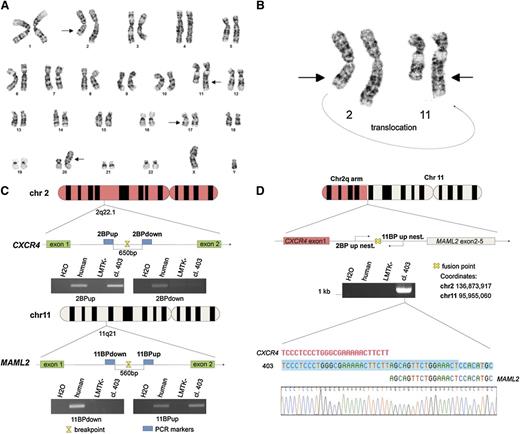
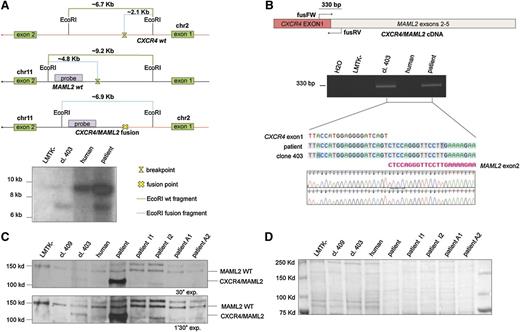
Twenty metaphase B cells from a CLL patient were examined by G-banding analysis. Nineteen cells presented an abnormal karyotype which showed a t(2;11) translocation (Figure 1A-B). These aberrations were not identified in previous studies of CLL and we decided to investigate this novel t(2;11)(q22.1;q21) translocation (Figure 1B). To better characterize this translocation, we used the LMTK− mouse fibroblast cell line to generate somatic mouse-human cell hybrids that contain a portion of the human karyotype from the patient’s CLL cells. Hybrid clones including the translocated chromosome t(2;11)(q22.1;q21) were isolated by PCR screening across the potential genomic breakpoint (supplemental Table 1). Among them the clone 403 was chosen for further studies. As shown in Figure 1C, we identified the region on the q arm of chromosomes 2 and 11 by chromosome walking. Our findings indicated that the breakpoint on chromosome 2 was within a region of 650 bp between “2BP up” and “2BP down” PCR markers; this region maps in the intron 1 of CXCR4 (Figure 1C top). We used the same approach to identify the breakpoint region on chromosome 11. In this case, the 560 (bp) region was located in the intron 1 of MAML2 (Figure 1C bottom). Next, we amplified the breakpoint region of the translocated chromosome t(2;11)(q22.1;q21) by a nested PCR, using 2 oligos annealing, respectively, on chromosome 2 “2BP up nest” and on chromosome 11 “11BP up nest” (Figure 1D upper). We analyzed the amplicon sequence and found a translocation breakpoint that links intron 1 of CXCR4 and intron 1 of MAML2 (Figure 1D lower). The t(2;11)(q22.1;q21) translocation breakpoint was also analyzed by Southern blot (Figure 2A). By digesting CXCR4 and MAML2 with EcoRI and using a probe that anneals in the intron 1 of human MAML2 we detected a ∼6.9-kb fragment corresponding to the sum of the fused CXCR4/MAML2 EcoRI-digested fragments (Figure 2A). We then decided to assess whether the CXCR4/MAML2 fusion gene could generate a chimeric protein. We carried out RT-PCR using RNA of the hybrid clone and the CLL sample. Complementary DNAs (cDNAs) were amplified with a set of oligos (fusFW) and (fusRW) annealing on exon 1 of CXCR4 and exon 2 of MAML2, respectively. As shown in Figure 2B, amplicon sequencing data demonstrate that the CXCR4/MAML2 chimeric mRNA is a fusion of CXCR4 exon 1 and MAML2 exon 2 in the hybrid clone 403 and in patient CLL cells. Studies on the chimera mRNA sequence revealed that this fusion is also in frame. To confirm that CXCR4/MAML2 chimeric protein is expressed in the patient’s CLL cells, we carried out a western blot using an anti-MAML2 antibody that recognizes an epitope within the exon 4 of MAML2. As shown in Figure 2C-D, this antibody detects 2 forms of MAML2: the first (wild type [WT]) has a size of ∼150 kDa and the second has a form of ∼125 kDa. The electrophoretic migration of CXCR4/MAML2 protein is different from MAML2 WT because the predicted fusion protein has 5 aa from CXCR4 instead of 168 aa. Clone GI-409 and other CLL samples (I1, I2, A1, A2) that do not express CXCR4/MAML2 were used as control (supplemental Figure 1C). Taken together, our results show a novel t(2;11)(q22.1;q21) translocation in a CLL patient that fuses CXCR4 and MAML2, 2 important genes in hematopoiesis. Exon 1 of CXCR4 translocates upstream of exon 2 of MAML2 and this event generates a CXCR4/MAML2 chimeric gene which is under the control of the CXCR4 promoter. In our patient’s CLL cells, MAML2 results were highly expressed if compared with other CLL cases that did not present t(2;11) translocation (supplemental Figure 1A-B). Characterization of this new translocation in CLL may result in a better understanding of the genetic basis of CLL.
CLL t(2;11) translocation characterization. (A) Giemsa banding of a CLL patient karyotype; chromosomal aberrations are marked with arrows. (B) Giemsa banding of chromosomes 2 and 11 of a CLL patient is reported; a portion of the chromosome 2 q arm is translocated on the q arm of chromosome 11; the curving arrow indicates the translocation orientation. (C) Schematic representation of chromosomes 11 and 2. The PCRs of the last 2 markers used to map the breakpoint on the chromosomes are represented in this figure. The region of 650 bp between the PCR markers 2BPdown and 2BPup contains the breakpoint and maps in the intron between exon 1 and exon 2 of CXCR4. The region of 560 bp between the PCR markers 11BPup and 11BPdown contains the breakpoint that maps in the intron between exon 1 and exon 2 of MAML2. (D) Schematic representation of translocated chromosome (2;11). The breakpoint region has been amplified by a nested PCR using (2BP up nest) and (11BP up nest) primers; the resulting amplicon has been sequenced and the fusion point has been characterized (sequence is shown in reverse). Primers sequences are reported in supplemental Table 2.
CLL t(2;11) translocation characterization. (A) Giemsa banding of a CLL patient karyotype; chromosomal aberrations are marked with arrows. (B) Giemsa banding of chromosomes 2 and 11 of a CLL patient is reported; a portion of the chromosome 2 q arm is translocated on the q arm of chromosome 11; the curving arrow indicates the translocation orientation. (C) Schematic representation of chromosomes 11 and 2. The PCRs of the last 2 markers used to map the breakpoint on the chromosomes are represented in this figure. The region of 650 bp between the PCR markers 2BPdown and 2BPup contains the breakpoint and maps in the intron between exon 1 and exon 2 of CXCR4. The region of 560 bp between the PCR markers 11BPup and 11BPdown contains the breakpoint that maps in the intron between exon 1 and exon 2 of MAML2. (D) Schematic representation of translocated chromosome (2;11). The breakpoint region has been amplified by a nested PCR using (2BP up nest) and (11BP up nest) primers; the resulting amplicon has been sequenced and the fusion point has been characterized (sequence is shown in reverse). Primers sequences are reported in supplemental Table 2.
CXCR4/MAML2 fusion gene is expressed in CLL. (A) Top panel, EcoRI restriction map around the breakpoint sites on chromosomes 2 and 11. The green line represents the full-length EcoRI-digested fragment without any break; the light blue line represents the fragment length after chromosome break and the EcoRI-digested fragment after CXCR4/MAML2 fusion. Bottom panel, Southern Blot EcoRI digested. The CXCR4/MAML2 gene fusion fragment of ∼7 kb is shown on lane 2 (GI-403 clone) and on lane 4 (patient B cells). The probe has been designed on the MAML2 gene and its amplification primers are reported in supplemental Table 2. (B) PCR on total cDNA performed with “fusFW” and “fusRV” oligos, designed to amplify a region of 330 bp across the exon1 CXCR4 and the exon2 of MAML2. Primer sequences are reported in supplemental Table 2. The amplicons obtained from this PCR were sequenced (reverse primer used). (C) Western blot using anti-MAML2 polyclonal antibody. The chimeric protein CXCR4/MAML2 (lanes 3 and 5) has a smaller size (∼20 kDa) than MAML2 wt. (D) Ponceau staining of the nitrocellulose membrane shows the total proteins are equally loaded in each lane. This control is relative to panel C.
CXCR4/MAML2 fusion gene is expressed in CLL. (A) Top panel, EcoRI restriction map around the breakpoint sites on chromosomes 2 and 11. The green line represents the full-length EcoRI-digested fragment without any break; the light blue line represents the fragment length after chromosome break and the EcoRI-digested fragment after CXCR4/MAML2 fusion. Bottom panel, Southern Blot EcoRI digested. The CXCR4/MAML2 gene fusion fragment of ∼7 kb is shown on lane 2 (GI-403 clone) and on lane 4 (patient B cells). The probe has been designed on the MAML2 gene and its amplification primers are reported in supplemental Table 2. (B) PCR on total cDNA performed with “fusFW” and “fusRV” oligos, designed to amplify a region of 330 bp across the exon1 CXCR4 and the exon2 of MAML2. Primer sequences are reported in supplemental Table 2. The amplicons obtained from this PCR were sequenced (reverse primer used). (C) Western blot using anti-MAML2 polyclonal antibody. The chimeric protein CXCR4/MAML2 (lanes 3 and 5) has a smaller size (∼20 kDa) than MAML2 wt. (D) Ponceau staining of the nitrocellulose membrane shows the total proteins are equally loaded in each lane. This control is relative to panel C.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grant PO1-CA81534 of the CLL Research Consortium (L.Z.R., T.J.K., C.M.C.).
Authorship
Contribution: M.A., G.R., and C.M.C. designed research; T.J.K. and L.Z.R. provided patient samples; M.A., G.R., D.W., V.B., M.d'A., and Y.P. performed research and analyzed data; M.A., G.R., Y.P., and C.M.C. wrote the manuscript; and all authors critically reviewed and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carlo M. Croce, The Ohio State University, Comprehensive Cancer Center, Biomedical Research Tower, Room 1082, 460W 12th Ave, Columbus, OH 43210; e-mail: carlo.croce@osumc.edu.
References
Author notes
M.A. and G.R. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal