To the editor:
Biological subclasses of T-cell acute lymphoblastic leukemia (T-ALL) can be defined by recurrent gene expression patterns, which typically segregate with specific chromosomal anomalies. The HOXA+ subgroup is characterized by deregulated homeobox A (HOXA) gene expression and is associated with translocations involving the mixed lineage leukemia (MLL) and/or MLLT10 loci, SET-NUP214, or TCRB-HOXA.1,2 Nevertheless, the genetic basis for many HOXA+ cases remains unexplained.
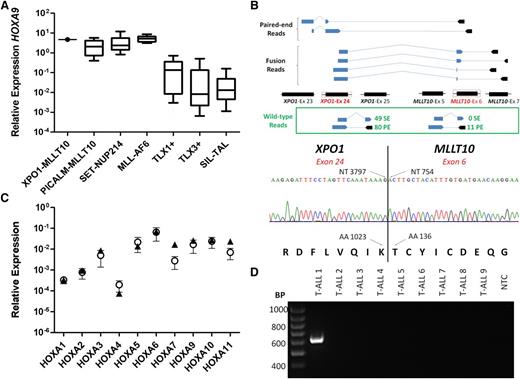
Diagnostic assessment of a 33-year-old man with T-ALL revealed high leukemic blast expression of HOXA9 at levels comparable to those in known HOXA+ cases (Figure 1A). Tests for PICALM-MLLT10, SET-NUP214, MLL-AF6, and TCRB-HOXA were negative. Leukemic cells exhibited a complex karyotype (46,XY,add(2)(p14),-10,-17,+2mars,inc[11]), which led us to speculate that HOXA positivity might be caused by a structural genetic abnormality. We therefore performed poly(A)-enriched sequencing (RNA-sequencing) of diagnostic RNA, analysis of which revealed fusion of exon 24 of XPO1 to exon 6 of MLLT10 (Figure 1B, upper panel). Expression of an in-frame XPO1-MLLT10 fusion transcript was confirmed by reverse transcriptase polymerase chain reaction (RT-PCR) and direct sequencing (Figure 1B, lower panel).
XPO1-MLLT10 fusion detected by RNA-sequencing is associated with deregulation of HOXA gene locus expression. (A) Expression of HOXA9 in genetic subgroups of T-ALL. Levels were determined by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) and calculated relative to an ABL housekeeping gene control. Boxes encompass the 25th through 75th percentiles, with the horizontal bar denoting the median expression level. Whiskers indicate the 10th and 90th percentiles. The numbers of cases tested in each group were as follows: XPO1-MLLT10, n = 1; PICALM-MLLT10, n = 36; SET-NUP214, n = 18; MLL-AF6, n = 9; TLX1+, n = 86; TLX3+, n = 67; SIL-TAL, n = 40. (B) Upper panel: Genomic mapping of the XPO1-MLLT10 fusion by poly(A)-enriched strand-specific RNA-sequencing using the SOLiD HQ5500xl system (Life Technologies). Mapping, coverage, and fusion discovery were determined by using LifeScope (Life Technologies), with reference to version hg19 of the human genome. A schematic representation of paired-end and fusion-spanning reads that revealed fusion between exon 24 of XPO1 (chr2:61708320-61708416) and exon 6 of MLLT10 (chr10:21901277-21901380) is shown. Solid lines indicate split reads spanning 2 exons, and dotted lines indicate 2 reads of the same fragment. The numbers of unique reads for the wild-type XPO1 (exons 24 and 25) and MLLT10 (exons 5 and 6) transcripts are also depicted. Lower panel: Confirmation of expression of an in-frame XPO1-MLLT10 fusion transcript by direct (Sanger) sequencing. The positions of the nucleotide (NT) and amino acid (AA) at the breakpoint of each gene are annotated. (C) Expression of HOXA genes in XPO1-MLLT10+ (n = 1; denoted by triangles) and PICALM-MLLT10+ (n = 4; mean levels denoted by circles with error bars indicating standard error of the mean) blasts. Transcript quantification was determined by qPCR using a TaqMan Low-Density Array, and the results of 2 experimental replicates were combined. Expression was calculated relative to a GAPDH housekeeping gene control. (D) RT-PCR for XPO1-MLLT10; 84 cases of HOXA+ T-ALL lacking known explicatory genetic anomalies were screened by using primers specific for the XPO1-MLLT10 fusion transcript (product size, 618 bp). A representative PCR result is shown. T-ALL 1 is the index XPO1-MLLT10+ case. NTC, no template control; PE, paired-end; SE, single-end.
XPO1-MLLT10 fusion detected by RNA-sequencing is associated with deregulation of HOXA gene locus expression. (A) Expression of HOXA9 in genetic subgroups of T-ALL. Levels were determined by quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) and calculated relative to an ABL housekeeping gene control. Boxes encompass the 25th through 75th percentiles, with the horizontal bar denoting the median expression level. Whiskers indicate the 10th and 90th percentiles. The numbers of cases tested in each group were as follows: XPO1-MLLT10, n = 1; PICALM-MLLT10, n = 36; SET-NUP214, n = 18; MLL-AF6, n = 9; TLX1+, n = 86; TLX3+, n = 67; SIL-TAL, n = 40. (B) Upper panel: Genomic mapping of the XPO1-MLLT10 fusion by poly(A)-enriched strand-specific RNA-sequencing using the SOLiD HQ5500xl system (Life Technologies). Mapping, coverage, and fusion discovery were determined by using LifeScope (Life Technologies), with reference to version hg19 of the human genome. A schematic representation of paired-end and fusion-spanning reads that revealed fusion between exon 24 of XPO1 (chr2:61708320-61708416) and exon 6 of MLLT10 (chr10:21901277-21901380) is shown. Solid lines indicate split reads spanning 2 exons, and dotted lines indicate 2 reads of the same fragment. The numbers of unique reads for the wild-type XPO1 (exons 24 and 25) and MLLT10 (exons 5 and 6) transcripts are also depicted. Lower panel: Confirmation of expression of an in-frame XPO1-MLLT10 fusion transcript by direct (Sanger) sequencing. The positions of the nucleotide (NT) and amino acid (AA) at the breakpoint of each gene are annotated. (C) Expression of HOXA genes in XPO1-MLLT10+ (n = 1; denoted by triangles) and PICALM-MLLT10+ (n = 4; mean levels denoted by circles with error bars indicating standard error of the mean) blasts. Transcript quantification was determined by qPCR using a TaqMan Low-Density Array, and the results of 2 experimental replicates were combined. Expression was calculated relative to a GAPDH housekeeping gene control. (D) RT-PCR for XPO1-MLLT10; 84 cases of HOXA+ T-ALL lacking known explicatory genetic anomalies were screened by using primers specific for the XPO1-MLLT10 fusion transcript (product size, 618 bp). A representative PCR result is shown. T-ALL 1 is the index XPO1-MLLT10+ case. NTC, no template control; PE, paired-end; SE, single-end.
We hypothesized that the common involvement of MLLT10 would result in similar deregulation of HOXA locus expression in XPO1-MLLT10+ and PICALM-MLLT10+ T-ALL. We tested the expression of a range of HOX genes by quantitative RT-PCR. As predicted, the pattern of HOXA gene transcription in the XPO1-MLLT10+ case was very similar to that in the PICALM-MLLT10+ cases (Figure 1C). A targeted RT-PCR screen of 84 HOXA+ T-ALL samples that lacked known explicatory genetic anomalies identified no further XPO1-MLLT10+ cases (Figure 1D), suggesting rarity and/or breakpoint heterogeneity.
Each of the genes involved in this fusion has been previously implicated in leukemia. Notably, MLLT10 (which encodes the AF10 protein) is involved in the recurrent PICALM-MLLT103 and MLL-MLLT104 translocations in both T-ALL and acute myeloblastic leukemia. Recently reported results of RNA-sequencing have identified HNRNPH1 and DDX3X as MLLT10 fusion partners in HOXA+ T-ALL.5 Our data provide further evidence of shared fusion partner–independent mechanisms of AF10-mediated transcriptional dysregulation, and this case adds to the repertoire of MLL and/or AF10-rearranged T-ALL that might be candidates for targeted DOT1L-directed therapy.6 MLLT10 breakpoints are heterogeneous, and increasing truncation of the transcript was reported to correlate with an earlier maturation block in T-ALL, although this was not confirmed in a later series.3,7 In this case, detailed characterization of T-cell receptor (TR) gene configuration revealed monoallelic TRG and TRD and incomplete TRB diversity-joining rearrangements (data not shown), consistent with an immature pre-β-selection immunogenotype.8
XPO1 (also CRM1) encodes exportin 1, a transport protein that mediates nuclear export of multiple tumor suppressor and growth regulatory molecules (eg, P53 and RB1). Pharmacologic XPO1 inhibition has shown promising antileukemic activity in preclinical models via a mechanism that is believed to involve either nuclear retention of XPO1 cargo upon which the leukemic cells depend for survival,9 and/or reactivation of nuclear protein phosphatase 2A.10 It is tempting to speculate that HOXA-independent activity of the XPO1-AF10 fusion protein could also contribute to leukemogenesis in this case, for example through aberrant transport of proteins that mediate proliferation and survival and/or by dominant negative inhibition of wild-type XPO1.
Authorship
Acknowledgments: This work was supported by a Kay Kendall Leukaemia Fund Intermediate Research Fellowship (J.B.), grants from INSERM, Groupement d'Intérêt Scientifique Infrastructures en Biologie Santé et Agronomie, Aix-Marseille Université, and grant No. ANR-10-INBS-0009-10 (for high throughput sequencing at the Transcriptomic and Genomic Marseille-Luminy Platform); a grant from the European Union’s FP7 Program (agreement No. 282510-BLUEPRINT) (S.S.); the Association pour la Recherche sur le Cancer (project No. SFI20111203756), by the Aix-Marseille Initiative d'Excellence project (No. ANR-11-IDEX-0001-02); the Cancer Banking Network of the Institut National du Cancer (E.A.M.); the 2007 and 2012 (Caracteristique Moléculaire et Épigenetique des Leucémies Aiguës Myéloïdes de l'Enfant) Translational Research Programs in Immature T/Myeloid Acute Leukemias; and the Association Laurette Fugain.
Contribution: J.B., A.B., A.D., and I.T. performed research; X.T. performed patient management; J.B., A.B., V.A., S.S., and E.A.M. analyzed and interpreted data; and J.B., A.B., S.S., and E.M. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Elizabeth A. Macintyre, Laboratoire Hématologie Biologique, Tour Pasteur 2ème étage, Hôpital Necker-Enfants Malades, 149 rue de Sèvres, 75743 Paris Cedex 15, France; e-mail: elizabeth.macintyre@nck.aphp.fr.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal