Key Points
In pediatric T-ALL, MLLT10 emerged as a promiscuous gene, maintaining the critical leukemogenic OM-LZ domain in all fusions.
MLLT10 gene fusions were associated with a specific expression profile within the HOXA subgroup of pediatric T-ALL.
Abstract
The MLLT10 gene, located at 10p13, is a known partner of MLL and PICALM in specific leukemic fusions generated from recurrent 11q23 and 11q14 chromosome translocations. Deep sequencing recently identified NAP1L1/12q21 as another MLLT10 partner in T-cell acute lymphoblastic leukemia (T-ALL). In pediatric T-ALL, we have identified 2 RNA processing genes, that is, HNRNPH1/5q35 and DDX3X/Xp11.3 as new MLLT10 fusion partners. Gene expression profile signatures of the HNRNPH1- and DDX3X-MLLT10 fusions placed them in the HOXA subgroup. Remarkably, they were highly similar only to PICALM-MLLT10–positive cases. The present study showed MLLT10 promiscuity in pediatric T-ALL and identified a specific MLLT10 signature within the HOXA subgroup.
Introduction
New genomic technologies, including whole-genome analysis and gene expression profiling (GEP), dramatically improved cytogenetic-molecular classification of T-cell acute lymphoblastic leukemia (T-ALL) which affects ∼15% of children with ALL.1 At least 6 main genetic categories (ie, TAL/LMO, TLX1, TLX3, NKX2-1/NKX2-2, MEF2C, and HOXA) have been identified. The HOXA group includes cases with TCRB-HOXA, SET-NUP214, MLL translocations, and PICALM-MLLT10.1,2 In a case of early T-cell precursor-ALL (ETP-ALL), the NAP1L1/12q21 gene, a member of the nucleosome assembly protein family, was recently identified as another MLLT10 partner.3
Interestingly, PICALM-MLLT10, MLL-MLLT10, and NAP1L1-MLLT10 fusions all retained the OM-LZ domain at their C terminus.3-5 It exerts a leukemogenic effect by interacting with chromatin modifying proteins such as the H3K79 methyltransferase hDOT1L.6,7
The present study focuses on 2 new MLLT10 fusion genes in pediatric T-ALL, placing them within the HOXA subgroup.
Study design
A combined interphase (CI) fluorescence in situ hybridization (FISH) test (supplemental Table 1, available on the Blood website) was applied in 42 pediatric T-ALL patients enrolled in the Associazione Italiana Ematologia Oncologia Pediatrica (AIEOP) ALL protocol. Informed consent of our work was obtained from the Bioethic Committee, University of Perugia (Prot.1.X.2011). Informed consent statements were provided according to the Declaration of Helsinki (AIEOP ALL protocol number NCT 00613457). To identify the new MLLT10 fusions, we used a 5′-rapid amplification of cDNA ends–polymerase chain reaction (5′-RACE-PCR) (Invitrogen) and Thermoscript reverse transcription PCR (RT-PCR) system (Invitrogen) according to the manufacturer’s instructions. Primers are listed in supplemental Table 2. PCR products were subcloned into pGEM-T easy vector (Promega) and sequenced with the AB3500 Genetic analyzer (Applied Biosystems). Single-nucleotide polymorphism (SNP) analysis was performed using the whole-genome cytogenetic 2.7M array (Affymetrix).
Statistical methods for microarray data (Affymetrix hgu 133 plus 2 arrays) were analyzed using the Bioconductor package for R (version 2.14.1). Data were deposited at GEO repository (series accession number GSE42765; http://www.ncbi.nlm.nih.gov/geo/). Heatmaps were created using the Ward method and Euclidean distance. The heatmap for the unsupervised analysis was created using the probe sets with the highest variances (threshold 90%), while the heatmap for the supervised analysis was created with differentially expressed probe sets. Arrays were normalized using robust multiple-array average.8 Batch effects were removed using ComBat.9 Differentially expressed genes were identified by the shrinkage T-statistic.10 False-positive findings were prevented by the local false discovery rate (lFDR). Probe sets with lFDR below 0.05 were considered significant.11,12
Results and discussion
CI-FISH identified MLLT10 rearrangements in 6 of 42 (14.3%) patients. Four of the total cohort (9.5%) showed the PICALM-MLLT10 fusion and 2 (4.8%) MLLT10 translocations to unknown partner(s). Clinical, hematologic, cytogenetic, and molecular data of these 2 patients are shown in Table 1. The MLLT10 FISH probe gave 3 signals in 55% of nuclei in case 1 and in 60% in case 2. In case 1, metaphase-FISH confirmed the split between the short and the long arms of 1 chromosome 10, resulting in the der(10)inv(10)(p12q25) seen at karyotype. The 10q breakpoint localized to band 25.3 in an ∼12-kb region without genes (supplemental Figure 1A).
Clinical, immunophenotypic, hematologic, molecular, and cytogenetic data
| Patient . | Sex/age, y . | Diagnosis . | WBC, ×109/L . | PICALM MLLT10 . | MLL-ENL . | MLL-AF4 . | SIL-TAL1 . | NOTCH1 . | FLT3 . | Karyotype . | CI-FISH . | SNPs . | MRD +33/+78 . | Chemotherapy* . | Follow-up, mo . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 (UPN 1036616) | F/7 | ETP-ALL | 10.55 | na | na | neg | neg | mut† | neg | 46,XX,inv(10) (p12q25)[2] /46,XX[17] | Split: MLLT10/10p12 (55%) | Normal | +/+ | High risk, no transplant donor | 33+ |
| Loss: IKZF1/7p12 (10%) | |||||||||||||||
| 2 (UPN 1023865) | M/11 | Cortical T-ALL | 152.8 | neg | neg | n.a. | neg | mut‡ | neg | Failed | Split: MLLT10/10p12 | Loss: 9p24.3-p11.2: from 0 to 47 508 608 bp | +/− | Standard risk | 73+ |
| Loss: PAX5,CDKN2A/B, JAK2/9p13-24 | Gain: 17q21.32-q25.3: from 45 273 751 to 78 786 769 bp | ||||||||||||||
| Gain: RP11-501C14/17q21.31 |
| Patient . | Sex/age, y . | Diagnosis . | WBC, ×109/L . | PICALM MLLT10 . | MLL-ENL . | MLL-AF4 . | SIL-TAL1 . | NOTCH1 . | FLT3 . | Karyotype . | CI-FISH . | SNPs . | MRD +33/+78 . | Chemotherapy* . | Follow-up, mo . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 (UPN 1036616) | F/7 | ETP-ALL | 10.55 | na | na | neg | neg | mut† | neg | 46,XX,inv(10) (p12q25)[2] /46,XX[17] | Split: MLLT10/10p12 (55%) | Normal | +/+ | High risk, no transplant donor | 33+ |
| Loss: IKZF1/7p12 (10%) | |||||||||||||||
| 2 (UPN 1023865) | M/11 | Cortical T-ALL | 152.8 | neg | neg | n.a. | neg | mut‡ | neg | Failed | Split: MLLT10/10p12 | Loss: 9p24.3-p11.2: from 0 to 47 508 608 bp | +/− | Standard risk | 73+ |
| Loss: PAX5,CDKN2A/B, JAK2/9p13-24 | Gain: 17q21.32-q25.3: from 45 273 751 to 78 786 769 bp | ||||||||||||||
| Gain: RP11-501C14/17q21.31 |
y, years; m, months; F, female; M, male; MRD, minimal residual disease; mut, mutated; n.a., data not available; neg., negative; UPN, unique patent number; WBC, white blood cell; [ ], number of cells analyzed.
AIEOP LLA 2000 (supplemental reference 1).
c.4766_4767insAGCAGAACCGGAGCAGCTGCGCAACAGCTC; p.S1589_F1590insAEPEQLRNSS.
c.G4793C; p.R1598P; NOTCH1 mutation numbers refer to CCDS 43905.1 and NP_060087.3.
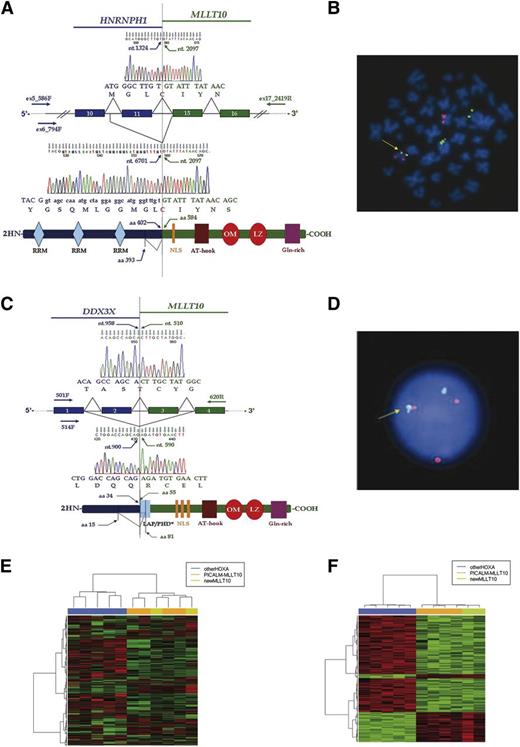
The MLLT10 gene breakpoint was narrowed to between exons 14 and 15 in patient 1 and between exons 1 and 3 in patient 2 (supplemental Figure 1B-C). 5′ RACE-PCR and sequencing showed an HNRNPH1-MLLT10 in-frame transcript in the first case and a DDX3X-MLLT10 fusion transcript in the second (Figure 1A,C). These results were confirmed by RT-PCR, cloning, and sequencing. Subsequently, a diagnostic double-color double-fusion (DCDF) test was developed in both cases (Figure 1B,D). The only common additional genetic lesion was a mutated NOTCH1 gene (Table 1).
Molecular characterization of the two new MLLT10 fusions. (A) Two HNRNPH1-MLLT10 splicing isoforms were identified in patient 1. (Top) Direct sequencing showed an in-frame HNRNPH1-MLLT10 isoform joining nucleotide 1324 (HNRNPH1 exon 11) to nucleotide 2097 (MLLT10 exon 15). (Bottom) Cloning and sequencing showed nucleotide 6701 (HNRNPH1 intron 10) fused in-frame with nucleotide 2097 (MLLT10 exon 15). Hypothetical fusion protein was shown in which HNRNPH1 maintained all 3 RRM at the N terminus and MLLT10 lost 2 of 3 NLS. MLLT10 maintained the critical OM-LZ domain at the C terminus. Primer and sequence numbers refer to GenBank accession NC_000005.9, NM_005520.2, NP_005511.1 for HNRNPH1 and NM_004641.3, NP_004632.1 for MLLT10. (B) DCDF test: Probes for MLLT10 (RP11-418C1 and RP11-249M6) in orange and for HNRNPH1 (CTD-3223H16 and RP11-410B18) in green showed 1 fusion signal on der(10) (arrow). (C) Two DDX3X-MLLT10 splicing isoforms were identified in patient 2. (Top) Sequencing showed an in-frame DDX3X-MLLT10 isoform joining nucleotide 958 (DDX3X exon 2) to nucleotide 510 (MLLT10 exon 3). (Bottom) Cloning and sequencing showed an in-frame isoform with nucleotide 900 (DDX3X exon 1) fused with nucleotide 590 (MLLT10 exon 4). The hypothetical fusion protein lost the DDX3X DEAD box domain at the N terminus and maintained part of the PHD, all 3 NLS and the OM-LZ domain at the C terminus. Primer and sequence numbers refer to GenBank accession NM_001356.3, NP_001347.3 for DDX3X and NM_004641.3, NP_004632.1 for MLLT10. (D) DCDF-FISH with probes for DDX3X in green (RP11-1058N11, flanking 5′, and RP11-10K13, flanking 3′) and MLLT10 in red (RP11-418C1 and RP11-249M6), showed 1 fusion signal (arrow). (E) Unsupervised analysis of 11 T-ALL HOXA patients. In such unsupervised analysis, patients bearing MLLT10 rearrangements and those without MLLT10 rearrangements (1 MLL-ENL, 1 MLL-AF6, 2 TCRB-HOXA, and 1 SET-NUP214) are naturally clustered in 2 distinct groups. PICALM-MLLT10 patients are indicated in orange; patient 1 and patient 2 (HNRNPH1-MLLT10, DDX3X-MLLT10) in green and patients without MLLT10 rearrangements in blue. (F) Supervised analysis was created using the significative probe sets from the comparison of HOXA patients with MLLT10 rearrangements (4 with PICALM-MLLT10 and the 2 new cases with HNRNPH1-MLLT10, DDX3X-MLLT10) vs patients without MLLT10 rearrangements (1 MLL-ENL, 1 MLL-AF6, 2 TCRB-HOXA, and 1 SET-NUP214). Patients bearing MLLT10 recombinations are indicated in orange or green while patients without MLLT10 rearrangements are indicated in blue. DCDF, double-color double-fusion; LAP, Leukemia Associated Protein; NLS, nuclear localization signal; PHD, plant homeo domain; RRM, RNA recognition motif.
Molecular characterization of the two new MLLT10 fusions. (A) Two HNRNPH1-MLLT10 splicing isoforms were identified in patient 1. (Top) Direct sequencing showed an in-frame HNRNPH1-MLLT10 isoform joining nucleotide 1324 (HNRNPH1 exon 11) to nucleotide 2097 (MLLT10 exon 15). (Bottom) Cloning and sequencing showed nucleotide 6701 (HNRNPH1 intron 10) fused in-frame with nucleotide 2097 (MLLT10 exon 15). Hypothetical fusion protein was shown in which HNRNPH1 maintained all 3 RRM at the N terminus and MLLT10 lost 2 of 3 NLS. MLLT10 maintained the critical OM-LZ domain at the C terminus. Primer and sequence numbers refer to GenBank accession NC_000005.9, NM_005520.2, NP_005511.1 for HNRNPH1 and NM_004641.3, NP_004632.1 for MLLT10. (B) DCDF test: Probes for MLLT10 (RP11-418C1 and RP11-249M6) in orange and for HNRNPH1 (CTD-3223H16 and RP11-410B18) in green showed 1 fusion signal on der(10) (arrow). (C) Two DDX3X-MLLT10 splicing isoforms were identified in patient 2. (Top) Sequencing showed an in-frame DDX3X-MLLT10 isoform joining nucleotide 958 (DDX3X exon 2) to nucleotide 510 (MLLT10 exon 3). (Bottom) Cloning and sequencing showed an in-frame isoform with nucleotide 900 (DDX3X exon 1) fused with nucleotide 590 (MLLT10 exon 4). The hypothetical fusion protein lost the DDX3X DEAD box domain at the N terminus and maintained part of the PHD, all 3 NLS and the OM-LZ domain at the C terminus. Primer and sequence numbers refer to GenBank accession NM_001356.3, NP_001347.3 for DDX3X and NM_004641.3, NP_004632.1 for MLLT10. (D) DCDF-FISH with probes for DDX3X in green (RP11-1058N11, flanking 5′, and RP11-10K13, flanking 3′) and MLLT10 in red (RP11-418C1 and RP11-249M6), showed 1 fusion signal (arrow). (E) Unsupervised analysis of 11 T-ALL HOXA patients. In such unsupervised analysis, patients bearing MLLT10 rearrangements and those without MLLT10 rearrangements (1 MLL-ENL, 1 MLL-AF6, 2 TCRB-HOXA, and 1 SET-NUP214) are naturally clustered in 2 distinct groups. PICALM-MLLT10 patients are indicated in orange; patient 1 and patient 2 (HNRNPH1-MLLT10, DDX3X-MLLT10) in green and patients without MLLT10 rearrangements in blue. (F) Supervised analysis was created using the significative probe sets from the comparison of HOXA patients with MLLT10 rearrangements (4 with PICALM-MLLT10 and the 2 new cases with HNRNPH1-MLLT10, DDX3X-MLLT10) vs patients without MLLT10 rearrangements (1 MLL-ENL, 1 MLL-AF6, 2 TCRB-HOXA, and 1 SET-NUP214). Patients bearing MLLT10 recombinations are indicated in orange or green while patients without MLLT10 rearrangements are indicated in blue. DCDF, double-color double-fusion; LAP, Leukemia Associated Protein; NLS, nuclear localization signal; PHD, plant homeo domain; RRM, RNA recognition motif.
HNRNPH1 and DDX3X are involved in RNA processing. HNRNPH1 encodes for a member of the ubiquitous heterogeneous nuclear ribonucleoprotein subfamily (hnRNPs). It is an RNA binding protein that is involved in pre-mRNA processing, and mRNA metabolism and transport.13 A HNRNPH1 frameshift mutation was previously described in gastric cancer14 and a HNRNPH1 splice variant with protein truncation was identified in murine breast cancer cells.15 Interestingly, a variant HNRNPH1 protein, covalently modified by O-linked acetyl hexosamine (GlcnaC), was isolated in acute leukemia with 11q23 cytogenetic changes.16
DDX3X is a member of the large family of RNA helicases with a DEAD box domain (Asp-Glu-Ala-Asp) that is involved in RNA transcription, splicing, mRNA transport, translation initiation, and cell-cycle regulation.17 DEAD box RNA helicases were implicated in diverse forms of leukemia.18,19 Recently, mutations inside and outside the DEAD box domain were detected in ∼3% of patients with chronic lymphocytic leukemia.20
Structural analysis of HNRNPH1-MLLT10 and DDX3X-MLLT10 fusions showed HNRNPH1 maintained 3 RNA recognition motifs while DDX3X lost the DEAD box domain, at the N terminus. At the C terminus, MLLT10 lost 2 of 3 NLS domains in patient 1 but maintained all 3 in patient 2 (Figure 1A,C). As in other MLLT10 fusions,3-5 both cases retained the OM-LZ domain, which is needed to induce acute myeloid leukemia in mice bearing PICALM-MLLT10 or MLL-MLLT10.6,7 Interestingly, DOT1L inhibitors binding the OM-LZ domain were successful in controlling MLL-MLLT10 and PICALM-MLLT10 murine leukemias.21
Whether these rare MLLT10 partners are part of a functional complex or share a common regulation pathway remains to be investigated. The Net View Tools software (http://netview.tigem.it/netview_project/netview_tools.html)22 showed DDX3X was significantly coexpressed and directly linked (mutual information >0.1) to HNRNPH1, PICALM, and NAP1L1 (supplemental Table 3).
We applied GEP to determine whether HNRNPH1-MLLT10 and DDX3X-MLLT10 shared leukemogenic properties with other MLLT10 recombinations within the HOXA category of T-ALL.23,24 In an unsupervised analysis of 11 T-ALL samples with the HOXA signature, the 6 cases with MLLT10 rearrangements (4 PICALM-MLLT10, 1 HNRNPH1-MLLT10, and 1 DDX3X-MLLT10) clustered separately from the other 5 cases (1 MLL-ENL, 1 MLL-AF6, 2 TCRB-HOXA, and 1 SET-NUP214) (Figure 1E). t test analysis revealed significant (lFDR < 0.05) differences in expression of 280 probe sets (supplemental Table 4). Supervised analysis with these probe sets confirmed 2 subgroups (Figure 1F). In the HOXA patients with MLLT10 rearrangements, HHEX gene expression was higher (>1.5 fold-change) than in those without. HHEX is highly expressed in normal hematopoietic stem cells and downregulated during normal T-cell development.25 HHEX overexpression was found in ETP-ALL (as seen in our case no. 1) and linked to upregulation of MEF2C which directly binds the HHEX promoter.2 Interestingly, HHEX is a member of the Nirenberg and Kim–like family of class II homeobox genes. In the other subgroup of 5 patients without MLLT10 rearrangements, gene set enrichment analysis showed enrichment of HOXA class I homeobox genes and target genes (supplemental Figure 2). Although these findings need to be confirmed in a larger series of T-ALL, the present results suggest that MLLT10 recombinations underlie a specific signature, within the HOXA category of T-ALL.
The present study provides insights into the biological pathways involved in MLLT10 recombinations in pediatric T-ALL. Finding 2 new MLLT10 fusion genes, involving HNRNPH1 and DDX3X, highlights the role of the MLLT10 gene, and particularly of its OM-LZ domain in this type of leukemia.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Diego Di Bernardo (University of Naples “Federico II”, Naples, Italy) for assistance in the NetViewTools analysis. F.N. was on leave from the Laboratory of Clinical Research, Istituto di Ricovero e Cura a Carattere Scientifico- Centro di Riferimento Oncologico della Basilicata Rionero in Vulture, Potenza, Italy.
L.B. is supported by a grant from Beat Leukemia Organizzazione Non Lucrativa di Utilita’ Sociale (ONLUS) and Società Italiana di Ematologia Sperimentale (SIES). M.G. is supported by Fondazione Città della Speranza ONLUS. G.t.K. is supported by Fondazione Cariparo progetto d’eccellenza. C.M. is partially supported by Associazione Italiana per la Ricerca sul Cancro (AIRC; IG-11512) and Fondo per gli Investimenti della Ricerca di Base (FIRB 2011 RBAP11TF7Z_005).
Authorship
Contribution: F.N. and V.P. performed karyotype analysis; V.P. selected DNA clones and performed FISH experiments; R.L.S. supervised FISH experiments and drafted the paper; L.B. designed and performed molecular experiments and drafted the paper; D.D.G. and P.G. were involved in cloning and sequencing; C.B., M.G., and G.t.K. performed GEP studies; G.C. performed SNP analysis; and C.M. was responsible for the conception of the study, supervision, and manuscript preparation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Cristina Mecucci, Haematology and Bone Marrow Transplantation Unit, University Hospital, 06132 Perugia, Italy; e-mail: crimecux@unipg.it.