Key Points
UBAP2L interacts with BMI1 as part of a novel Polycomb subcomplex.
UBAP2L regulates HSC activity via a mechanism unrelated to the repression of the Ink4a/Arf locus.
Abstract
Multipotent long-term repopulating hematopoietic stem cells (LT-HSCs) can self-renew or differentiate into the less primitive short-term repopulating stem cells (ST-HSCs), which themselves produce progenitors that ensure the daily supply of all essential blood components. The Polycomb group (PcG) protein BMI1 is essential for the activity of both HSCs and progenitor cells. Although BMI1 operates by suppressing the Ink4a/Arf locus in progenitors and ST-HSCs, the mechanisms through which this gene regulates the activity of LT-HSCs remain poorly understood. Toward this goal, we isolated BMI1-containing protein complexes and identified UBAP2L as a novel BMI1-interacting protein. We also showed that UBAP2L is preferentially expressed in mouse and human HSC-enriched populations when compared with more mature cell types, and that this gene is essential for the activity of LT-HSCs. In contrast to what is observed for Bmi1 knockdown, we found that UBAP2L depletion does not affect the Ink4a/Arf locus. Given that we demonstrated that BMI1 overexpression is able to rescue the deleterious effects of Ubap2l downregulation on LT-HSC activity and that UBAP2L is part of a PcG subcomplex comprising BMI1, we propose a model in which at least 2 different BMI1-containing PcG complexes regulate HSC activity, which are distinguishable by the presence of UBAP2L.
Introduction
Bmi1 is a well-known determinant of hematopoietic stem cell (HSC) function. Bmi1−/− mice display severe hematopoietic defects, including progressive loss of hematopoietic cells from the bone marrow (BM).1 The frequency of long-term repopulating HSCs (LT-HSCs) is normal in Bmi1−/− fetal livers (FLs), but is decreased in adult Bmi1−/− BM, suggesting that Bmi1 is dispensable for HSC specification, but essential for their maintenance.2,3 Bmi1−/− FL cells fail to repopulate recipient mice, highlighting the cell autonomous nature of the Bmi1−/− phenotype in HSCs.2,3 Retroviral introduction of Bmi1 in these cells rescues this defect, implying that Bmi1 is essential for HSC self-renewal.2 Bmi1 has been shown to prevent premature senescence by repressing the Ink4a/Arf locus encoding the cell-cycle inhibitors p16Ink4a and p19Arf; however, concomitant deletion of these genes, or deletion of Trp53 in Bmi1−/− mice, does not completely restore the hematopoietic defects observed in these mice.4-10 Silencing of the BMI1-interacting protein E4F1 by RNA interference rescues the short-term repopulation activity of Bmi1−/− FL cells, but this is not sufficient to bring back the long-term repopulation activity of these cells to normal levels.8 More recently, BMI1 has been shown to regulate the DNA damage response pathway.11,12 Although some aspects of the Bmi1−/− hematopoietic phenotype can be alleviated by disruption of the DNA damage response pathway via Chk2 deletion, the long-term repopulation activity of HSCs remained impaired.11 It thus appears that the mechanism by which BMI1 regulates self-renewal of LT-HSCs remains to be determined.
Methods
Ethics approval
Human leukemia samples were collected with informed consent and cryopreserved by the Leukemia Cell Bank of Quebec. RNA sequencing was performed on all acute myeloid leukemia (AML) samples that were transplanted into NSG mice as part of the Leucégène Project at the Institute for Research in Immunology and Cancer (IRIC). The Leucégène Project was approved by the research ethics board of Hôpital Maisonneuve-Rosemont and Université de Montréal; animal manipulations were approved by the research ethics board of Université de Montréal.
Large-scale purification of BMI1-containing protein complexes
BMI1-PC-Flag expression was induced in HEK 293 cells with 1 µg/mL doxycycline for 48 hours. Cells were lysed in buffer A (25 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid [HEPES] pH 7.0, 25 mM KCl, 0.05 mM EDTA, 5 mM MgCl2, 10% glycerol, 0.1% Nonidet P-40 [NP-40]) on ice for 30 minutes. Lysate was centrifuged at 850g for 5 minutes and pellet was resuspended in buffer B (50 mM HEPES pH 7.6, 50 mM KCl, 0.1 mM EDTA, 10% glycerol, 0.3 M ammonium sulfate) and incubated with rotation for 30 minutes. Lysate was centrifuged at 400 000g for 12 minutes and supernatant was dialyzed against buffer C (50 mM HEPES 7.6, 300 mM KCl, 1 mM EDTA, 0.1% NP-40, 10% glycerol). Anti-Flag resin was added to the extract and incubated with rotation for 5 hours. Beads were washed with buffer C and Flag rising buffer (50 mM NH4HCO3 pH 8.0, 75 mM KCl, 2 mM EDTA). Proteins were eluted with Flag elution buffer (0.5 M NH4OH pH 11.5, 0.5 mM EDTA) and analyzed by mass spectrometry (MS).
MS analysis
Samples were dried in a speed-vac and reconstituted in 50 mM ammonium bicarbonate. Tris (2-carboxyethyl) phosphine hydrochloride was added at the final concentration of 5 mM. The digestion was performed by adding 1 µg of trypsin in 50 mM ammonium bicarbonate overnight at 37°C. Samples were dried in a speed-vac and reconstituted in 50 µL of 0.2% aqueous formic acid. Samples were then separated by online 2-dimensional (2D) separation (strong cation exchange/reverse-phase chromatography [SCX/C18]) using an Eksigent nanoLC-2D system and diluted in H2O/2% acetonitrile/0.2% formic acid before liquid chromatography–MS analyses. The Optimize Technologies SCX column (0.3 mm internal diameter [i.d.] × 45 mm) was connected directly to the switching valve. The sample was loaded on an SCX column at 10 µL per minute for 5 minutes. Peptides were eluted onto the C18 precolumn (0.3 mm i.d. × 45 mm) using pulsed fractions of 0, 250, 500, 750, 1 M, and 2 M ammonium acetate (pH 3.0). A 56-minute gradient from 10% to 60% acetonitrile (0.2% formic acid [FA]) was used to elute peptides from a homemade reversed-phase column (150 µm i.d. × 100 mm) with a flow rate set at 600 nL per minute. The column was directly connected to a nanoprobe interfaced with an LTQ-Orbitrap XL mass spectrometer from Thermo Fisher. Each full MS spectrum was followed by 6 MS/MS spectra (7 scan events), where the 6 most abundant multiply charged ions were selected for MS/MS sequencing. Tandem MS experiments were performed using collision-induced dissociation in the linear ion trap. The data were processed using the 2.4 Mascot (Matrix Science) search engine with tolerance parameters set to 15 ppm and 0.5 Da for the precursor and the fragment ions, respectively, to achieve a false discovery rate of <1% (P < .01). The selected variable modifications were carbamidomethyl (C), deamidation (NQ), oxidation (M), and phosphorylation (STY). Tandem mass spectra were searched against the Uniprot human database (301 754 sequences).
Immunoprecipitations and western blotting
Cellular extracts were prepared as described for the BMI1-containing complex purification section. Immunoprecipitations and western blotting were performed according to published methods using the following antibodies: α-UBAP2L (Sigma-Aldrich), α-Flag (Sigma-Aldrich), α-BMI1 (EMD Millipore), α-RNF2 (MBL International), and α-TUBA (Cell Signaling Technology).
Generation of conditional and knockout Ubap2l mutant mice
A plasmid containing a 10-kb genomic insert including exons 2 to 4 of Ubap2l was obtained by homologous recombination in bacteria using the RP23-16L4 BAC clone (BACPAC) and the recombineering system from the National Cancer Institute. A LoxP flanked selection cassette was introduced upstream of the start codon (ATG)-containing exon 2 and recombined using a Cre recombinase-expressing bacterial strain to generate a single LoxP site. A second selection cassette was then introduced downstream of exon 4 to generate the final targeting vector. The plasmid backbone of the construct was cut with restriction enzymes prior to electroporation into a C2 embryonic stem cell (ES) line. ES clones were selected with G418 for 7 to 9 days, randomly picked, and analyzed by Southern blotting to ensure proper integration into the endogenous Ubap2l locus. Selected clones were further expanded and electroporated with a Cre- or Flpe-expressing plasmid to generate either a null or a conditional allele, respectively, and reanalyzed by Southern blotting to confirm correct rearrangement. A clone of each modified allele was microinjected into blastocysts obtained from C57BL/6J albino mice and introduced into foster mothers of the same strain. Chimeras from each litter were then crossed with C57BL/6J animals, and progeny carrying each modification were maintained on a pure C57BL/6J genetic background. Mice were housed and treated in accordance with institutional and governmental regulations. C57BL/6J-Pep3b mice were purchased from The Jackson Laboratory. Ubap2l mutant mice were maintained on a pure C57BL/6J genetic background.
HSC isolation, infection, qRT-PCR, CFC assays, and transplantations
BM cells were isolated from C57BL/6J-Ly5.1-Pep3b mice and stained with allophycocyanin (APC)–conjugated lineage antibodies (GR1, B220, TER119 [AbLab]). Cells were then stained with anti-APC magnetic MicroBeads according to the manufacturer’s instructions (Miltenyi Biotec) and depletion of lineage-positive cells was performed using the AutoMACS magnetic cell separator system (Miltenyi Biotec). The lineage-negative population was subsequently stained with a phycoerythrin (PE)–conjugated anti-SCA1 antibody (BD Biosciences) and the Lin−SCA1+ cell population was sorted using a FACSAria cell sorter (BD Biosciences). Isolation of CD150+CD48−Lin−SCA1+cKIT+ populations was performed as previously described.13 Short hairpin RNAs (shRNAs) targeting Ubap2l were generated as previously described14 and generation of retroviruses producing GP+E-86 using shRNA vectors was performed using a published method.15 Freshly sorted Lin−SCA1+ cells were prestimulated for 24 hours in Dulbecco modified Eagle medium supplemented with 15% fetal bovine serum, 10 ng/mL interleukin-3 and interleukin-6, 100 ng/mL steel factor, 50 µg/mL gentamycin, 10 µg/mL ciprofloxacin, and 10−4 M β-mercaptoethanol, and cocultured with GP+E-86 cells for 3 days in the presence of 6 µg/mL polybrene. Following infection, cells were either kept in culture to perform quantitative reverse transcription–polymerase chain reaction (qRT-PCR) and colony forming cell (CFC) assays as previously described8 or transplanted into sublethally irradiated (800 cGy 137Cs γ irradiation) C57BL/6J-Ly5.2 recipients along with a radioprotective dose of C57BL/6J-Ly5.1-Pep3b total BM cells. Competitive repopulation experiments using Ubap2l conditional knockout mice were performed as follows: 1 million donor (Mx1Cre;Rosa26YFP;Ubap2l+/+ or Mx1Cre;Rosa26YFP;Ubap2lFloxed/Floxed) Ly5.2 BM cells were transplanted into sublethally irradiated (800 cGy 137Cs γ irradiation) mice along with 1 million competitor Ly5.1 BM cells. One month posttransplantation, to activate the Mxcre transgene in vivo, mice were injected intraperitoneally with polyinosinic-polycytidylic acid (pIpC; GE Healthcare), 4 times every other day (a first dose of 150 µg followed by 3 doses of 125 µg) and the repopulation activity of the deleted cells was monitored in the peripheral blood and BM.
Flow cytometry
Evaluation of donor-derived reconstitution was performed as previously described.14 For the staining of the myeloid and lymphoid populations of the BM, the following antibodies were used: PE-Cy5–conjugated α-MAC1 (CD11b) (Abcam), APC-conjugated α-B220 (AbLab), APC-conjugated α-GR1 (AbLab).
Isolation of UBAP2L-BMI1-RNF2-PHC1 complex
Cellular extracts were prepared as described for the BMI1-containing complex purification section. Extracts were loaded on a 10% to 20% glycerol gradient (37.5 mM HEPES-KOH pH 7.9, 90 mM KCl, 6.25 mM MgCl2, 0.05 mM EDTA) and centrifuged at 130 000g for 24 hours in a SW41 rotor. Fractions were collected using a FoxyR1 fraction collector (Teledyne Isco).
Results
UBAP2L is a novel BMI1-interacting protein
To identify proteins potentially involved in the regulation of HSC activity by BMI1, we searched for novel BMI1-interacting proteins using affinity purification (Figure 1A). We expressed BMI1-PC-Flag protein under an inducible promoter and selected clones for which levels of tagged BMI1 were similar to those of the endogenous protein (Figure 1A). We then isolated protein complexes comprising BMI1 from cellular extracts using an anti-Flag resin and identified BMI1 interaction partners by MS. BMI1 is a member of the PcG protein family, whose members assemble into protein complexes that establish and maintain a repressed chromatin state,16 and as expected, many PcG proteins, such as RNF2 for example, copurified with BMI1 (Figure 1B). Interestingly, the protein UBAP2L (Ubiquitin-associated protein 2-like), which has never been shown to associate with BMI1 and for which no link with PcG protein function has been described, was consistently found in BMI1-containing protein complexes (Figure 1B). To confirm the interaction between UBAP2L and BMI1, we used the reverse approach and performed immunoprecipitations in HEK 293 cells expressing Flag-UBAP2L. Using an anti-Flag antibody, we observed that BMI1 and RNF2 copurify with Flag-UBAP2L (Figure 1C, lanes 1-4). Moreover, immunoprecipitations performed using an antibody against the known BMI1-interacting protein RNF2 revealed the presence of BMI1 and Flag-UBAP2L in RNF2 immunoprecipitates (Figure 1C, lanes 5-8). Altogether, these results suggest that UBAP2L associates with BMI1 and RNF2.
UBAP2L is a novel BMI1-interacting protein. (A) Strategy for large-scale purification of BMI1-containing protein complexes using a Tet-On system to induce expression of BMI1-PC-Flag. *The clone used to perform complex purification. (B) Proteins identified by MS in eluates from Flag-resin following complex purification. PcG proteins are in bold. Three independent experiments were performed and results from a representative experiment are shown. Proteins identified with a minimum of 2 unique peptides and present in at least 2 of the 3 experiments were included in the table (false discovery rate <1%). UBAP2L was found in all 3 experiments. (C) Immunoprecipitations performed using extracts from HEK 293 control or Flag-UBAP2L–expressing cells. The antibodies used to perform the immunoprecipitations (top right) and the immunoblottings (bottom right) are indicated. *The position of full-length UBAP2L protein.
UBAP2L is a novel BMI1-interacting protein. (A) Strategy for large-scale purification of BMI1-containing protein complexes using a Tet-On system to induce expression of BMI1-PC-Flag. *The clone used to perform complex purification. (B) Proteins identified by MS in eluates from Flag-resin following complex purification. PcG proteins are in bold. Three independent experiments were performed and results from a representative experiment are shown. Proteins identified with a minimum of 2 unique peptides and present in at least 2 of the 3 experiments were included in the table (false discovery rate <1%). UBAP2L was found in all 3 experiments. (C) Immunoprecipitations performed using extracts from HEK 293 control or Flag-UBAP2L–expressing cells. The antibodies used to perform the immunoprecipitations (top right) and the immunoblottings (bottom right) are indicated. *The position of full-length UBAP2L protein.
Ubap2l is preferentially expressed in HSCs and AML specimens with high stem cell content
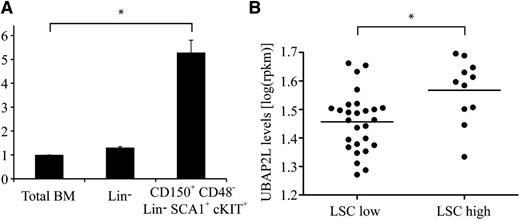
To evaluate the possibility that UBAP2L might be involved in the regulation of HSC activity, we first quantified Ubap2l expression in mouse HSCs. Ubap2l transcripts were found to be ∼5 times more abundant in phenotypic LT-HSCs (CD150+CD48−Lin−SCA1+cKIT+ cells, tested competitive repopulating unit (CRU) frequency of 1:3 to 1:417 ) compared with total BM, as opposed to lineage-depleted (Lin−) cells, for which Ubap2l levels were comparable to those of total BM (Figure 2A). These results suggest a higher requirement for Ubap2l in primitive HSC populations, a tendency also observed for human cord blood HSC populations (supplemental Figure 1, available on the Blood Web site). Given that Bmi1 is essential for the maintenance of leukemic stem cells (LSCs), we monitored UBAP2L messenger RNA (mRNA) levels in a panel of human primary AML specimens. We observed increased UBAP2L levels in AML samples with high LSC frequency (LSC high) compared with specimens showing low LSC frequency (LSC low) (Figure 2B and supplemental Table 1, tested LSC frequency ≥1:50 000 for LSC-high specimens and ≤1:2 000 000 for LSC-low specimens),18 suggesting that Ubap2l might play an important role in LSC activity.
Ubap2l is preferentially expressed in primitive HSCs and AML specimens with high stem cell frequency. (A) Ubap2l mRNA levels in total mouse BM, Lin− cells and primitive HSCs as assessed by qRT-PCR. The values are expressed relative to Gapdh, Tbp, and Hprt. The average of 2 independent experiments is presented with the standard error of the mean. The values for total BM were set to 1. *Statistically significant difference (total BM vs CD150+CD48−Lin−SCA1+cKIT+: P = .0012). (B) UBAP2L mRNA levels in various human primary AML specimens as determined by RNA sequencing. LSC low refers to AML specimens with low stem cell frequency, and LSC high, to specimens with high stem cell frequency. *Statistically significant difference (P = .0057).
Ubap2l is preferentially expressed in primitive HSCs and AML specimens with high stem cell frequency. (A) Ubap2l mRNA levels in total mouse BM, Lin− cells and primitive HSCs as assessed by qRT-PCR. The values are expressed relative to Gapdh, Tbp, and Hprt. The average of 2 independent experiments is presented with the standard error of the mean. The values for total BM were set to 1. *Statistically significant difference (total BM vs CD150+CD48−Lin−SCA1+cKIT+: P = .0012). (B) UBAP2L mRNA levels in various human primary AML specimens as determined by RNA sequencing. LSC low refers to AML specimens with low stem cell frequency, and LSC high, to specimens with high stem cell frequency. *Statistically significant difference (P = .0057).
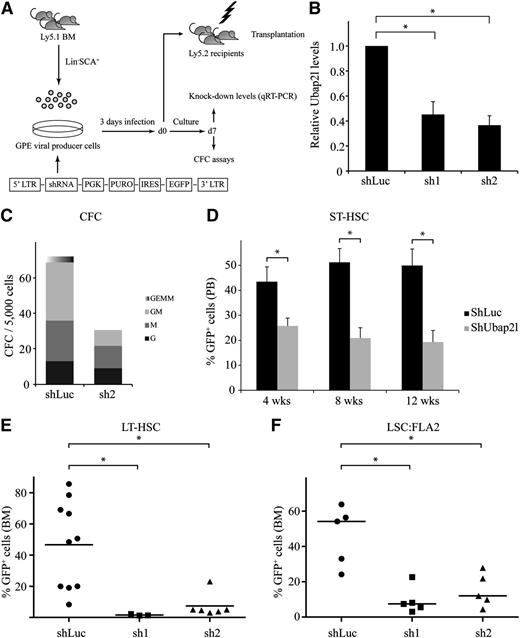
shRNA-mediated Ubap2l downregulation results in decreased progenitor and HSC activity
The above results prompted us to investigate whether manipulating Ubap2l levels would impact on HSC and LSC activity. We first generated shRNA retroviral vectors targeting different regions of the mouse Ubap2l coding sequence, and infected Lin−SCA1+ mouse BM cells with these constructs (Figure 3A). Ubap2l knockdown levels in these cells ranged from 55% to 65% (Figure 3B). CFC assays performed with the most effective shRNA (Figure 3B sh2) revealed that Ubap2l knockdown causes a reduction of progenitor activity, with multipotent and bipotent progenitors being more affected than unipotent progenitors (Figure 3C). We transplanted cells infected with the different shRNAs in sublethally irradiated mice along with a radioprotective dose of host BM cells and monitored the repopulation activity of these cells in the peripheral blood of the recipients at different times following transplantation. This allowed the retrospective evaluation of the effects of Ubap2l knockdown on the activity of short-term repopulating stem cells (ST-HSCs). A decrease in ST-HSC activity was noted, as determined by the reduction in the percentage of green fluorescent protein-positive (GFP+) cells (donor-derived and expressing Ubap2l shRNA) in the recipient’s peripheral blood (Figure 3D). We then tested the effects of Ubap2l knockdown on the activity of LT-HSCs by monitoring the reconstitution activity of Lin−SCA1+ cells infected with the different Ubap2l shRNAs in the recipient’s BM 16 weeks posttransplantation and observed that almost all GFP+ cells were lost in the BM of these mice for both Ubap2l shRNAs (Figure 3E). Altogether, these results suggest that manipulating Ubap2l levels impacts on progenitor and HSC activity. We next tested the effect of Ubap2l silencing on LSC activity by assessing the impact of the different Ubap2l shRNAs on the reconstitution potential of FLA2 leukemia, a FL-derived leukemia generated by the retroviral overexpression of Meis1 and Hoxa9 with an LSC frequency of 1 in 1.4.19 A reduction in Ubap2l levels in these cells had a dramatic impact on their ability to repopulate recipient mice (Figure 3F), suggesting that Ubap2l also plays an important role in LSC activity.
shRNA-mediated Ubap2l downregulation results in reduced progenitor and HSC activity. (A) Schematic representation of the shRNA-mediated gene knockdown approach. (B) Ubap2l knockdown levels in Lin−SCA1+ cells infected with 2 different shRNAs targeting Ubap2l (sh1 and sh2) as determined by qRT-PCR. The values for Ubap2l mRNA levels are expressed relative to Tbp and Hprt. The average of 2 independent experiments is presented with the standard error of the mean. The values for shLuc were set to 1. *Statistically significant differences (shLuc vs sh1: P = .0060, shLuc vs sh2: P = .0011). (C) CFC content of Lin−SCA1+ cells infected with sh2 determined by morphological analysis at day 7 of the culture. Colonies were grown in the presence of puromycin. Representative of 2 independent experiments performed in duplicate. (D) Reconstitution activity of Lin−SCA1+ cells infected with the different Ubap2l shRNAs (see panel B) as assessed by the percentage of GFP+ cells in the peripheral blood of the recipients 4, 8, and 12 weeks posttransplantation. The average of 2 independent experiments (n = 6-9 mice per condition) is shown with the standard error of the mean. Results for sh1 and sh2 were combined. *Statistically significant differences (4 weeks: P = .0154, 8 weeks: P = .0006, 12 weeks: P = .0017). (E) GFP levels in BM of mice transplanted with Lin−SCA1+ cells expressing sh1 and sh2 16 weeks posttransplantation. The average of 2 independent experiments is presented with the standard error of the mean. *Statistically significant differences (shLuc vs sh1: P = .0210, shLuc vs sh2: P = .0053). (F) Effect of sh1 and sh2 on the repopulation activity of FLA2 leukemia cells as determined by the percentage of GFP+ cells in recipients’ BM when signs of leukemia appearance was noted. Representative of 2 independent experiments with the mean of 5 mice per condition shown with the standard error of the mean. *Statistically significant differences (shLuc vs sh1: P = .0021, shLuc vs sh2: P = .0070).
shRNA-mediated Ubap2l downregulation results in reduced progenitor and HSC activity. (A) Schematic representation of the shRNA-mediated gene knockdown approach. (B) Ubap2l knockdown levels in Lin−SCA1+ cells infected with 2 different shRNAs targeting Ubap2l (sh1 and sh2) as determined by qRT-PCR. The values for Ubap2l mRNA levels are expressed relative to Tbp and Hprt. The average of 2 independent experiments is presented with the standard error of the mean. The values for shLuc were set to 1. *Statistically significant differences (shLuc vs sh1: P = .0060, shLuc vs sh2: P = .0011). (C) CFC content of Lin−SCA1+ cells infected with sh2 determined by morphological analysis at day 7 of the culture. Colonies were grown in the presence of puromycin. Representative of 2 independent experiments performed in duplicate. (D) Reconstitution activity of Lin−SCA1+ cells infected with the different Ubap2l shRNAs (see panel B) as assessed by the percentage of GFP+ cells in the peripheral blood of the recipients 4, 8, and 12 weeks posttransplantation. The average of 2 independent experiments (n = 6-9 mice per condition) is shown with the standard error of the mean. Results for sh1 and sh2 were combined. *Statistically significant differences (4 weeks: P = .0154, 8 weeks: P = .0006, 12 weeks: P = .0017). (E) GFP levels in BM of mice transplanted with Lin−SCA1+ cells expressing sh1 and sh2 16 weeks posttransplantation. The average of 2 independent experiments is presented with the standard error of the mean. *Statistically significant differences (shLuc vs sh1: P = .0210, shLuc vs sh2: P = .0053). (F) Effect of sh1 and sh2 on the repopulation activity of FLA2 leukemia cells as determined by the percentage of GFP+ cells in recipients’ BM when signs of leukemia appearance was noted. Representative of 2 independent experiments with the mean of 5 mice per condition shown with the standard error of the mean. *Statistically significant differences (shLuc vs sh1: P = .0021, shLuc vs sh2: P = .0070).
Ubap2l-deleted BM cells show reduced repopulation activity
As an alternative approach to confirm the role of UBAP2L in HSC activity, we generated conditional and knockout Ubap2l mice by targeting exons 2, 3, and 4 of the Ubap2l gene (Figure 4A) and by taking advantage of the Mx1Cre;Rosa26YFP system for the conditional knockout mouse, in which yellow fluorescent protein (YFP) expression allows for the monitoring of excised cells.20,21 Southern blot analysis of genomic DNA isolated from selected ES clones (Figure 4B) and mouse genotyping (supplemental Figure 2) confirmed proper targeting of the Ubap2l locus, and deletion of UBAP2L was confirmed by qRT-PCR (Figure 4C) and western blotting (Figure 4D). To evaluate the impact of Ubap2l deletion on HSC function, BM cells from Mx1Cre;Rosa26YFP;Ubap2l+/+ and Mx1Cre;Rosa26YFP;Ubap2lFloxed/Floxed mice were transplanted into sublethally irradiated mice along with an equivalent dose of competitor BM cells. One month posttransplantation, mice were treated with pIpC and the repopulation activity of the deleted cells was monitored at different times following pIpC treatment. We found that Ubap2l deletion causes an important decrease in the percentage of donor cells (Figure 4E, left panel) and YFP+ cells (donor-derived and Ubap2l excised, Figure 4E, right panel) in the recipient’s peripheral blood at all time points tested. Similar effects were also observed in recipient’s BM (Figure 4F). We observed a small reduction in repopulation activity for the Ubap2lFloxed/Floxed BM cells compared with wild-type cells before pIpC treatment (Figure 4E, left panel). We believe that this results from Cre activation following irradiation of the mice before transplantation. This hypothesis is supported by our ability to detect YFP+ cells in recipient’s peripheral blood before pIpC treatment (Figure 4E, right panel). Analysis of the deleted cells revealed a small reduction in all cell subpopulations of the BM, spleen and thymus, reminiscent of a stem cell phenotype (data can be found in the supplemental Tables). Altogether, these results confirm that UBAP2L plays an important role in HSC function. Of note is the observation that although the knockdown was incomplete, a greater impact on LT-HSCs was noted for Ubap2l shRNAs (Figure 3) compared with conditional deletion (Figure 4). This could be explained by the infection procedure of the Lin−SCA1+ cells, which implies keeping the cells in culture for at least 3 days. As it is the case for Bmi1 deletion,12 this situation might have induced stress to the cells, suggesting that the more severe phenotype observed with shRNAs in Lin−SCA1+ cells is likely to be the result of cumulative effects rather than being attributable only to the knockdown of Ubap2l.
Conditional Ubap2l deletion results in loss of HSC activity. (A) Targeting strategy to generate the conditional and knockout Ubap2l mice. 2-5, Ubap2l exons; LoxP, Cre-recombinase recognition site; Frt, Flpe recombinase recognition site; Neo, neomycin selection cassette. (B) Southern blot analysis of genomic DNA isolated from selected ES clones to confirm the proper targeting of the Ubap2l locus. The introduction of an extra EcoRI site located immediately before the 5′ LoxP site allowed for the detection of an additional genomic restriction fragment of 3.59 kb when using radiolabeled probe hybridizing to a region located upstream of the targeted region (left panel). The correct recombination of the targeted region was verified at the 3′ end by the detection of a 8.58-kb fragment corresponding to the insertion of the FRT-pgk-NEO-pA-FRT-LoxP cassette using a probe hybridizing downstream of the targeted region (right panel). Single integration of the targeting fragment was confirmed by the detection of the same band using a NEO probe (data not shown). Efficient recombination between the LoxP and FRT sites was monitored in ES clones after transfection with either Flpe- or Cre-expressing plasmid by the detection of restriction fragments of the appropriate length using the indicated probes. (C) Relative Ubap2l mRNA expression in MEFs. 3-4, 9-10, 14-15, and 21-22 refer to the exons targeted by the primers used for qRT-PCR. The values are expressed relative to Hprt. The average of 3 independent experiments is presented with the standard deviation. The values for MEFs +/+ were set to 1. (D) Western blot validating the absence of UBAP2L protein in Ubap2l−/− MEFs. (E) Competitive repopulation activity of conditional Ubap2l knockout BM in recipient’s peripheral blood at different time points (4 weeks posttransplantation and 18 days, 4 weeks, 8 weeks, 12 weeks, 16 weeks, and 20 weeks post-pIpC treatment). Shown are means with standard deviation (n = 15 mice per genotype). The Mann-Whitney test was used for statistical analysis. *P < .05. (F) Competitive repopulation activity of conditional Ubap2l knockout BM in recipient’s BM 28 weeks post-pIpC treatment. Shown are means with standard deviation (n = 6 mice per genotype). The Mann-Whitney test was used for statistical analysis. *P < .05.
Conditional Ubap2l deletion results in loss of HSC activity. (A) Targeting strategy to generate the conditional and knockout Ubap2l mice. 2-5, Ubap2l exons; LoxP, Cre-recombinase recognition site; Frt, Flpe recombinase recognition site; Neo, neomycin selection cassette. (B) Southern blot analysis of genomic DNA isolated from selected ES clones to confirm the proper targeting of the Ubap2l locus. The introduction of an extra EcoRI site located immediately before the 5′ LoxP site allowed for the detection of an additional genomic restriction fragment of 3.59 kb when using radiolabeled probe hybridizing to a region located upstream of the targeted region (left panel). The correct recombination of the targeted region was verified at the 3′ end by the detection of a 8.58-kb fragment corresponding to the insertion of the FRT-pgk-NEO-pA-FRT-LoxP cassette using a probe hybridizing downstream of the targeted region (right panel). Single integration of the targeting fragment was confirmed by the detection of the same band using a NEO probe (data not shown). Efficient recombination between the LoxP and FRT sites was monitored in ES clones after transfection with either Flpe- or Cre-expressing plasmid by the detection of restriction fragments of the appropriate length using the indicated probes. (C) Relative Ubap2l mRNA expression in MEFs. 3-4, 9-10, 14-15, and 21-22 refer to the exons targeted by the primers used for qRT-PCR. The values are expressed relative to Hprt. The average of 3 independent experiments is presented with the standard deviation. The values for MEFs +/+ were set to 1. (D) Western blot validating the absence of UBAP2L protein in Ubap2l−/− MEFs. (E) Competitive repopulation activity of conditional Ubap2l knockout BM in recipient’s peripheral blood at different time points (4 weeks posttransplantation and 18 days, 4 weeks, 8 weeks, 12 weeks, 16 weeks, and 20 weeks post-pIpC treatment). Shown are means with standard deviation (n = 15 mice per genotype). The Mann-Whitney test was used for statistical analysis. *P < .05. (F) Competitive repopulation activity of conditional Ubap2l knockout BM in recipient’s BM 28 weeks post-pIpC treatment. Shown are means with standard deviation (n = 6 mice per genotype). The Mann-Whitney test was used for statistical analysis. *P < .05.
Bmi1 and Ubap2l genetically interact to regulate LT-HSC activity
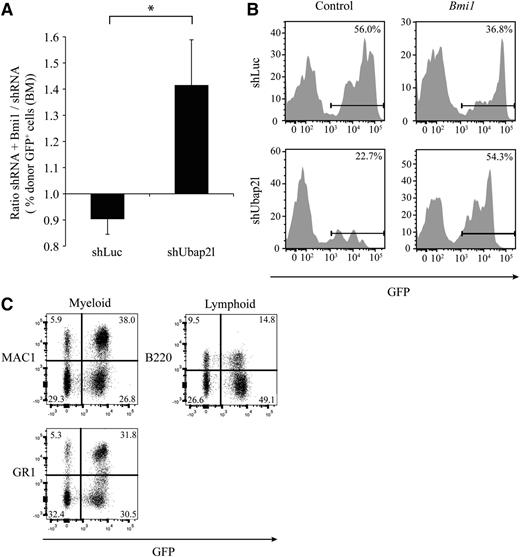
To determine whether the mechanism by which Ubap2l regulates LT-HSC activity is related to Bmi1 function, we simultaneously introduced Bmi1 complementary DNA (cDNA) and Ubap2l sh2 in Lin−SCA1+ BM cells and evaluated the effects of BMI1 overexpression on the reconstitution activity of cells expressing control shLuc or Ubap2l shRNA 16 weeks posttransplantation. Bmi1 overexpression conferred a slight disadvantage to control cells (ie, expressing shLuc) as determined by the small reduction in the percentage of GFP+ cells observed in the recipient’s BM (Figure 5A-B). In contrast, Bmi1 was able to relieve the reduction in repopulation activity imposed by Ubap2l shRNA (Figure 5A-B). ShUbap2l + Bmi1 BM cells isolated from mice had normal morphology (supplemental Figure 3A) and were able to differentiate into myeloid and lymphoid lineages as revealed by the presence of GFP+MAC1+ and GFP+GR1+, as well as GFP+B220+ cells, in the BM of the recipients, respectively (Figure 5C). qRT-PCR analyses of GFP+ sorted BM cells confirmed an overexpression of Bmi1 in rescued cells compared with control cells (supplemental Figure 3B). Importantly, these analyses also revealed that Bmi1 overexpression in these cells allowed them to tolerate lower levels of Ubap2l expression (supplemental Figure 3C). Altogether, these results demonstrate that Bmi1 is able to at least partially rescue the long-term repopulation defect imposed by Ubap2l knockdown in BM cells.
Bmi1 rescues the long-term repopulation defect caused by Ubap2l knockdown. (A) Effect of Bmi1 overexpression on the reconstitution activity of Lin−SCA1+ cells infected with shLuc or shUbap2l, 16 weeks posttransplantation. Shown is the average of 2 independent experiments with 3 different infections (n = 13 mice per condition) with the standard error of the mean. *Statistically significant difference (P = .0486). (B) Representative FACS profiles of donor cells in recipient’s BM 16 weeks posttransplantation of Lin−SCA1+ cells infected with different combinations of shUbap2l and Bmi1 cDNA. (C) Assessment of the contribution of rescued shUbap2l + Bmi1 BM cells (GFP+) to myeloid and lymphoid populations 16 weeks posttransplantation as determined by FACS using MAC1 and GR1, and B220 antibodies, respectively. The results are from 1 mouse and are representative of all rescued BM analyzed. FACS, fluorescence-activated cell sorting.
Bmi1 rescues the long-term repopulation defect caused by Ubap2l knockdown. (A) Effect of Bmi1 overexpression on the reconstitution activity of Lin−SCA1+ cells infected with shLuc or shUbap2l, 16 weeks posttransplantation. Shown is the average of 2 independent experiments with 3 different infections (n = 13 mice per condition) with the standard error of the mean. *Statistically significant difference (P = .0486). (B) Representative FACS profiles of donor cells in recipient’s BM 16 weeks posttransplantation of Lin−SCA1+ cells infected with different combinations of shUbap2l and Bmi1 cDNA. (C) Assessment of the contribution of rescued shUbap2l + Bmi1 BM cells (GFP+) to myeloid and lymphoid populations 16 weeks posttransplantation as determined by FACS using MAC1 and GR1, and B220 antibodies, respectively. The results are from 1 mouse and are representative of all rescued BM analyzed. FACS, fluorescence-activated cell sorting.
UBAP2L silencing does not affect the Ink4a/Arf locus and BMI1 and RNF2 protein levels
One hypothesis to explain the rescue of the effects of Ubap2l knockdown by Bmi1 could be that UBAP2L stabilizes BMI1 protein. To test this, we silenced Ubap2l expression in Lin− cells from mouse BM using Ubap2l shRNA and analyzed BMI1 protein levels by western blotting. shRNA-mediated reduction of Ubap2l levels did not affect BMI1 and RNF2 expression in Lin− cells (Figure 6A) or in HEK 293 and HeLa cells (data not shown). We also monitored BMI1 and RNF2 protein levels in Ubap2l−/− mouse embryonic fibroblasts (MEFs). We observed that BMI1 and RNF2 expression in these cells is similar to that of wild-type MEFs (Figure 6B). We then analyzed the impact of manipulating Ubap2l levels on the expression of the known BMI1 target p16Ink4a, and did not detect major changes in the transcript levels of this gene in Lin−SCA1+ cells infected with Ubap2l shRNA (Figure 6C) or in Ubap2l−/− MEFs (Figure 6D). Similar results were also observed in NIH 3T3 cells (supplemental Figure 4). These results suggest that UBAP2L is not involved in the regulation of the Ink4a/Arf locus, and that an additional BMI1-dependent mechanism, linked to UBAP2L and unrelated to repression of the Ink4a/Arf locus, regulates HSC activity.
UBAP2L does not affect BMI1 and RNF2 protein levels, and p16Ink4a expression, and forms a distinct PcG complex with BMI1. (A) Effect of Ubap2l silencing on BMI1 and RNF2 protein levels in Lin− cells. Following infection of Lin− cells with shUbap2l, GFP+ cells were sorted. Ubap2l knockdown was determined by qRT-PCR (left panel). The values for Ubap2l mRNA levels are expressed relative to Hprt with the standard error of the mean shown and the value for shLuc was set to 1. BMI1 and RNF2 protein levels were monitored by western blotting (right panel). (B) Effect of Ubap2l deletion on BMI1 and RNF2 protein levels determined by western blotting in Ubap2l+/+, Ubap2l+/−, and Ubap2l−/− MEFs. (C) Impact of Ubap2l knockdown on p16Ink4a levels. Lin−SCA1+ cells were infected with shUbap2l or shBmi1 and the mRNA levels of p16Ink4a were determined by qRT-PCR immediately after infection. For all genes, the values are expressed relative to Hprt and the value for shLuc was set to 1. The average of 2 infections is shown with the standard error of the mean. Representative of 2 independent experiments. (D) Effect of Ubap2l deletion on p16Ink4a levels. p16Ink4a mRNA levels were determined by qRT-PCR in Ubap2l+/+ and Ubap2l−/− MEFs. The values are expressed relative to Hprt and the value for Ubap2l+/+ MEFs was set to 1. Shown are results for MEFs from 1 Ubap2l−/− mouse with the standard error of the mean. Representative of 2 independent experiments. (E) Identification of a BMI1 PcG subcomplex containing UBAP2L. HEK 293 cells were transfected with Flag-UBAP2L and cell extracts were fractionated on glycerol gradients. The presence of Flag-UBAP2L, BMI1, RNF2, and PHC1 in the fractions of the gradient was determined by western blotting with the antibodies indicated to the right. *The position of full-length Flag-UBAP2L protein. (F) Model for the role of UBAP2L in the regulation of HSC activity.
UBAP2L does not affect BMI1 and RNF2 protein levels, and p16Ink4a expression, and forms a distinct PcG complex with BMI1. (A) Effect of Ubap2l silencing on BMI1 and RNF2 protein levels in Lin− cells. Following infection of Lin− cells with shUbap2l, GFP+ cells were sorted. Ubap2l knockdown was determined by qRT-PCR (left panel). The values for Ubap2l mRNA levels are expressed relative to Hprt with the standard error of the mean shown and the value for shLuc was set to 1. BMI1 and RNF2 protein levels were monitored by western blotting (right panel). (B) Effect of Ubap2l deletion on BMI1 and RNF2 protein levels determined by western blotting in Ubap2l+/+, Ubap2l+/−, and Ubap2l−/− MEFs. (C) Impact of Ubap2l knockdown on p16Ink4a levels. Lin−SCA1+ cells were infected with shUbap2l or shBmi1 and the mRNA levels of p16Ink4a were determined by qRT-PCR immediately after infection. For all genes, the values are expressed relative to Hprt and the value for shLuc was set to 1. The average of 2 infections is shown with the standard error of the mean. Representative of 2 independent experiments. (D) Effect of Ubap2l deletion on p16Ink4a levels. p16Ink4a mRNA levels were determined by qRT-PCR in Ubap2l+/+ and Ubap2l−/− MEFs. The values are expressed relative to Hprt and the value for Ubap2l+/+ MEFs was set to 1. Shown are results for MEFs from 1 Ubap2l−/− mouse with the standard error of the mean. Representative of 2 independent experiments. (E) Identification of a BMI1 PcG subcomplex containing UBAP2L. HEK 293 cells were transfected with Flag-UBAP2L and cell extracts were fractionated on glycerol gradients. The presence of Flag-UBAP2L, BMI1, RNF2, and PHC1 in the fractions of the gradient was determined by western blotting with the antibodies indicated to the right. *The position of full-length Flag-UBAP2L protein. (F) Model for the role of UBAP2L in the regulation of HSC activity.
UBAP2L, BMI1, RNF2, and PHC1 define a novel Polycomb subcomplex
One explanation for the 2 BMI1-dependent mechanisms observed in hematopoietic cells could be that BMI1 is part of 2 separate protein complexes, each regulating different aspects of hematopoietic cell function. To test this hypothesis, we fractionated cellular extracts from HEK 293 cells expressing Flag-UBAP2L on glycerol gradients and analyzed the distribution of UBAP2L, BMI1, and other PcG proteins by western blotting across the fractions. The Flag-UBAP2L signal was restricted to 5 fractions at the top of the gradient (Figure 6E). Interestingly, BMI1 and RNF2 colocalized with Flag-UBAP2L in these fractions, but were also found in heavier fractions that did not contain UBAP2L. Another PcG protein, PHC1, specifically colocalized with UBAP2L, and was excluded from the heavier BMI1/RNF2 complex (Figure 6E). These results suggest that more than 1 BMI1-containing protein complex exist, distinguishable by the presence of UBAP2L and PHC1.
Discussion
We report here the identification of a novel protein, UBAP2L, which associates with the PcG protein BMI1. We demonstrate that Ubap2l transcripts are enriched in primitive mouse and human HSC populations and using a shRNA-based approach and a conditional knockout mouse, we show that this gene is essential for HSC activity. We also found that UBAP2L depletion does not cause a derepression of the Ink4a/Arf locus, as opposed to what is observed for Bmi1 knockout cells. Given that Bmi1 overexpression is able to rescue the loss of LT-HSC activity caused by Ubap2l silencing, these observations raise the possibility that at least 2 Bmi1-dependent mechanisms are at play in hematopoietic cells. This hypothesis is supported by our ability to resolve 2 different BMI1-containing protein complexes from cellular extracts, and by the known observation that PcG proteins can form distinct Polycomb-repressive complexes, each comprising a distinct PCGF paralog, a ubiquitin ligase (RNF2, RING1A), and a unique set of associated proteins.22 Based on our results, we propose a model in which at least 2 different BMI1-containing protein complexes regulate hematopoietic cell function (Figure 6F): a UBAP2L-independent complex, which is most likely involved in the repression of the Ink4a/Arf locus, and the UBAP2L-BMI1-RNF2-PHC1 complex, which operates via a mechanism unrelated to the repression of the Ink4a/Arf locus. These results position UBAP2L as a key gene for HSC activity and provide insight into the regulation of HSC function by BMI1.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Mélanie Fréchette, Evelyn Andrea Mejia Alfaro, Valérie Blouin-Chagnon as well as Jessica Simard for their help with animal care and transplantation experiments. The authors acknowledge Simon Fortier and Jana Krosl for help with BM extractions, and Danièle Gagné as well as Gaël Dulude for assistance with flow cytometry and cell sorting. The authors thank Céline Moison for critical comments on the manuscript.
This work was supported by a Canadian Cancer Society Research Institute fellowship (M.-E.B.) and grant (700849; G.S.), and grants from the Canadian Institutes of Health Research (MOP 14168 [S.M.], MOP 15064 [G.S.]).
C.B. is recipient of a fellowship award from the Cole Foundation. S.M. holds the Canada Research Chair in Cellular Signaling.
Authorship
Contribution: M.-E.B. designed and performed the experiments under the supervision of G.S.; R.A. and S.G. generated the conditional and knockout Ubap2l mice; R.A. characterized the Ubap2l mutant mice, generated the shUbap2l vectors, and performed the qPCR experiment with NIH 3T3 cells; J.C., M.S., S.G., and M.-E.B. isolated the CD150+CD48−Lin−SCA1+cKIT+ population and carried out qRT-PCR on these cells; N.M. assisted M.-E.B. with BM extractions, cell culture, and the in vivo rescue experiment; É.B. and P.T. executed the MS experiments; C.P., A.B., F.B., and J.H. performed the RNA sequencing and LSC frequency experiments with the different human normal and AML specimens; C.B. and S.M. generated the pREV-TRE-PC-TEV-Flag vector; and M.-E.B., R.A., and G.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guy Sauvageau, Université de Montréal, C.P.6128, Succursale Centre-Ville, Montréal, QC, Canada, H3C 3J7; e-mail: guy.sauvageau@umontreal.ca.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal