Key Points
Loss of STAT3 in NK cells enhances the expression of granzyme B, perforin, and DNAM-1, resulting in enhanced tumor surveillance.
STAT3 binds the IFN-γ promoter and interferes with cytokine-induced IFN-γ production in NK cells.
Abstract
The members of the signal transducer and activator of transcription (STAT) family of transcription factors modulate the development and function of natural killer (NK) cells. NK cell–mediated tumor surveillance is particularly important in the body’s defense against hematological malignancies such as leukemia. STAT3 inhibitors are currently being developed, although their potential effects on NK cells are not clear. We have investigated the function of STAT3 in NK cells with Stat3Δ/ΔNcr1-iCreTg mice, whose NK cells lack STAT3. In the absence of STAT3, NK cells develop normally and in normal numbers, but display alterations in the kinetics of interferon-γ (IFN-γ) production. We report that STAT3 directly binds the IFN-γ promoter. In various in vivo models of hematological diseases, loss of STAT3 in NK cells enhances tumor surveillance. The reduced tumor burden is paralleled by increased expression of the activating receptor DNAM-1 and the lytic enzymes perforin and granzyme B. Our findings imply that STAT3 inhibitors will stimulate the cytolytic activity of NK cells against leukemia, thereby providing an additional therapeutic benefit.
Introduction
The Janus kinase/signal transducer and activator of transcription (STAT) signaling pathway is involved in many cellular processes, including development, differentiation, and proliferation.1 STAT transcription factors may induce or repress transcription and have been implicated in natural killer (NK) cell development and function.2-6 STAT1 is known to regulate NK cell cytotoxicity and cytokine production.5,7 STAT4 is highly expressed in resting NK cells and regulates interferon-γ (IFN-γ) production upon interleukin-12 (IL-12) stimulation by activating the T-box transcription factor T-bet.8,9 STAT5 is essential for NK cell development and survival mediating IL-2 and IL-15 signaling,10 and STAT6 has been reported to be involved in the differentiation of NK cells.11
STAT3 is constitutively activated in many cancers and has been reported to mediate the crosstalk between tumor and immune cells.12,13 Cytokines produced by the tumor activate STAT3 in infiltrating immune cells and suppress their activity.13 As a result, NK cell functions may be altered: inhibitory as well as activating effects have been reported.2-4
Inhibiting STAT3 is a potential approach for treating various forms of cancer. However, our knowledge of STAT3’s role in NK cells is limited. Kortylewski and colleagues studied Stat3Δ/ΔMx1-Cre mice lacking STAT3 in all Mx1-expressing cells. They reported improved tumor surveillance against B16F10 melanoma cells, which was attributed to T cells and neutrophils. Stat3Δ/ΔMx1-Cre NK cells killed YAC-1 cells better, albeit only after prior B16F10 challenge.14 The complex interaction of the immune system does not allow any conclusion on the cell-intrinsic role of STAT3 in NK cells.
To assess the role of STAT3 in NK cells, we crossed Stat3fl/fl with Ncr1-iCreTg mice.10,15 Stat3Δ/ΔNcr1-iCreTg mice lack Stat3 in the NKp46+ NK cell compartment and are viable, fertile, and show no obvious signs of disease. The use of Stat3Δ/ΔMx1-Cre mice, in which STAT3 is deleted in all cells including the entire hematopoietic compartment, enabled us to compare the phenotype of Stat3Δ/ΔNcr1-iCreTg mice with the consequences of global STAT3 loss.
Material and methods
Mice
Mice were bred on a C57BL/6N background and maintained at the University of Veterinary Medicine Vienna under pathogen-free conditions according to Federation of Laboratory Animal Science Associations guidelines. The following mice were studied: Stat3fl/flMx1-Cre (inducible Cre expression activate after type 1 interferon response, eg, after poly(I:C) injection, expressing 1 allele of the Cre-transgene),16 Stat3Δ/ΔNcr1-iCreTg and Stat3fl/fl littermates,15 and C57BL/6 mice expressing 1 allele of the Cre-transgene (Ncr1-iCreTg),10 and wild-type littermates. All experiments were carried out with age-matched 6- to 12-week-old mice and were approved by the institutional ethics committee and conform to Austrian law (license 66.009/0019-II/10b/2010 14.1.10 and 68.205/0218-II/3b/2012).
Cell culture
Splenic NK cells were isolated using DX5-labeled MACS beads according to the manufacturer’s instructions (Miltenyi) and cultured with 5000 U/mL rhIL-2 (Proleukin, Novartis). NK cell stimulations were performed in the presence of 5000 U/mL rhIL-2 ± 5 ng/mL recombinant murine IL-12 (rmIL-12) (R&D), 50 ng/mL rmIL-15 (PeproTech), 100 U/mL rmIFN-β (Sigma), 5 ng/mL rmIL-10 (R&D), 100 ng/mL rmIL-21 (Immunotools), 50 ng/mL rmIL-23 (R&D), 100 ng/mL rmIL-18 (R&D), or 50 ng/mL rmIL-6 (R&D) and 50 ng/mL rmIL6Rα (R&D). Tumor cell lines were cultured as previously published.5 Both v-abl+ transformed cell lines used in this study were derived from C57BL6/N mice.
Poly(I:C) treatment
Stat3fl/fl and Stat3fl/flMx1-Cre mice were treated 3 times by IP injection of 300 μg poly(I:C) (Sigma) for 14 days. Seven days after the last injection, mice were analyzed. Deletion of Stat3 in Stat3fl/flMx1-Cre results in a global Stat3 deletion in this mouse model, including the entire hematopoietic compartment and is denoted as Stat3Δ/ΔMx1-Cre.
NK cell cytotoxicity
In vitro cytotoxicity assays were performed as previously published.5,17 For the DNAM-1 blockade, NK cells were incubated with anti-CD226 for 1 hour at 4°C and washed. Blocking efficiency was checked by flow cytometry and the blocked cells were incubated with the tumor cells at different effector:target (E:T) ratios.
In vivo tumor challenge
For the B16F10 melanoma model, mice were injected IV with 5 × 104 B16F10 cells. After 24 days, mice were euthanized and lung tissue was analyzed. Tumor nodules in the lung were counted by 3 independent researchers in a blinded manner. During disease progression, blood was analyzed by flow cytometry. For Kaplan-Meier plots, mice were euthanized at the first signs of dyspnea and health detractions.
In the v-abl leukemic tumor model, 106v-abl+ tumor cells were injected IV or subcutaneously into each flank of the mice. After subcutaneous injection, mice were euthanized at day 12 and tumor weight was determined. For the survival assay (IV injection), mice were euthanized at the first signs of paralysis and health detractions. During the disease progression, the blood was analyzed by flow cytometry. In the A-MuLV model, newborn mice were injected with 100 μL of replication-incompetent ecotropic retrovirus encoding for v-abl by IP injection as described previously.18 Mice were checked daily for disease onset.
Western blotting
Preparation of protein lysates and western blot was done as previously reported.5 The following antibodies were used: STAT3 (Cell Signaling CS#9132), pSTAT3-Y705 (CS#9131), STAT4 (CS#2653), pSTAT4-Y693 (BD-612738), pSTAT5-Y694 (BD-611964), STAT5 (sc-836) (Santa Cruz), Granzyme B (CS#4275), Perforin (CS#3693), and β-actin (sc-69879). Immunoreactive bands were visualized by chemiluminescence detection (LumiGLO, Cell Signaling) by the ChemiDoc MP Imaging System (Bio-Rad).
STAT3 inhibitors
STAT3 Inhibitor VI, S31-201-Calbiochem (#573130), and STAT3 Inhibitor XIV, LLL12-Calbiochem (#573131) were purchased from Merck Millipore. NK cells were treated for 4 hours with inhibitors at the concentration indicated or with dimethylsulfoxide (DMSO) control before stimulation with the respective cytokines.
Chromatin immunoprecipitation
MACS-purified and IL-2–cultured NK cells (107) were either left untreated or stimulated with IL-12 for 30 minutes followed by cross-linking using 1% formaldehyde for 10 minutes at 37°C. The reaction was stopped by the addition of 0.5 M glycine for 5 minutes. Cell nuclei were prepared and lysed in 1 mL of lysis buffer on 4°C overnight. Chromatin was sheared by sonication yielding chromatin fragments between 100 and 500 bp and diluted 2.5-fold in chromatin immunoprecipitation dilution buffer. Immunoprecipitation was performed at 4°C overnight with an anti-STAT3 antibody (Cell Signaling CS#9132). Chromatin was precleared using 25 µL salmon sperm DNA/protein A-agarose beads and incubated with the antibody overnight. Immune complexes were collected with 25 µL beads for 3 hours and washed with RIPA buffer, high salt buffer, LiCl buffer, and TE buffer. Samples were eluted 2× in elution buffer (2% sodium dodecyl sulfate, 10 mM dithiothreitol, and 100 mM NaHCO3). DNA cross-linking was reversed by heating at 65°C overnight followed by proteinase K digestion. DNA was extracted with phenol-chloroform, precipitated in isopropanol, and resuspended in TE buffer. The predicted binding site was obtained using Ensemble for the sequence of the IFN-γ promoter and blasting for the potential STAT binding motif TTC(N)2-4GAA (supplemental Figure 4F, available on the Blood Web site). Obtained DNA fragments were analyzed by quantitative polymerase chain reaction (qPCR) using the following primer pairs: Ifng: forward: 5′-GTGCTGTGCTCTGTGGATGAG-3′ and reverse: 5′-GAAGGCTCCTCGGGATTACGT-3′ CD19down: forward: 5′-CCCTCTTCTCATTCGTTTTCCA-3′and reverse: 5′-CCAGGAAAGAATTTGAGAAAAATCA-3′. N-fold enrichment was calculated relative to a negative region downstream of the CD19 gene (CD19down).
Histology
Paraffin-embedded sections (3 μm) were stained with hematoxylin and eosin according to standard histological procedures. Blood smears were fixated and stained using Hemacolor staining kit (Merck). All stainings were scanned and photographed with an Olympus IX71 microscope using CellSens Dimension Software (Olympus).
Statistical analysis
Unpaired t test, Mann-Whitney (nonparametrical), 1-way analysis of variance with Tukey’s post hoc test and log-rank tests were performed using GraphPad Prism Software, version 5.0 (San Diego, CA), where appropriate. The level of significance is indicated for each experiment (*P < .05; **P < .01; ***P < .001).
Results
NK cells develop and mature normally in the absence of Stat3
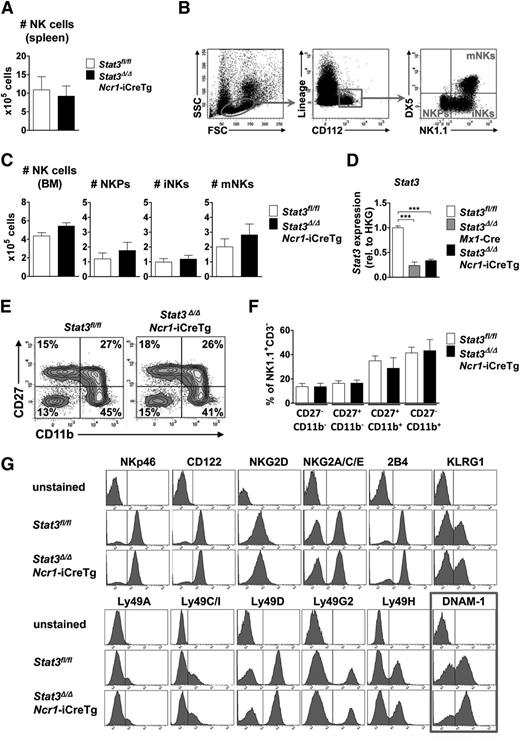
The numbers of NK cells in spleen and bone marrow were comparable in Stat3Δ/ΔMx1-Cre (where STAT3 is absent from the entire hematopoietic compartment) and Stat3Δ/ΔNcr1-iCreTg mice (lacking STAT3 only in the NKp46+ NK cell compartment) and their Stat3fl/fl littermates (Figure 1A,C; supplemental Figure 1A-B). NK cells differentiate from NK precursors (NKPs) to immature NK cells (iNKs) before becoming fully mature and migrating to the periphery.19 NK cells at all stages of development (a gating scheme is presented in Figure 1B) are present at comparable numbers in Stat3Δ/ΔNcr1-iCreTg mice (Figure 1C; supplemental Figure 1B). In Stat3Δ/ΔMx1-Cre animals, we detected an increase in the NKP population, which does not translate to altered numbers of mature NK cells but most likely reflects changes in the cytokine milieu in the bone marrow of these animals (supplemental Figure 1B). qPCR analysis verified the successful deletion and the significant decrease in levels of Stat3 messenger RNA (mRNA) (Figure 1D). Although NK cells develop in the bone marrow, NK cell maturation occurs in the spleen. We failed to detect any significant changes in NK cell maturation in Stat3Δ/ΔNcr1-iCreTg and Stat3Δ/ΔMx1-Cre mice compared with their Stat3fl/fl littermates (Figure 1E-F; supplemental Figure 1F-G). Similarly, when we expanded NK cells derived from Stat3Δ/ΔNcr1-iCreTg mice and Stat3fl/fl controls in vitro, no alterations in cell growth or cell-cycle distribution were detected (supplemental Figure 1H). Confirmation that NK cell proliferation is not altered came from in vivo 5-bromo-2′-deoxyuridine incorporation experiments; 5-bromo-2′-deoxyuridine incorporation in splenic and bone marrow NK cells was comparable (supplemental Figure 1I). In summary, the data led us to conclude that STAT3 is not required for survival, proliferation, or development of NK cells.
Lack of STAT3 does not influence NK cell development, maturation, and proliferation. (A) Bar graphs depict absolute numbers of splenic NK cells (CD3−NK1.1+NKp46+) of Stat3Δ/ΔNcr1-iCreTg mice and Stat3fl/fl controls as determined by flow cytometry. Five independently conducted experiments with n ≥ 14 per genotype are pooled. Data represent mean ± standard error of the mean (SEM). (B) Flow cytometric gating scheme for identification of NKPs, iNKs, and mature NKs (mNKs) in the bone marrow. (C) For the analysis of NK cell development, BM cells of Stat3Δ/ΔNcr1-iCreTg mice and Stat3fl/fl controls were counted and analyzed via flow cytometry by gating on Lin− (CD3, CD19, Ter119, Ly6C/G) CD122+ cells. NKPs are defined as Lin−CD122+NK1.1−DX5−, iNKs as Lin−CD122+NK1.1+DX5−, and mNKs as Lin−CD122+NK1.1+DX5+. Bar graphs depict mean ± SEM (n ≥ 6 per genotype). (D) Stat3 deletion efficiency of MACS-purified NK cells was analyzed ex vivo by qPCR. Data represent mean ± SEM (3 replicates per group); gene expression is calculated relative to the housekeeping gene (HKG) Gapdh and normalized to Stat3fl/fl. (E-F) For detection of NK cell maturation stages, splenocytes of Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were analyzed for CD27 and CD11b expression after previous gating on CD3−NK.1.1+ cells. Statistical analyses are summarized (F) (n ≥ 8 per genotype). (G) Splenocytes of Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were stained for individual NK cell receptors after gating on NK cells (CD3−NK1.1+). One representative histogram is shown (4 independent experiments, with n ≥ 10 in total per genotype). There are significantly more Stat3Δ/ΔNcr1-iCreTg NK cells expressing DNAM-1: 57 ± 0.8 (Stat3fl/fl) vs 66 ± 1.5 (Stat3Δ/ΔNcr1-iCreTg) %DNAM-1+ NK cells. Statistics are included in supplemental Figure 1D.
Lack of STAT3 does not influence NK cell development, maturation, and proliferation. (A) Bar graphs depict absolute numbers of splenic NK cells (CD3−NK1.1+NKp46+) of Stat3Δ/ΔNcr1-iCreTg mice and Stat3fl/fl controls as determined by flow cytometry. Five independently conducted experiments with n ≥ 14 per genotype are pooled. Data represent mean ± standard error of the mean (SEM). (B) Flow cytometric gating scheme for identification of NKPs, iNKs, and mature NKs (mNKs) in the bone marrow. (C) For the analysis of NK cell development, BM cells of Stat3Δ/ΔNcr1-iCreTg mice and Stat3fl/fl controls were counted and analyzed via flow cytometry by gating on Lin− (CD3, CD19, Ter119, Ly6C/G) CD122+ cells. NKPs are defined as Lin−CD122+NK1.1−DX5−, iNKs as Lin−CD122+NK1.1+DX5−, and mNKs as Lin−CD122+NK1.1+DX5+. Bar graphs depict mean ± SEM (n ≥ 6 per genotype). (D) Stat3 deletion efficiency of MACS-purified NK cells was analyzed ex vivo by qPCR. Data represent mean ± SEM (3 replicates per group); gene expression is calculated relative to the housekeeping gene (HKG) Gapdh and normalized to Stat3fl/fl. (E-F) For detection of NK cell maturation stages, splenocytes of Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were analyzed for CD27 and CD11b expression after previous gating on CD3−NK.1.1+ cells. Statistical analyses are summarized (F) (n ≥ 8 per genotype). (G) Splenocytes of Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were stained for individual NK cell receptors after gating on NK cells (CD3−NK1.1+). One representative histogram is shown (4 independent experiments, with n ≥ 10 in total per genotype). There are significantly more Stat3Δ/ΔNcr1-iCreTg NK cells expressing DNAM-1: 57 ± 0.8 (Stat3fl/fl) vs 66 ± 1.5 (Stat3Δ/ΔNcr1-iCreTg) %DNAM-1+ NK cells. Statistics are included in supplemental Figure 1D.
NK cells bind their target cells via specific receptors. By integrating signals from activating and inhibitory receptors, NK cells ultimately lyse a target cell or leave it unaffected. Levels of CD122, of the activating receptors NKp46, NKG2C/E, NKG2D, 2B4, Ly49D, and H and of the inhibitory receptors KLRG1, NKG2A, Ly49A, C, G2, and I, are superimposable in Stat3fl/fl, Stat3Δ/ΔNcr1-iCreTg, and Stat3Δ/ΔMx1-Cre splenic NK cells (Figure 1G; supplemental Figure 1C). However, loss of STAT3 is associated with a consistently and significantly increased number of NK cells expressing the activating receptor DNAM-1 (CD226) (Figure 1G; supplemental Figure 1D), which suppresses tumors by binding its ligands CD155 and CD112 on target cells.20,21 The enhanced expression of DNAM-1 in the absence of STAT3 was confirmed in NK cells derived from Stat3Δ/ΔMx1-Cre mice (supplemental Figure 1C,E).
Cytokine stimulation in splenic and intestinal NKp46+ cells
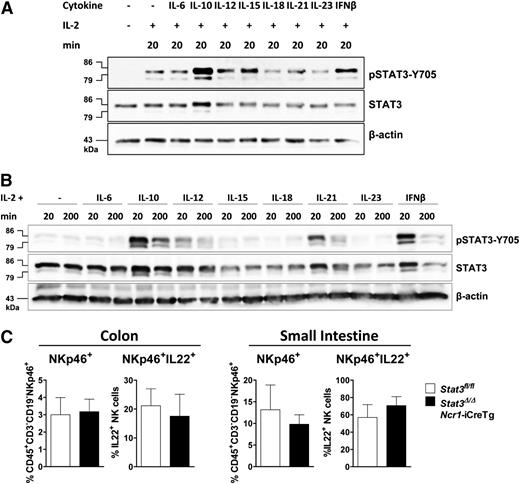
We next investigated which cytokines induce STAT3 activation in primary (Figure 2A) and IL-2 expanded NK cells (Figure 2B). IL-2 treatment is required to keep the cells viable. It induces STAT3-Y705 phosphorylation that is further increased upon stimulation with IL-10, IL-12, IL-15, or IFN-β (Figure 2A). In contrast, in IL-2–cultured NK cells stimulation with IL-15 fails to induce STAT3 phosphorylation. Remarkably, IL-21 was able to provoke a strong pSTAT3 response under these experimental conditions (Figure 2B).
Different cytokines activate STAT3 in primary and IL-2–expanded NK cells. (A) NK cells were MACS-purified, FACS-sorted, and stimulated with IL-2 alone or together with IL-6 (+IL6Rα), IL-10, IL-12, IL-15, IL-18, IL-21, IL-23, or IFN-β for 20 minutes. Western blot analysis show levels of pSTAT3-Y705 and STAT3. β-actin was detected as loading control. (B) IL-2–expanded NK cells were stimulated with indicated cytokines for 20 or 200 minutes. Western blots show levels of pSTAT3-Y705 and STAT3. β-actin was used as loading control. (C) Lamina propria cells from the colon or small intestine of Stat3fl/fland Stat3Δ/ΔNcr1-iCreTg mice. Cells were stimulated with IL-23, fixed, and permeabilized, and IL-22 production of CD45+CD3−CD19−NKp46+ cells was analyzed by flow cytometry.
Different cytokines activate STAT3 in primary and IL-2–expanded NK cells. (A) NK cells were MACS-purified, FACS-sorted, and stimulated with IL-2 alone or together with IL-6 (+IL6Rα), IL-10, IL-12, IL-15, IL-18, IL-21, IL-23, or IFN-β for 20 minutes. Western blot analysis show levels of pSTAT3-Y705 and STAT3. β-actin was detected as loading control. (B) IL-2–expanded NK cells were stimulated with indicated cytokines for 20 or 200 minutes. Western blots show levels of pSTAT3-Y705 and STAT3. β-actin was used as loading control. (C) Lamina propria cells from the colon or small intestine of Stat3fl/fland Stat3Δ/ΔNcr1-iCreTg mice. Cells were stimulated with IL-23, fixed, and permeabilized, and IL-22 production of CD45+CD3−CD19−NKp46+ cells was analyzed by flow cytometry.
IL-23 fails to trigger activation of STAT3 in splenic NK cells but this does not necessarily imply that the IL-23–induced IL-22 production22 in intestinal NKp46+ ILC3 cells is independent of STAT3. To clarify this point, we stimulated lamina propria cells of the small intestine and colon of Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg animals with IL-23. No changes in the numbers of intestinal NKp46+ or NKp46+ IL-22+ cells were detectable (Figure 2C).
To investigate whether the increase in DNAM-1+ NK cells is regulated by a distinct cytokine, we stimulated splenocytes of Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice with various cytokines as indicated. No changes were observed; significantly more Stat3Δ/ΔNcr1-iCreTg NK cells expressed DNAM-1 (supplemental Figure 2A). Under identical experimental conditions, we uncovered a slight decrease of IFN-γ expression in cells lacking STAT3 regardless of the stimulus (supplemental Figure 2B), whereas perforin levels were elevated upon stimulation with IL-2, IL-15, IL-12/IL-15, IL-21 and IFN-β (supplemental Figure 2C). Numbers of granzyme B+ cells were only slightly increased after combined stimulation with IL-12 and IL-15 (supplemental Figure 2D). This finding prompted us to investigate the production of perforin, granzyme B and IFN-γ in more detail.
Increased levels of perforin and granzyme B in the absence or upon inhibition of STAT3
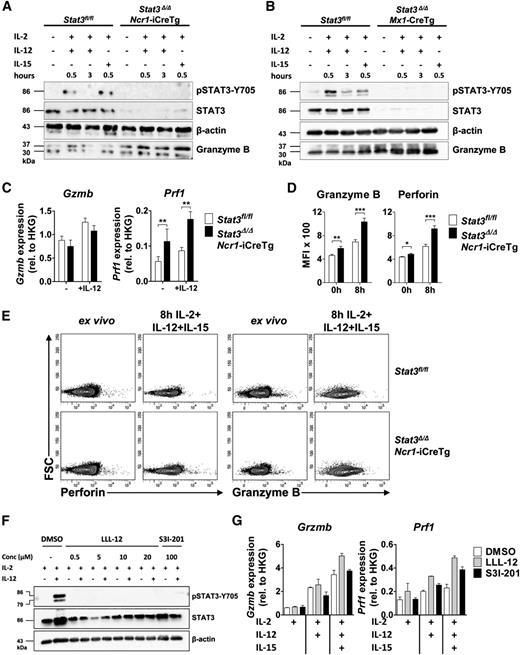
Levels of granzyme B protein and mRNA were investigated using freshly purified NK cells of Stat3fl/fl, Stat3Δ/ΔNcr1-iCreTg mice (Figure 3A,C) and Stat3Δ/ΔMx1-Cre NK cells (Figure 3B). Interestingly, despite elevated protein levels, granzyme B mRNA was unaltered under comparable experimental conditions (Figure 3C). In contrast, we readily detected increased levels of perforin mRNA expression upon stimulation with IL-2 and IL-12 (Figure 3C). Intracellular fluorescence-activated cell sorter (FACS) staining verified that the levels of perforin and granzyme B were significantly enhanced in cells derived from Stat3Δ/ΔNcr1-iCreTg mice, both in nonstimulated NK cells ex vivo as well as when the cell were stimulated with IL-2, IL-12, and IL-15 (Figure 3D-E). In contrast, levels of granzyme A remained unaffected by loss of STAT3 (supplemental Figure 3A-B).
Loss of STAT3 in NK cells is accompanied by increased expression of perforin and granzyme B. (A-B) One representative western blot (n = 2) of (A) ex vivo MACS-purified and CD3−NKp46+ sorted Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg NK cells and (B) MACS-purified and IL-2–expanded Stat3fl/fl and Stat3Δ/ΔMx1-Cre NK cells without any stimulus or after stimulation for 30 minutes with IL-2 + IL-12, 3 hours with IL-2 + IL-12, or 30 minutes with IL-2 + IL-15. STAT3 activation, STAT3 deletion in Stat3Δ/ΔNcr1-iCreTg NK cells, and granzyme B expression were detected. (C) MACS-purified and FACS-sorted (CD3−NK1.1+) Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg NK cells were stimulated ex vivo with IL-2 alone or together with IL-12. Gzmb and Prf1 mRNA expressions were determined by qPCR. Data represent mean ± SEM of 2 independent experiments (n ≥ 5); RNA levels are calculated relative to that of the HKG Rplp0. (D-E) Splenocytes from Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were analyzed directly ex vivo or after stimulation for 8 hours with IL-2, IL-12, and IL-15 and stained for CD3, NKp46 (CD3−NKp46+), and perforin or granzyme B. Levels were analyzed by flow cytometry. Bar graphs depict mean fluorescence intensities (MFI) (n ≥ 7 per genotype). (F) MACS-purified and FACS-sorted (CD3−NK1.1+) NK cells were treated with DMSO (control), with 0.5, 5, 10, or 20 µM of the STAT3 inhibitor LLL-12 or with 100 µM of the STAT3 inhibitor S31-201 for 4 hours. NK cells were either left untreated (IL-2 media only) or stimulated with IL-12 for 20 minutes. Western blot analysis shows levels of pSTAT3-Y705 and STAT3. β-actin was used as loading control. (G) MACS-purified and FACS-sorted (CD3−NK1.1+) NK cells were treated with DMSO, with 0.5 µM of the inhibitor LLL-12, or with 100 µM of the inhibitor S31-201 for 4 hours after stimulation with IL-2, IL-2 + IL-12, or IL-2 + IL-12 + IL-15 for 3 hours. Perforin and granzyme B mRNA levels were analyzed by qPCR. (Data represent mean ± SEM; levels are calculated relative to the HKG Rplp0.)
Loss of STAT3 in NK cells is accompanied by increased expression of perforin and granzyme B. (A-B) One representative western blot (n = 2) of (A) ex vivo MACS-purified and CD3−NKp46+ sorted Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg NK cells and (B) MACS-purified and IL-2–expanded Stat3fl/fl and Stat3Δ/ΔMx1-Cre NK cells without any stimulus or after stimulation for 30 minutes with IL-2 + IL-12, 3 hours with IL-2 + IL-12, or 30 minutes with IL-2 + IL-15. STAT3 activation, STAT3 deletion in Stat3Δ/ΔNcr1-iCreTg NK cells, and granzyme B expression were detected. (C) MACS-purified and FACS-sorted (CD3−NK1.1+) Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg NK cells were stimulated ex vivo with IL-2 alone or together with IL-12. Gzmb and Prf1 mRNA expressions were determined by qPCR. Data represent mean ± SEM of 2 independent experiments (n ≥ 5); RNA levels are calculated relative to that of the HKG Rplp0. (D-E) Splenocytes from Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were analyzed directly ex vivo or after stimulation for 8 hours with IL-2, IL-12, and IL-15 and stained for CD3, NKp46 (CD3−NKp46+), and perforin or granzyme B. Levels were analyzed by flow cytometry. Bar graphs depict mean fluorescence intensities (MFI) (n ≥ 7 per genotype). (F) MACS-purified and FACS-sorted (CD3−NK1.1+) NK cells were treated with DMSO (control), with 0.5, 5, 10, or 20 µM of the STAT3 inhibitor LLL-12 or with 100 µM of the STAT3 inhibitor S31-201 for 4 hours. NK cells were either left untreated (IL-2 media only) or stimulated with IL-12 for 20 minutes. Western blot analysis shows levels of pSTAT3-Y705 and STAT3. β-actin was used as loading control. (G) MACS-purified and FACS-sorted (CD3−NK1.1+) NK cells were treated with DMSO, with 0.5 µM of the inhibitor LLL-12, or with 100 µM of the inhibitor S31-201 for 4 hours after stimulation with IL-2, IL-2 + IL-12, or IL-2 + IL-12 + IL-15 for 3 hours. Perforin and granzyme B mRNA levels were analyzed by qPCR. (Data represent mean ± SEM; levels are calculated relative to the HKG Rplp0.)
STAT3 inhibitors are currently being developed and hold great promise for the treatment of cancer. We tested the effects of inhibitors LLL-12 and S31-201 on levels of perforin and granzyme B in primary NK cells. At concentrations in which the inhibitors efficiently block STAT3 activation (Figure 3F), they induced increased levels of perforin mRNA upon IL-2/IL-12/IL-15 treatment. The effects on granzyme B mRNA were not consistent with the unaltered mRNA regulation upon Stat3 deletion (Figure 3G).
NK cells lacking Stat3 show altered kinetics of IFN-γ production
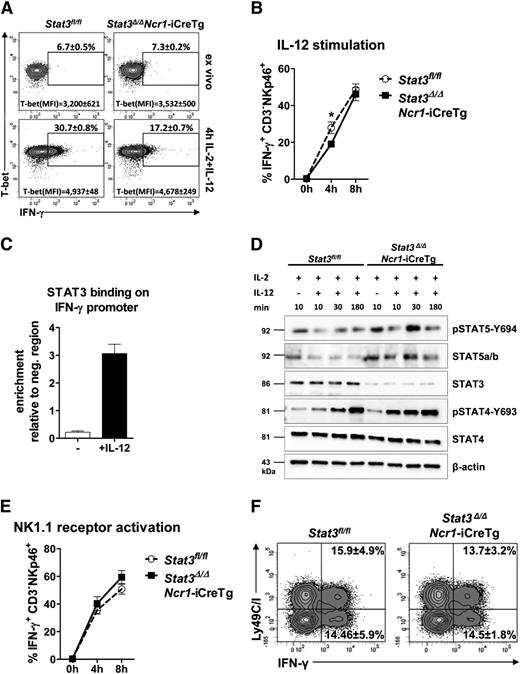
STAT3 is involved in the response to and the secretion of cytokines.13,23,24 Intracellular FACS staining revealed a decrease in IFN-γ expression after 4 hours of IL-12 stimulation in Stat3-deficient NK cells. Interestingly, after stimulation for 8 hours, the difference disappeared (Figure 4A-B). Regulation of IFN-γ production is complex and involves the transcriptional regulator T-bet.8,9 Intracellular FACS staining showed unaltered levels of T-bet after 4 hours of IL-12 stimulation despite the reduction of IFN-γ+ NK cells in Stat3Δ/ΔNcr1-iCreTg and in Stat3Δ/ΔMx1-Cre mice (Figure 4A; supplemental Figure 4B-D). Similarly, expression of IL-12 receptor was unaltered (supplemental Figure 4E). IFN-γ production is also controlled by members of the STAT family, and STAT4 has a key role in IL-12–induced production of IFN-γ.25 It is thus conceivable that STAT3 binds the IFN-γ promoter and interferes with STAT-driven transcription. Chromatin immunoprecipitation assays in primary NK cells indeed showed that IL-12 induced binding of STAT3 to the IFN-γ promoter (Figure 4C), with binding occurring at a STAT binding region illustrated in supplemental Figure 4F.26 Absence of STAT3 may also alter the activation of other members of the STAT family: Levels of pSTAT4 and pSTAT5 were enhanced in ex vivo–derived Stat3Δ/ΔNcr1-iCreTg NK cells (Figure 4D).
STAT3 regulates NK cell–dependent IFN-γ production by directly binding to the IFN-γ promoter. (A) Splenocytes from Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were analyzed directly ex vivo or after 4 hours stimulation with IL-2 + IL-12. Cells were stained for CD3 and NKp46 (CD3−NKp46+) followed by fixation, permeabilization, and intracellular staining of T-bet and IFN-γ. Levels were analyzed by flow cytometry. % IFN-γ+ NK cells and mean fluorescence intensities (MFI) of T-bet are depicted in representative FACS plots. Statistics are included in supplemental Figure 4D. (B) Splenocytes from Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were analyzed directly ex vivo or after stimulation for 4 or 8 hours with IL-2 + IL-12. Intracellular IFN-γ expression of NK cells were analyzed by flow cytometry. Stat3Δ/ΔNcr1-iCreTg NK cells show a decreased IFN-γ production after 4 hours that is no longer detectable after 8 hours. Data represent mean ± SEM of 3 independent experiments (n ≥ 13 in total). (C) Primary IL-2 cultured NK cells were stimulated for 30 minutes with IL-2 or IL-2 + IL-12. The reaction was stopped by addition of formaldehyde. Chromatin immunoprecipitation was performed using an anti-STAT3 antibody and n-fold enrichment was calculated relative to the expression of a negative region (“CD19 down”). (D) NK cells of Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were MACS-purified and FACS-sorted (CD3−NK1.1+) and stimulated with IL-2 and IL-12 for the indicated time. Cell lysates were used for western blot analysis of pSTAT5-Y694, STAT5a/b, STAT3, pSTAT4-Y693, and STAT4. β-actin was used as loading control. Stat3Δ/ΔNcr1-iCreTg NK cells show an increase in STAT4 and STAT5 activation compared with Stat3fl/fl controls. (E) Splenocytes from Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were stimulated with anti-NK1.1 (PK136) for 4 or 8 hours before staining for CD3 and NKp46 (CD3−NKp46+) followed by fixation, permeabilization, and intracellular staining and detection of IFN-γ expression by flow cytometry. (F) Splenocytes from Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were stimulated with anti-NK1.1 (PK136) for 6 hours before staining for CD3, NKp46, and Ly49C/I. Cells were fixed and permeabilized, and the intracellular IFN-γ expression of the Ly49C/I negative and positive NK cell fraction (CD3−NKp46+ cells) was analyzed. Statistics are included in supplemental Figure 4A.
STAT3 regulates NK cell–dependent IFN-γ production by directly binding to the IFN-γ promoter. (A) Splenocytes from Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were analyzed directly ex vivo or after 4 hours stimulation with IL-2 + IL-12. Cells were stained for CD3 and NKp46 (CD3−NKp46+) followed by fixation, permeabilization, and intracellular staining of T-bet and IFN-γ. Levels were analyzed by flow cytometry. % IFN-γ+ NK cells and mean fluorescence intensities (MFI) of T-bet are depicted in representative FACS plots. Statistics are included in supplemental Figure 4D. (B) Splenocytes from Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were analyzed directly ex vivo or after stimulation for 4 or 8 hours with IL-2 + IL-12. Intracellular IFN-γ expression of NK cells were analyzed by flow cytometry. Stat3Δ/ΔNcr1-iCreTg NK cells show a decreased IFN-γ production after 4 hours that is no longer detectable after 8 hours. Data represent mean ± SEM of 3 independent experiments (n ≥ 13 in total). (C) Primary IL-2 cultured NK cells were stimulated for 30 minutes with IL-2 or IL-2 + IL-12. The reaction was stopped by addition of formaldehyde. Chromatin immunoprecipitation was performed using an anti-STAT3 antibody and n-fold enrichment was calculated relative to the expression of a negative region (“CD19 down”). (D) NK cells of Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were MACS-purified and FACS-sorted (CD3−NK1.1+) and stimulated with IL-2 and IL-12 for the indicated time. Cell lysates were used for western blot analysis of pSTAT5-Y694, STAT5a/b, STAT3, pSTAT4-Y693, and STAT4. β-actin was used as loading control. Stat3Δ/ΔNcr1-iCreTg NK cells show an increase in STAT4 and STAT5 activation compared with Stat3fl/fl controls. (E) Splenocytes from Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were stimulated with anti-NK1.1 (PK136) for 4 or 8 hours before staining for CD3 and NKp46 (CD3−NKp46+) followed by fixation, permeabilization, and intracellular staining and detection of IFN-γ expression by flow cytometry. (F) Splenocytes from Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice were stimulated with anti-NK1.1 (PK136) for 6 hours before staining for CD3, NKp46, and Ly49C/I. Cells were fixed and permeabilized, and the intracellular IFN-γ expression of the Ly49C/I negative and positive NK cell fraction (CD3−NKp46+ cells) was analyzed. Statistics are included in supplemental Figure 4A.
We used an anti-NK1.1 antibody to investigate whether changes in IFN-γ production also occur upon receptor stimulation. We failed to detect any significant differences in IFN-γ secretion between Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg NK cells (Figure 4E) and no significant alterations in the production of IFN-γ by Ly49C/I−/+ NK cells were found (Figure 4F; supplemental Figure 4A). These experiments indicate that STAT3 contributes to IFN-γ production upon IL-12 stimulation, although it is of minor importance upon NK cell receptor stimulation.
Enhanced NK cell–dependent tumor surveillance in the absence of STAT3
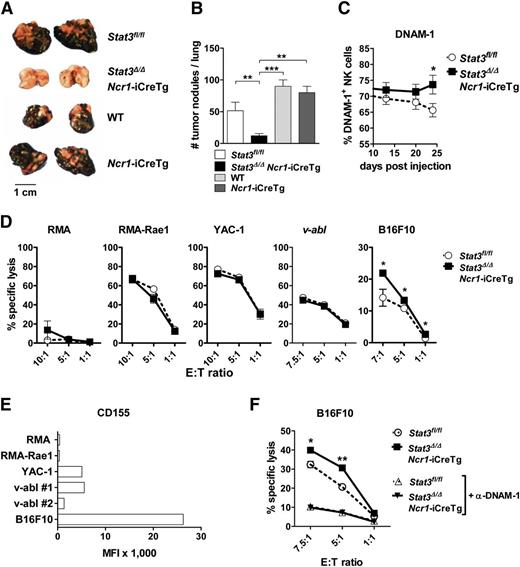
We next asked whether the enhanced levels of perforin, granzyme B, and DNAM-1 in Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice result in an enhanced NK cell–mediated tumor surveillance. We used B16F10 cells, a well-established tumor model controlled by NK cells.10,27,28 B16F10 cells were injected IV and tumor burden was analyzed after 3 weeks. Disease latency was significantly enhanced and accompanied by a drastic reduction of tumor nodules on day 24 in Stat3Δ/ΔNcr1-iCreTg mice (Figure 5A-B; supplemental Figure 5A). B16F10-induced tumor formation was even more suppressed in Stat3Δ/ΔMx1-Cre mice, although deletion of STAT3 in the NK cell compartment alone largely sufficed to improve tumor surveillance (supplemental Figure 5D). The improved tumor surveillance was accompanied by consistently elevated numbers of DNAM-1+ NK cells in the blood (Figure 5C), whereas the levels of CMKLR1 recognizing the chemoattractant chemerin and the exhaustion marker PD-1 were unaltered (supplemental Figure 5B-C). Consistently, in vitro cytotoxicity assays showed that the melanoma cell line B16F10 was killed more efficiently by NK cells lacking STAT3 (Figure 5D). No differences were observed in cytotoxicity assays using other target cell lines. One major difference between target cell lines is the expression of the DNAM-1 ligand CD155, which is only found at high levels in B16F10 cells (Figure 5E). Blocking DNAM-1 using antibodies significantly reduced cytotoxicity and abolished the differences between Stat3Δ/ΔNcr1-iCreTg and control cells (Figure 5F). Cytotoxicity against other target cell lines depends on other recognition receptors, activation of which ultimately leads to the release of granzymes and perforin. In vitro cytotoxicity assays employ NK cells expanded in IL-2 for a week. Western blot experiments showed that the changes in levels of perforin and granzyme B that we observe in freshly isolated NK cells lacking STAT3 are no longer detectable upon cultivation with IL-2 (supplemental Figure 5E), whereas the differences related to an increase in % DNAM-1+ cells persist (supplemental Figure 5F).
Stat3-deficient NK cells show enhanced in vitro cytotoxicity against B16F10 melanoma cells. (A-B) A total of 5 × 104 B16F10 melanoma cells were injected IV into Stat3fl/fl, Stat3Δ/ΔNcr1-iCreTg, Ncr1-iCreTg, and wild-type mice. After 24 days, the number of tumor nodules in the lung was assessed by 3 independent researchers in a blinded manner. (B) Statistical analysis summarizes 4 independent experiments (n ≥ 11 in total). (C) DNAM-1 expression on Stat3fl/fl (n = 11) and Stat3Δ/ΔNcr1-iCreTg (n = 10) NK cells in the blood of B16F10-inoculated mice was analyzed twice a week using flow cytometry (gated on CD3−NKp46+NK cells). Graph represents mean ± SEM. (D) In vitro cytotoxicity assays of IL-2–cultured NK cells with RMA, RMA-Rae1, YAC-1, v-abl+, and B16F10 target cell lines. The E:T cell ratios ranged from 1:1 to 10:1. After 4 to 6 hours of incubation at 37°C, lysis of target cells was analyzed by flow cytometry. Graphs represent mean ± SEM (biological replicates: 2, technical replicates: 3 each). (E) RMA, RMA-Rae1, YAC-1, v-abl+ cell line #1 and cell line #2, and B16F10 target cell lines were analyzed for the expression of the DNAM-1 ligand CD155 by flow cytometry. (F) IL-2–cultured Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg NK cells were treated with an anti-DNAM-1 antibody for 1 hour. Control and DNAM-1–blocked NK cells of both genotypes were incubated with B16F10 target cells. The E:T cell ratios ranged from 1:1 to 7.5:1; after 4 hours’ incubation at 37°C, the lysis of the target cells was analyzed by flow cytometry. Symbols represent means and error bars indicate SEM of triplicates. Data are representative for 2 independent experiments.
Stat3-deficient NK cells show enhanced in vitro cytotoxicity against B16F10 melanoma cells. (A-B) A total of 5 × 104 B16F10 melanoma cells were injected IV into Stat3fl/fl, Stat3Δ/ΔNcr1-iCreTg, Ncr1-iCreTg, and wild-type mice. After 24 days, the number of tumor nodules in the lung was assessed by 3 independent researchers in a blinded manner. (B) Statistical analysis summarizes 4 independent experiments (n ≥ 11 in total). (C) DNAM-1 expression on Stat3fl/fl (n = 11) and Stat3Δ/ΔNcr1-iCreTg (n = 10) NK cells in the blood of B16F10-inoculated mice was analyzed twice a week using flow cytometry (gated on CD3−NKp46+NK cells). Graph represents mean ± SEM. (D) In vitro cytotoxicity assays of IL-2–cultured NK cells with RMA, RMA-Rae1, YAC-1, v-abl+, and B16F10 target cell lines. The E:T cell ratios ranged from 1:1 to 10:1. After 4 to 6 hours of incubation at 37°C, lysis of target cells was analyzed by flow cytometry. Graphs represent mean ± SEM (biological replicates: 2, technical replicates: 3 each). (E) RMA, RMA-Rae1, YAC-1, v-abl+ cell line #1 and cell line #2, and B16F10 target cell lines were analyzed for the expression of the DNAM-1 ligand CD155 by flow cytometry. (F) IL-2–cultured Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg NK cells were treated with an anti-DNAM-1 antibody for 1 hour. Control and DNAM-1–blocked NK cells of both genotypes were incubated with B16F10 target cells. The E:T cell ratios ranged from 1:1 to 7.5:1; after 4 hours’ incubation at 37°C, the lysis of the target cells was analyzed by flow cytometry. Symbols represent means and error bars indicate SEM of triplicates. Data are representative for 2 independent experiments.
Loss of STAT3 in NK cells improves tumor surveillance against leukemia
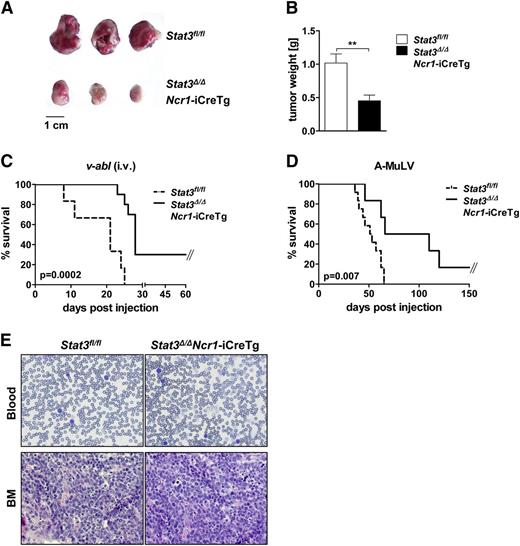
An increasing body of evidence describes control of leukemogenesis in an NK cell–dependent manner.29-31 We used the well-established Bcr/Abl leukemia model to test the effect of deleting STAT3. We subcutaneously injected v-abl+ leukemic cell lines into Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice. Twelve days later, large tumors had evolved in Stat3fl/fl animals, whereas a pronounced reduction of tumor mass was observed in Stat3Δ/ΔNcr1-iCreTg mice (Figure 6A,B). Further evidence that the absence of STAT3 in the NK cell compartment significantly enhances NK cell–mediated tumor surveillance was obtained by injecting 2 individually derived leukemic cell lines intravenously into Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice (Figure 6C; supplemental Figure 6A). Again, Stat3Δ/ΔNcr1-iCreTg animals survived significantly longer than the control animals. Finally we injected newborn mice with a replication-incompetent ecotropic retrovirus encoding for v-abl. This model system more closely mimics the development of human disease: a mono- or oligoclonal disease evolves slowly and is under the tight control of NK cells.31 Again, disease incidence and disease latency differed significantly. Whereas all Stat3fl/fl mice and control mice expressing only Cre recombinase succumbed to disease, 20% of Stat3Δ/ΔNcr1-iCreTg mice survived and disease latency was significantly delayed (Figure 6D; supplemental Figure 6B). There were no visible differences in the phenotype of the disease and all mice were densely infiltrated with leukemic cells at the end stage of disease (Figure 6E). We also detected constantly increased numbers of DNAM-1+ NK cells in the blood of Stat3Δ/ΔNcr1-iCreTg mice during disease progression (supplemental Figure 6C).
Stat3Δ/ΔNcr1-iCreTg mice show enhanced tumor surveillance in leukemia and lymphoma models in vivo. (A-B) A total of 106v-abl+ leukemic cells were injected subcutaneously in the flanks of Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice. After 12 days, tumor weight was determined (A). (B) Bar graphs summarize data of 3 independent experiments with n ≥ 14. (C) Kaplan-Meier plot of Stat3fl/fl (n = 7) and Stat3Δ/ΔNcr1-iCreTg (n = 9) mice after IV injection of 106v-abl+ leukemic cells. One representative experiment of 2 independent experiments is shown. Mice were euthanized at the first signs of paralysis and poor health. (D) Newborn Stat3fl/fl (n = 12) and Stat3Δ/ΔNcr1-iCreTg (n = 7) mice were infected with a replication-incompetent ecotropic retrovirus encoding for v-abl by subcutaneous injection. Mice were euthanized at the first signs of paralysis and health detractions. One representative experiment of 3 independent experiments is shown. (E) Hematoxylin and eosin stains of blood and bone marrow of Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice suffering from v-abl–induced leukemia.
Stat3Δ/ΔNcr1-iCreTg mice show enhanced tumor surveillance in leukemia and lymphoma models in vivo. (A-B) A total of 106v-abl+ leukemic cells were injected subcutaneously in the flanks of Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice. After 12 days, tumor weight was determined (A). (B) Bar graphs summarize data of 3 independent experiments with n ≥ 14. (C) Kaplan-Meier plot of Stat3fl/fl (n = 7) and Stat3Δ/ΔNcr1-iCreTg (n = 9) mice after IV injection of 106v-abl+ leukemic cells. One representative experiment of 2 independent experiments is shown. Mice were euthanized at the first signs of paralysis and poor health. (D) Newborn Stat3fl/fl (n = 12) and Stat3Δ/ΔNcr1-iCreTg (n = 7) mice were infected with a replication-incompetent ecotropic retrovirus encoding for v-abl by subcutaneous injection. Mice were euthanized at the first signs of paralysis and health detractions. One representative experiment of 3 independent experiments is shown. (E) Hematoxylin and eosin stains of blood and bone marrow of Stat3fl/fl and Stat3Δ/ΔNcr1-iCreTg mice suffering from v-abl–induced leukemia.
Discussion
We report a pronounced suppressive effect of STAT3 on NK cell–mediated tumor surveillance. In the absence of STAT3, murine NK cells develop and mature normally. Challenging the mice with tumor cells uncovered the key role of STAT3 in NK cell–mediated tumor surveillance. Experiments with the B16F10 melanoma model revealed a significantly delayed tumor formation in Stat3Δ/ΔNcr1-iCreTg mice. The improved tumor cell rejection extends to hematopoietic malignancies: Bcr/Abl+ leukemic cell lines are significantly better controlled by Stat3-deficient NK cells regardless of the experimental model used. Most importantly, work with a slowly evolving mono- or oligoclonal leukemia provoked by the injection of a retrovirus encoding A-MuLV—which closely resembles the development of human disease—confirmed the significant delay in tumorigenesis in Stat3Δ/ΔNcr1-iCreTg mice. The experiments illustrate the power and significance of NK cell–mediated tumor surveillance and reveal that STAT3 has a tumor promoting role in NK cells. NK cells lacking STAT3 are significantly better equipped to eliminate tumor targets.
Yu and Kortylewski were the first to show that deletion of STAT3 in the hematopoietic compartment improves tumor immune surveillance.14 The comparison of B16F10-dependent tumor cell rejection in Stat3Δ/ΔNcr1-iCreTg and Stat3Δ/ΔMx1-Cre mice confirms the effect of Stat3 deletion in the NK cell compartment. Although tumor surveillance is further improved in Stat3Δ/ΔMx1-Cre mice, a substantial portion of the beneficial effect stems from deletion of Stat3 in NK cells.
NK cells are important producers of IFN-γ, which is a critical determinant of tumor surveillance. We discovered that STAT3 regulates IFN-γ production by binding the IFN-γ promoter upon IL-12 stimulation. In the absence of STAT3, the onset of IFN-γ secretion upon cytokine stimulation is delayed. However, the changed kinetics does not translate to impaired tumor surveillance. It is attractive to speculate that other members of the STAT transcription factor family compensate for the absence of STAT3.
In the absence of STAT3, there is a consistent increase in levels of perforin and granzyme B in NK cells. The difference is abrogated upon cultivation in IL-2. Because perforin is a target of STAT5,32,33 it is likely that the persistent strong IL-2 stimulation that triggers STAT5 activation blurs the differences observed in freshly isolated cells. As a consequence, studies of cytotoxicity in vivo and in vitro give different results. The level of perforin is an important determinant of NK cell cytotoxicity, and NK cells lacking 1 allele of perforin have an impaired capacity to kill.34 The comparable levels of perforin and granzyme B in Stat3-deficient NK cells maintained in culture explain why the NK cell–dependent cytotoxicity against hematopoietic tumor cell lines in vitro are superimposable with those of wild-type NK cells. In vivo, where perforin and granzyme B are increased in Stat3Δ/ΔNcr1-iCreTg mice, NK cell–dependent tumor surveillance is clearly enhanced. Differences in NK cell–dependent cytotoxicity are only seen in vitro when target cells express DNAM-1 ligands at high levels. In contrast to perforin, %DNAM-1+ cells remain elevated in Stat3-deficient NK cells, even upon prolonged culture in vitro.21 Recent evidence links DNAM-1 to perforin: DNAM-1 is required for IL-2–activated perforin-mediated antitumor responses.20 There is a need for caution when comparing in vitro and in vivo data because prolonged culture of NK cells in IL-2 changes intracellular signaling patterns and consequently gene expression.
The increased numbers of DNAM-1+ NK cells are accompanied by elevated levels of perforin and granzyme B, which may contribute to the enhanced NK cell–mediated tumor cell killing. To date there are no reports on gene dosage effects regarding granzyme B, although it has been suggested that altered granzyme B expression contributes to increased tumor cell surveillance.35,36 Interestingly, the amount of granzyme B protein was increased without a concomitant change in the level of mRNA, suggesting the involvement of microRNAs. Both perforin and granzyme B have been described as targets of microRNA-dependent regulation in NK cells.37-39 Further research will be required to investigate whether STAT3 interferes with the control of microRNAs in NK cells.
In summary, our in vivo data define an inhibitory role for STAT3 in NK cell–dependent tumor surveillance. The evidence is compelling and unambiguous. The increase in cytolytic capacity involves more than 1 downstream molecular target; the increased expression levels of DNAM-1, perforin, and granzyme B contribute to the effect. Our conclusions are based on several independent tumor models and it appears likely that they will be relevant to additional systems, including human cancers. The mechanism is still not fully understood and we cannot exclude the possibility that other mechanisms are involved. Further research is required to discover which molecular players contribute to the in vivo effects of deleting STAT3 in the NK cell compartment.
Our observations improve the prospects of patients suffering from cancer controlled by NK cell–dependent surveillance, such as many hematological diseases. Recent evidence has highlighted the contribution of NK cells to the control of minimal residual disease.40 Although STAT3 has a dual role in tumor formation and acts both as a tumor suppressor and as a tumor promoter, in hematological malignancies it functions predominantly as tumor promoter. Mutations in STAT3 have been proposed to be drivers of hematological disorders,41,42 and inhibitors of STAT3 are being developed as novel anti-cancer therapeutics.43-45 Although we are unaware of any STAT3 inhibitors that have entered clinical trials, clinical use is expected in the near future. Our study shows that inhibiting STAT3 in NK cells will not only affect the tumor cells themselves, but will also have the benefit of improving tumor surveillance.
Leukemia and melanoma are among the types of cancer known to be controlled by NK cells. In planning tumor therapy, it is therefore important to consider the beneficial effects of STAT3 inhibitors on NK cell–mediated tumor surveillance: it may be possible to kill 2 birds with 1 stone.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank G. Tebb, S. Fajmann, P. Jodl, and Z. Horvath-Bago for their help. The authors are also grateful to the mouse facility.
The work was supported by an Austrian Science Fund (SFB F28) (M.M., B.S., and V.S.; http://www.fwf.ac.at/en/projects/sfb.html) and the Herzfelder’sche Familienstiftung (V.S.).
Authorship
Contribution: D.G., E.M.P., E.S., P.K., and M.B. performed the research; D.G., E.M.P., B.S., M.M., and V.S. designed the research and analyzed data; V.P. provided reagents; and V.S. and D.G. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Veronika Sexl, Institute of Pharmacology and Toxicology, University of Veterinary Medicine Vienna, Veterinärplatz 1, A-1210 Vienna, Austria; e-mail: veronika.sexl@vetmeduni.ac.at.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal