Key Points
A subset of lymphomas with gene expression and pathological characteristics of Burkitt lymphomas but absence of MYC translocation does exist.
These lymphomas carry chr 11q proximal gains and telomeric losses, suggesting co-deregulation of oncogenes and tumor suppressor genes.
Abstract
The genetic hallmark of Burkitt lymphoma (BL) is the t(8;14)(q24;q32) and its variants leading to activation of the MYC oncogene. It is a matter of debate whether true BL without MYC translocation exists. Here, we identified 59 lymphomas concordantly called BL by 2 gene expression classifiers among 753 B-cell lymphomas. Only 2 (3%) of these 59 molecular BL lacked a MYC translocation, which both shared a peculiar pattern of chromosome 11q aberration characterized by interstitial gains including 11q23.2-q23.3 and telomeric losses of 11q24.1-qter. We extended our analysis to 17 MYC-negative high-grade B-cell lymphomas with a similar 11q aberration and showed this aberration to be recurrently associated with morphologic and clinical features of BL. The minimal region of gain was defined by high-level amplifications in 11q23.3 and associated with overexpression of genes including PAFAH1B2 on a transcriptional and protein level. The recurrent region of loss contained a focal homozygous deletion in 11q24.2-q24.3 including the ETS1 gene, which was shown to be mutated in 4 of 16 investigated cases. These findings indicate the existence of a molecularly distinct subset of B-cell lymphomas reminiscent of BL, which is characterized by deregulation of genes in 11q.
Introduction
Burkitt lymphoma (BL) is an aggressive B-cell lymphoma characterized by typical morphological, immunophenotypic, and molecular features.1 The t(8;14)(q24;q32) hallmark translocation or its variants, which juxtapose the MYC oncogene to one of the 3 immunoglobulin (IG) loci, is detectable by cytogenetic or, currently, molecular cytogenetic techniques in almost all cases of BL.2-4
There is an ongoing discussion about whether BL is without exception defined by the presence of an IG-MYC translocation as the driving mutation or whether a small subset of true BL that lacks an IG-MYC translocation or even a MYC breakpoint does exist.1,5 In the latter case, it could be speculated that alternative mechanisms substitute for the hallmark MYC activation, pathogenomic for the vast majority of BL.4,6 A recent study on MYC-negative endemic BL has suggested that downregulation of microRNA hsa-mir-34b could be such a pathogenetic mechanism substituting MYC translocations.5 Alternatively, a common cell of origin could determine the typical features of BL in MYC-positive and -negative cases.
Various studies have reported lymphomas with clinical, histomorphologic, immunophenotypic, or gene expression features of BL lacking a detectable MYC translocation by fluorescence in situ hybridization (FISH).5,7-9 Nevertheless, caution is warranted before concluding that these indeed represent true MYC-negative BL, because the scattering of breakpoints in the MYC and IG loci along with small insertions of one locus into the other can render MYC breaks undetectable even if an extensive set of FISH probes is applied.10 Moreover, there is no gold standard to define BL in the absence of an MYC break. Prototypic BL that fulfills all characteristics of the World Health Organization (WHO) classification are rare.1,7 This in turn makes the distinction between BL and so called “intermediate lymphomas between Burkitt and diffuse large B-cell lymphomas (DLBCL)” difficult.1 In a recent cytogenetic study, Pienkowska-Grela et al described 4 lymphomas in young adult patients displaying typical BL morphology that bona fide lacked an MYC rearrangement by FISH and chromosome analyses. Remarkably, these cases shared a recurrent abnormality within chromosome 11 described as dup(11)(q23q13). This result led them to reclassify the cases as intermediate between DLBCL and BL because of the immunophenotypical and genetic features.8
Similarly, Poirel et al observed by cytogenetic review of childhood mature B-cell non-Hodgkin lymphomas that the 13 cases classified as BL without a detectable MYC translocation shared alterations with the MYC-positive BL except for the existence of more frequent aneuploidy and the presence of a derivative chromosome 11q.11
In the present study, we investigated the incidence of MYC-negative BL defined by gene expression analysis using 2 different classifiers. These cases were analyzed by histologic, immunophenotypic, and various molecular means. We show that the very rare cases of these MYC-negative BL display a recurrent pattern of alterations on chromosome 11q. We subsequently show that this pattern was also present in a subset of high-grade B-cell lymphomas with features of, but not ultimately fulfilling, the diagnostic criteria of BL. Clinical, morphologic, and molecular characterizations of these lymphomas with the typical 11q abnormality patterns suggest that these represent a distinct subset of MYC-negative high-grade B-cell lymphomas with features resembling but not identical to BL, and identify candidate genes potentially involved in their pathogenesis.
Material and methods
Lymphoma materials
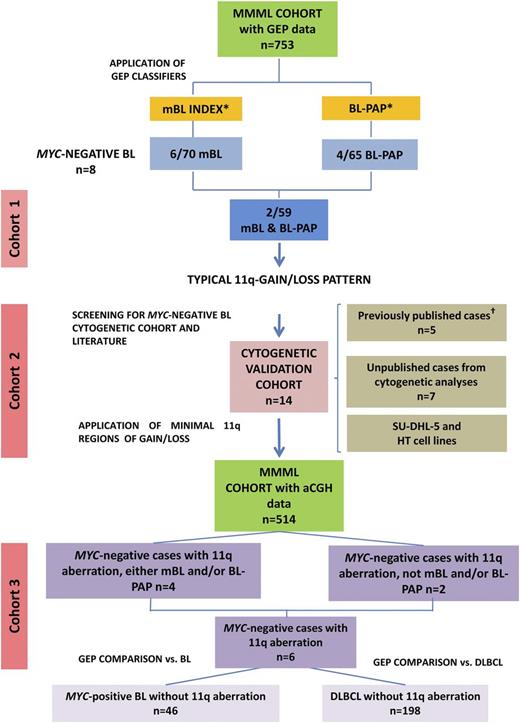
A global overview on the cohorts studied and the selection of cases included in the present study is given in Figure 1. This study was performed as part of the Molecular Mechanisms in Malignant Lymphoma (MMML) Network Project of the Deutsche Krebshilfe, for which approval was obtained by the Institutional Review Board of the Medical Faculty Kiel under 403/05 as well as the study central. The protocols of the Berlin-Frankfurt-Münster (BFM) clinical trials have also been approved by central and local review boards. This study was conducted in accordance with the Declaration of Helsinki.
Diagram of the strategy used for the identification of potential MYC-negative BL. One case from cohort 2 is also part of cohort 3 (MMML series). *mBL7 and BL-PAP GEP classifiers.12 †Previously published cases.8,13
Cohort 1: lymphomas called by both GEP classifiers (mBL and BL-PAP classifiers) BLs (n = 59)
To identify MYC-negative lymphomas with a BL gene expression profiling (GEP), 2 independently developed gene expression classifiers, the molecular-BL (mBL) index7 and the BL-pathway activation pattern (BL-PAP classifier)12 were applied to a cohort of 753 previously characterized lymphomas from the MMML Network. The 2 algorithms concordantly selected a total of 59 BL by GEP (cohort 1). Two of these cases lacked a MYC translocation and were thus considered true MYC-negative BL.
Cohort 2: high-grade B-cell lymphomas and cell lines with features of BL, MYC negative (n = 14)
Twelve MYC-negative lymphomas, 5 of which were previously reported,8,13,14 and SU-DHL-5 and HT cell lines, derived from MYC-negative high-grade B-cell lymphomas (www.dsmz.de), were included for cytogenetic and genetic analyses. Information on case selection is detailed in the supplemental Methods and in supplemental Table 1, available on the Blood Web site. All biopsy specimens were evaluated by at least 2 hematopathologists according to the WHO classification (Table 1; supplemental Table 2).1 Case 10, although its cytology showed features of BL, was initially classified as follicular lymphoma grade 3 with simultaneous DLBCL (supplemental Figure 1).13,14
Immunohistochemical and clinical characteristics of the 12 MYC-negative B-cell lymphomas and SU-DHL-5 and HT cell lines (cohort 2)
| . | . | . | . | . | Immunohistochemistry . | . | . | Pathology panel . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case no. . | Age, gender . | Morphologic diagnosis . | Localization . | MYC . | CD20 . | CD10 . | CD5 . | BCL2 . | BCL6 . | MUM1 . | Ki67 . | EBER . | Treatment protocol . | Outcome, follow-up . | |
| Case 1 | 7, male | Aggressive B-NHL, BL | LN(c) | + | + | + | – | – | + | – | 90% | – | B-NHL BFM 04, treatment modified due to AT | In remission, alive, 62 mo | WK, IO, MMML panel |
| Case 2 | 7, male | BL | LN (colon) | – | + | + | – | – | + | na | 95% | – | NHL-BFM-95 | In remission, alive, 146 mo | WK, IO |
| Case 3 | 16, female | High-grade B-NHL, atypical BL | LN | – | + | + | na | + | + | + | 95% | – | NHL-BFM-95 | In remission, alive, 43 mo | WK, IO |
| Case 4 | 18, male | BL | LN(c) | + | + | – | – | + | na | >95% | – | GMALL 2008 | In complete remission, alive, 48 mo | WK, GR | |
| Case 5 | 22, male | BL | LN (ing) | + | + | – | – | – | na | >95% | – | GMALL 2002 | In complete remission, alive, 36 mo | WK, GR | |
| Case 6 | 25, male | BL | LN (ab) | + | + | – | – | + | na | >95% | – | CODOX-m 2003 | Dead (progression), 12 mo | WK, GR | |
| Case 7 | 37, female | BL | LN(c),T | + | + | – | – | + | na | >95% | – | GMALL 2002 | In complete remission, alive, 72 mo | WK, GR | |
| Case 8 | 11, male | Mature B-cell lymphoma/lymphoblastic B-lymphoma | LN(c) | + | + | – | – | + | na | >95% | na | EORTC 58951 | Alive, 3 mo | WK, EJ | |
| Case 9 | 12, male | BL | leg | + | + | + | – | – | + | – | >95% | – | B-NHL BFM 04 | Alive, 8 mo | WK, IO |
| Case 10 | 6, female | FL | LN (c) | + | + | na | – | + | – | >95% | – | Ritux + B-NHL BFM 04 | Alive, 24 mo | WK, IO | |
| Case 11 | 9, male | Atypical BL | LN(ing) | + | + | – | – | + | – | >95% | – | COG-ANHL01P1 | In complete remission, alive, 10 mo | WK, EJ | |
| Case 12 | 76, male | BL | LN (c) | – | + | + | na | w+ | na | na | >95% | na | Alive, na | WK, IO | |
| SU-DHL-5 | 17, female | B-NHL (DLBCL) | + | + | + | – | |||||||||
| HT | 70, male | B-NHL (DLBCL) | + | + | + | – | |||||||||
| . | . | . | . | . | Immunohistochemistry . | . | . | Pathology panel . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Case no. . | Age, gender . | Morphologic diagnosis . | Localization . | MYC . | CD20 . | CD10 . | CD5 . | BCL2 . | BCL6 . | MUM1 . | Ki67 . | EBER . | Treatment protocol . | Outcome, follow-up . | |
| Case 1 | 7, male | Aggressive B-NHL, BL | LN(c) | + | + | + | – | – | + | – | 90% | – | B-NHL BFM 04, treatment modified due to AT | In remission, alive, 62 mo | WK, IO, MMML panel |
| Case 2 | 7, male | BL | LN (colon) | – | + | + | – | – | + | na | 95% | – | NHL-BFM-95 | In remission, alive, 146 mo | WK, IO |
| Case 3 | 16, female | High-grade B-NHL, atypical BL | LN | – | + | + | na | + | + | + | 95% | – | NHL-BFM-95 | In remission, alive, 43 mo | WK, IO |
| Case 4 | 18, male | BL | LN(c) | + | + | – | – | + | na | >95% | – | GMALL 2008 | In complete remission, alive, 48 mo | WK, GR | |
| Case 5 | 22, male | BL | LN (ing) | + | + | – | – | – | na | >95% | – | GMALL 2002 | In complete remission, alive, 36 mo | WK, GR | |
| Case 6 | 25, male | BL | LN (ab) | + | + | – | – | + | na | >95% | – | CODOX-m 2003 | Dead (progression), 12 mo | WK, GR | |
| Case 7 | 37, female | BL | LN(c),T | + | + | – | – | + | na | >95% | – | GMALL 2002 | In complete remission, alive, 72 mo | WK, GR | |
| Case 8 | 11, male | Mature B-cell lymphoma/lymphoblastic B-lymphoma | LN(c) | + | + | – | – | + | na | >95% | na | EORTC 58951 | Alive, 3 mo | WK, EJ | |
| Case 9 | 12, male | BL | leg | + | + | + | – | – | + | – | >95% | – | B-NHL BFM 04 | Alive, 8 mo | WK, IO |
| Case 10 | 6, female | FL | LN (c) | + | + | na | – | + | – | >95% | – | Ritux + B-NHL BFM 04 | Alive, 24 mo | WK, IO | |
| Case 11 | 9, male | Atypical BL | LN(ing) | + | + | – | – | + | – | >95% | – | COG-ANHL01P1 | In complete remission, alive, 10 mo | WK, EJ | |
| Case 12 | 76, male | BL | LN (c) | – | + | + | na | w+ | na | na | >95% | na | Alive, na | WK, IO | |
| SU-DHL-5 | 17, female | B-NHL (DLBCL) | + | + | + | – | |||||||||
| HT | 70, male | B-NHL (DLBCL) | + | + | + | – | |||||||||
−, negative; +, positive; ab, abdomen; c, cervical; EBER, Epstein-Barr–encoded RNA test; FL, follicular lymphoma; ing, inguinal; LN, lymph node; na, not available; NHL, non-Hodgkin lymphoma; T, tonsil; w, weak.
Cases 4, 5, 6, and 7 correspond to cases 1, 2, 3, and 4 from Pienkowska-Grela et al.8 Case 10 displayed a simultaneously follicular and diffuse grown pattern. Although its cytology showed features of BL, it was classified as FL grade 3 (minor component) with simultaneous DLBCL (major compartment), and it was included in a previous series of pediatric lymphomas already published; corresponds to case 23 from Oschlies et al13 and pFL8 from Martin-Guerrero et al.14 Information about SU-DHL-5 and HT cell lines was obtained from DSMZ (www.DSMZ.de). Lymphoma samples were scored positive if >25% of the tumor cells stained positive except for MYC, where the cutoff was established in 40%.
Minimal regions of gain and loss in chromosome 11q were defined in this series.
Cohort 3: MMML cases with 11q gain/loss pattern determined by CGH-array (n = 6, including 2 cases from cohort 1 and 1 case from cohort 2)
A total of 514 MMML cases with available comparative genomic hybridization (CGH)-array data were screened for aberrations in the minimal regions previously described in cohort 2. Six cases showing at least a partial gain and a partial loss within these regions were selected for cohort 3. Cases with non-mBL GEP, relapsed samples, and DLBCL cases IG/MYC-positive, IGH/BCL2-positive DLBCL, and BCL6 break-positive were excluded. Gene expression and immunohistochemical and genetic features of these cases were compared with 2 different sets of reference samples. The first set included all samples that exhibited an mBL GEP and IG-MYC positivity (n = 46). The second set was composed of DLBCL samples (n = 198). All cases selected for these 2 sets had no imbalances in the minimal 11q regions.
Immunohistochemistry, conventional cytogenetics, and FISH
Copy number and mutational analyses
SNP 6.0 array (Affymetrix, Santa Clara, CA) was used in cases 1 through 3 and 8 and cell lines as previously described.18 Cases 4 through 7 and 9 through 12 were analyzed using Agilent Human CGH Microarray platforms owing to methodologic requirements for formalin fixed paraffin embedded tissues (GEO-Number GSE47508). Gains and losses were defined by using different software (see supplemental Methods). Mutational analysis of the ATM gene in case 1 was performed as previously described.19,20 FLI1 and ETS1 genes’ mutational analyses were carried out in selected samples and cell lines (supplemental Table 3). ID3 mutational analysis was performed as previously described.21
Bisulfite pyrosequencing analyses
The hsa-mir-34b methylation pattern was determined by bisulfite pyrosequencing on cases 1 through 7, selected IG-MYC cell lines (CA-46, DAUDI, and RAJI), and primary IG-MYC BL samples (n = 6) according to standard protocols. The results were evaluated with Pyro Q-CpG 1.0.9 software (Biotage AB, Uppsala, Sweden) (supplemental Table 3).
Custom exome sequencing analysis in cell lines
Custom exome sequencing was performed in SU-DHL-5 and HT cell lines outsourced to Aros (www.arosab.com/). Details on analysis are provided in the supplemental Methods.
Gene expression analyses
To evaluate the impact of 11q alterations in the GEP, differential gene expression analysis was performed as previously described. Details on the analysis are provided in the supplemental Methods.
Statistical methods
Comparison of immunohistochemical and genetic features between cases with aberrant 11q-gain/loss pattern and each reference set individually was performed using methods previously described.7 Genetic complexity was defined as the number of observed copy number (CN) aberrations per tumor sample. Complexity was compared between groups by the Kruskal-Wallis rank-sum test.
MicroRNAs target gene analysis
Test for overrepresentation of microRNA targets within a list of differentially expressed genes was performed in multiple hypergeometric tests as implemented in the HTSanalyzeR package (version 2.8.0).22
Western blot analysis
Protein extract preparation and western blot experiments were performed as previously described using standard procedures. Details are provided in the supplemental Methods.
Results
MYC-negative lymphomas with a gene expression profile of BL are rare (cohort 1)
To identify BL, we applied 2 different GEP classifiers for BL, the mBL index7 and the BL-PAP classifier12 , to a total of 753 aggressive lymphomas included in the cohort of the MMML network. Based on the mBL and BL-PAP indices, we classified 70 and 65 lymphomas, respectively, as molecularly defined BL. An IG-MYC fusion was identified in 63 of 70 (90%) mBL and 60 of 65 (92%) BL-PAP. One additional case identified by both classifiers carried a non–IG-MYC translocation, leaving 6 of 70 mBL and 4 of 65 BL-PAP lacking a MYC-translocation.
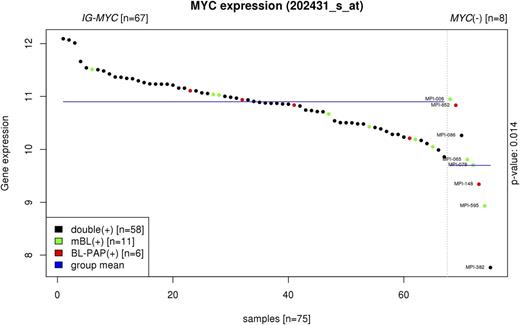
Supervised analysis comparing IG-MYC–positive and MYC-negative BL revealed 708 and 266 genes differentially expressed in the mBL and BL-PAP groups, respectively (false discovery rate = .05). The MYC gene itself was significantly lower expressed at the transcript level in both MYC-negative mBL and MYC-negative BL-PAPs compared with their MYC-positive counterparts (P = .014) (Figure 2).
Supervised analysis comparing MYC expression between IG-MYC–positive and MYC-negative lymphomas from mBL and BL-PAP groups. mBL: BL cases defined by mBL index (green); BL-PAP: BL cases defined by BL-PAP index (red); double: BL cases defined by both mBL and BL-PAP indices (black) (P = .014).
Supervised analysis comparing MYC expression between IG-MYC–positive and MYC-negative lymphomas from mBL and BL-PAP groups. mBL: BL cases defined by mBL index (green); BL-PAP: BL cases defined by BL-PAP index (red); double: BL cases defined by both mBL and BL-PAP indices (black) (P = .014).
The thorough gene expression analyses left only 2 MYC-negative lymphomas (MPI-086, MPI-382) from a total of 59 lymphomas, which were called “BL” by both GEP classifiers applied. Interestingly, both cases were diagnosed in young males aged 14 and 24 years, showed a germinal center B (GCB)-like cell of origin gene expression signature, and were initially diagnosed as BL but reclassified by the pathology panel as high-grade B-cell lymphoma, not otherwise specified (NOS), and DLBCL, centroblastic, respectively. They had nodal presentation and the immunophenotype was that typical of BL. Both cases lacked the t(14;18)/IGH-BCL2 fusion, BCL6 breaks, or MALT1 breaks by FISH. Moreover, both shared low MYC-expression with 1.9-fold lower expression than in IG-MYC–positive cases.
Despite the considerable CN alterations found in the 2 cases (12 and 9 alterations, respectively), both shared only one aberration, which was a peculiar pattern of 11q22-q24 gain and 11q24-qter loss (11q-gain/loss) (supplemental Figure 2).
Cytogenetic and array-based characterization of the chromosome 11 aberrations pattern in MYC-negative lymphomas resembling BL (cohort 2)
Cytogenetic and concomitant aberrations.
Because a recent cytogenetic study suggested a similar pattern appearing as partial trisomy 11 resulting from a dup(11)(q23q13) to characterize lymphomas with BL morphology without MYC gene rearrangement,8 an extensive genetic investigation of this chromosome 11 aberration pattern was initiated. In addition to the cases described by Pienkowska-Grela et al,8 we identified 8 lymphomas with features of BL predominantly in children and young adults. The mean age of the whole series was 20.5 years (range 6-76).
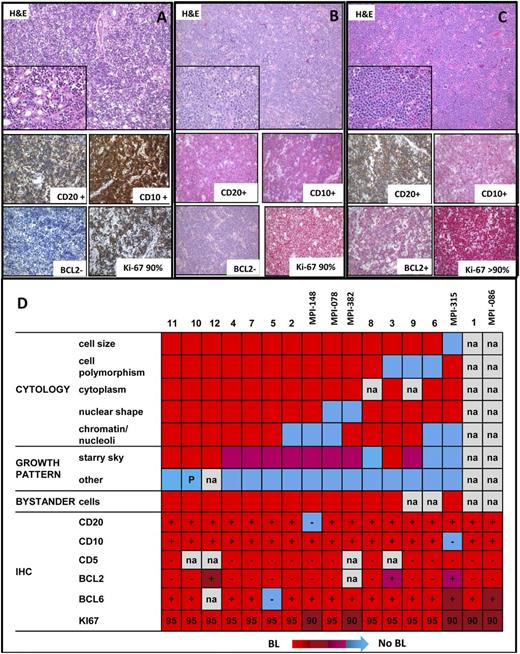
The median follow-up of the patients was 36 months. All patients but one (case 6) were alive at last follow-up (Table 1). The cytogenetic analyses confirmed the absence of a Burkitt translocation and other recurrent IG translocations such as the t(14;18)/BCL2 breaks. Despite the lack of any detectable MYC translocation, these cases resembled BL in many aspects including morphologic features (“starry sky” pattern, many mitoses, and very few infiltrating reactive lymphocytes) and immunophenotype (mostly CD20+/CD10+/BCL2– with Ki67 positivity close to 100%) (Figure 3; supplemental Table 2). Epstein-Barr virus RNA expression was not detectable in 10 cases analyzed (Table 1). Cytogenetically, the chromosome 11 aberration was mostly described as inverted duplication 11q13-q23 (5/9). In addition to the chromosome 11 aberrations, 6q14-q24 and 18q21-q22 changes were recurrent in the cases, being present in 2 and 3 of the 9 evaluable cases, respectively (Table 2).
Morphologic features of MYC-negative high-grade lymphomas. (A) Hematoxylin and eosin preparation showing the morphology and CD20, CD10, BCL2, and Ki67 expression by immunohistochemistry of (A) case 1, (B) case 2, and (C) case 3 in cohort 2. (D) Heatmap showing the morphologic and the immunohistochemical features of the 17 MYC-negative high-grade lymphomas. The morphologic characteristics included information of the cytology, growth pattern, and bystander cells. Red, pro-Burkitt: medium cell size; no cell polymorphism; narrow cytoplasm; round nuclear shape; multiple, small, and paracentric nucleoli; “starry-sky” growth pattern; scattered bystander cells; and sparse eosinophils. Blue, no BL-like: large cell size; cell polymorphism; abundant cytoplasm; irregular nuclear shape; multiple, small, eccentric (centroblast-like) nucleoli/single, large, central (immunoblast-like) nucleolus/finely, granular (lymphoblast-like)/other chromatin; absent “starry-sky” growth pattern; abundant bystander cells. Immunohistochemical analyses included data on CD20, CD10, CD5, BCL2, BCL6, and Ki67. –, negative; +, positive; IHC, immunohistochemistry; na, not available; P, partial.
Morphologic features of MYC-negative high-grade lymphomas. (A) Hematoxylin and eosin preparation showing the morphology and CD20, CD10, BCL2, and Ki67 expression by immunohistochemistry of (A) case 1, (B) case 2, and (C) case 3 in cohort 2. (D) Heatmap showing the morphologic and the immunohistochemical features of the 17 MYC-negative high-grade lymphomas. The morphologic characteristics included information of the cytology, growth pattern, and bystander cells. Red, pro-Burkitt: medium cell size; no cell polymorphism; narrow cytoplasm; round nuclear shape; multiple, small, and paracentric nucleoli; “starry-sky” growth pattern; scattered bystander cells; and sparse eosinophils. Blue, no BL-like: large cell size; cell polymorphism; abundant cytoplasm; irregular nuclear shape; multiple, small, eccentric (centroblast-like) nucleoli/single, large, central (immunoblast-like) nucleolus/finely, granular (lymphoblast-like)/other chromatin; absent “starry-sky” growth pattern; abundant bystander cells. Immunohistochemical analyses included data on CD20, CD10, CD5, BCL2, BCL6, and Ki67. –, negative; +, positive; IHC, immunohistochemistry; na, not available; P, partial.
CN and cytogenetic aberrations of the 12 cases of MYC-negative B-cell lymphomas and SU-DHL-5 and HT cell lines (cohort 2)
| Case no. . | Large CN alterations (>4Mb) . | Karyotype . | t(8;14) . | MYC BAP/ IGH BAP . | MYC BAP 1/2 . | BCL2/ MALT1 BAP . | CEP11/ATM/ FDX . | CEP11/11qter . | CCND1 BAP . |
|---|---|---|---|---|---|---|---|---|---|
| Case 1 | Gains (7q34-qter, 8q23.1-q24.22, 11q12.2-q23.3, 13q22.3-q33.2, 18q12.3-q21.1); ampl (12q12-q13.12, 18q21.1-q22.1); losses (4q21.21-q22.1, 6q13-q22.2, 11q23.3-qter, 13q33.2-qter, 18q22.1-q23) | 46,XY,del(6)(q14q24),add(7)(p21),dup(11) (q13q23),t(12;18)(q12;q23),ins(18;?)(q21;?)[18] | – | – | – (3 copies) | CEP11x2;ATM x3;FDXx3 | |||
| Case 2 | Gains (11q12.1-q23.2); ampl (11q23.3-qter); losses (10q23.2-q23.32, 11q23.3-qter) | 46,XY,?dup(11)(q12q23),der(18)?t(13;18)(q21;q21)[15] | – | – | –/– | – | CEP11x2; ATMx2;FDXx2 | ||
| Case 3 | Gains (3p26.3-q13.11, 3q22.1-q29, 10q24.32-q26.3, 18q21.1-q23); ampl(3q12.1-q13.11); losses (3q13.11-q22.1, 6q14.3-q24.2, 11q23.3-q25, Xp22.33-qter) | 47,X,-X,+3,add(3)(q11)x2,t(6;10)(p22;q22),der(6)t(6;10)(p22;q22)del(6)(q12q23),add(11) (q23),+mar [12]/46,idem,t(9;13)q10;p10),-mar[5] | – | – | –/– | CEP11x2; ATMx2;FDXx2 | |||
| Case 4 | Gains (11q13.4-q23.3); losses (11q23.3-qter) | 46,XY,dup(11)(q23q13) [20] | – | – | CEP11x2; ATMx3;FDXx3 | CEP11x2 11qterx1 | |||
| Case 5 | Gains (7q34-qter, 11q13.4-q24.1,12); losses (11q24.1-qter) | 46–47,XY,dup(11)(q23q13)-13,+1∼2mar[15]/46-47,sl, +add(8)(p21),+der(14)t(4;14)(q31;q13),add(18)(q22)[cp5] | – | – | |||||
| Case 6 | Gains (11q13.1-q23.3, 19p13.3-p13.2); ampl (11q23.3); losses (11q23.3-qter) | 46,XY,dup(11)(q23q13)[9] | –/+ | – | CEP11x2; ATMx3;FDXx3 | CEP11x2 11qterx1 | CCND1x3 | ||
| Case 7 | Gains (11q13.1-q24.1, 12); losses (11q24.1-qter) | 47,XX,dup(11)(q23q13), +12[cp21] | –/+ | – | CEP11x2 11qterx1 | CCND1x3 | |||
| Case 8 | Gain (7q31.1-qter; 11q12.1-q24.2; 12pter-p12.2; 12p12.1-qter); losses (9q21.11-q31.1; 11q24.2-qter) | 47,XY,del(9)(q22q34), der(11)(11pter->11q23::?11q23->11q11:?),+12[7]/47,sl,add(17)(p11)[2] | – | ||||||
| Case 9 | Gain (11q13.1-q23.3); ampl (11q14.3-q23.3); losses (2q14.3-q31.1; 11q23.3-qter) | na | – | – | –/– | ATMx4-5;FDXx4-5 | |||
| Case 10 | Gain (10; 11q12.1-q23.3; 11q23.3); losses (11q23.3-qter) | na | –/– | – | |||||
| Case 11 | Gain (8q24.13-q24.23; 11q13.1-q23.3); losses(7q11.21-q11.22; 7q11.22-q11.23; 11q23.3-qter) | 46, XY,add(11)(q23)[6]/46,XY[14] | – | – | –/– | – | CEP11X2 11qterx1 | ||
| Case 12 | Gain (11q12.1-q22.3); losses (11q22.3-qter) | na | – | – | – | CEP11X2 11qterx1 | |||
| SU-DHL-5 | Gains (11q23.2-q23.3, 12pter-q13, 19pter-p13.2); losses (6pter-p22.1, 6q13-qter, 11q23.3-qter) | 47,XX,add(6)(p21),del(6)(q13),+12,del(12)(q12) | – | – | –/– | – | |||
| HT | Gain (10q21.1-q22.2, 11q22.3-q23.2); ampl 11q22.3-q23.2; 11q23.2-q23.3); losses (2p25.3-p24.1, 2p22.3-p16.2, 2p16.1-p15, 2p15-p12, 2q13-q24.1, 4pter-p12, 11q23.3-qter) | 46(42-46)<2n>XY,+2, der(2;4)(p10;q10), der(2)(del)(2)(p11?q21), dup(10)(q11q22-23), dup(11)(?q23qter) | – | – | –/– | – |
| Case no. . | Large CN alterations (>4Mb) . | Karyotype . | t(8;14) . | MYC BAP/ IGH BAP . | MYC BAP 1/2 . | BCL2/ MALT1 BAP . | CEP11/ATM/ FDX . | CEP11/11qter . | CCND1 BAP . |
|---|---|---|---|---|---|---|---|---|---|
| Case 1 | Gains (7q34-qter, 8q23.1-q24.22, 11q12.2-q23.3, 13q22.3-q33.2, 18q12.3-q21.1); ampl (12q12-q13.12, 18q21.1-q22.1); losses (4q21.21-q22.1, 6q13-q22.2, 11q23.3-qter, 13q33.2-qter, 18q22.1-q23) | 46,XY,del(6)(q14q24),add(7)(p21),dup(11) (q13q23),t(12;18)(q12;q23),ins(18;?)(q21;?)[18] | – | – | – (3 copies) | CEP11x2;ATM x3;FDXx3 | |||
| Case 2 | Gains (11q12.1-q23.2); ampl (11q23.3-qter); losses (10q23.2-q23.32, 11q23.3-qter) | 46,XY,?dup(11)(q12q23),der(18)?t(13;18)(q21;q21)[15] | – | – | –/– | – | CEP11x2; ATMx2;FDXx2 | ||
| Case 3 | Gains (3p26.3-q13.11, 3q22.1-q29, 10q24.32-q26.3, 18q21.1-q23); ampl(3q12.1-q13.11); losses (3q13.11-q22.1, 6q14.3-q24.2, 11q23.3-q25, Xp22.33-qter) | 47,X,-X,+3,add(3)(q11)x2,t(6;10)(p22;q22),der(6)t(6;10)(p22;q22)del(6)(q12q23),add(11) (q23),+mar [12]/46,idem,t(9;13)q10;p10),-mar[5] | – | – | –/– | CEP11x2; ATMx2;FDXx2 | |||
| Case 4 | Gains (11q13.4-q23.3); losses (11q23.3-qter) | 46,XY,dup(11)(q23q13) [20] | – | – | CEP11x2; ATMx3;FDXx3 | CEP11x2 11qterx1 | |||
| Case 5 | Gains (7q34-qter, 11q13.4-q24.1,12); losses (11q24.1-qter) | 46–47,XY,dup(11)(q23q13)-13,+1∼2mar[15]/46-47,sl, +add(8)(p21),+der(14)t(4;14)(q31;q13),add(18)(q22)[cp5] | – | – | |||||
| Case 6 | Gains (11q13.1-q23.3, 19p13.3-p13.2); ampl (11q23.3); losses (11q23.3-qter) | 46,XY,dup(11)(q23q13)[9] | –/+ | – | CEP11x2; ATMx3;FDXx3 | CEP11x2 11qterx1 | CCND1x3 | ||
| Case 7 | Gains (11q13.1-q24.1, 12); losses (11q24.1-qter) | 47,XX,dup(11)(q23q13), +12[cp21] | –/+ | – | CEP11x2 11qterx1 | CCND1x3 | |||
| Case 8 | Gain (7q31.1-qter; 11q12.1-q24.2; 12pter-p12.2; 12p12.1-qter); losses (9q21.11-q31.1; 11q24.2-qter) | 47,XY,del(9)(q22q34), der(11)(11pter->11q23::?11q23->11q11:?),+12[7]/47,sl,add(17)(p11)[2] | – | ||||||
| Case 9 | Gain (11q13.1-q23.3); ampl (11q14.3-q23.3); losses (2q14.3-q31.1; 11q23.3-qter) | na | – | – | –/– | ATMx4-5;FDXx4-5 | |||
| Case 10 | Gain (10; 11q12.1-q23.3; 11q23.3); losses (11q23.3-qter) | na | –/– | – | |||||
| Case 11 | Gain (8q24.13-q24.23; 11q13.1-q23.3); losses(7q11.21-q11.22; 7q11.22-q11.23; 11q23.3-qter) | 46, XY,add(11)(q23)[6]/46,XY[14] | – | – | –/– | – | CEP11X2 11qterx1 | ||
| Case 12 | Gain (11q12.1-q22.3); losses (11q22.3-qter) | na | – | – | – | CEP11X2 11qterx1 | |||
| SU-DHL-5 | Gains (11q23.2-q23.3, 12pter-q13, 19pter-p13.2); losses (6pter-p22.1, 6q13-qter, 11q23.3-qter) | 47,XX,add(6)(p21),del(6)(q13),+12,del(12)(q12) | – | – | –/– | – | |||
| HT | Gain (10q21.1-q22.2, 11q22.3-q23.2); ampl 11q22.3-q23.2; 11q23.2-q23.3); losses (2p25.3-p24.1, 2p22.3-p16.2, 2p16.1-p15, 2p15-p12, 2q13-q24.1, 4pter-p12, 11q23.3-qter) | 46(42-46)<2n>XY,+2, der(2;4)(p10;q10), der(2)(del)(2)(p11?q21), dup(10)(q11q22-23), dup(11)(?q23qter) | – | – | –/– | – |
–, negative; +, positive; BAP, break apart.
Bold indicates chromosomal alterations that involve chromosome 11.
The cell lines SU-DHL-5 and HT were selected as potential models of this subgroup of lymphoma because they showed the same aberration pattern 11q-gain/loss (cytogenetics or arrays). None of the cases and cell lines carried ID3 mutations, which have been recently shown to be present in >60% of BL.21,23,24
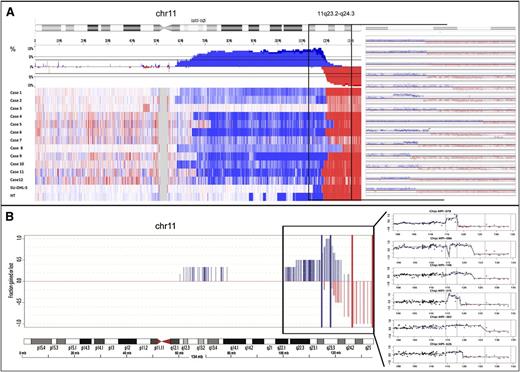
To define the pattern of chromosome 11 aberrations at high resolution, we performed single nucleotide polymorphism (SNP) array analyses on DNA extracted from fresh frozen cultured cells (cases 1-3, 8, SU-DHL-5, and HT) and oligonucleotide CGH-array on DNA from formalin fixed paraffin embedded tissue tumor samples (cases 4-7 and 9-12) (Figure 4). Common regions of alteration in addition to 11q-gain/loss pattern were gains of 7q34-qter (3 cases), 12pter-p12.2 (4 cases), 18q21.2 (3 cases), and 19pter-p13.2 (2 cases), and losses of 6q14.3-q22.2 (3 cases) (supplemental Table 4). No recurrent regions of CN neutral loss of heterozygosity (CNN-LOH) were observed.
11q alterations in MYC-negative high-grade lymphomas. (A) Chromosomal view of chromosome 11 analyzed by SNP arrays in cohort 2 (n = 14, including SU-DHL-5 and HT cell lines). (B) Chromosomal view of chromosome 11 analyzed by custom CGH-array in cohort 3 (n = 6).
11q alterations in MYC-negative high-grade lymphomas. (A) Chromosomal view of chromosome 11 analyzed by SNP arrays in cohort 2 (n = 14, including SU-DHL-5 and HT cell lines). (B) Chromosomal view of chromosome 11 analyzed by custom CGH-array in cohort 3 (n = 6).
Minimal gained/amplified 11q region.
With regard to chromosome 11q, interstitial gains in 11q23.2-q23.3 were detected in 13 of 14 samples analyzed in the cohort 2 (92.9%). Of those, 5 of 13 samples included focal high-level amplifications in 11q23.3 (38.5%) (cases 2, 6, 9, 10, and HT). The minimal region of gain was defined as chr11:114 530,818-117 939,359bp (hg18) and the minimal region of amplification as chr11:117 107,361-117 939,359bp (hg18) (supplemental Table 5). Remarkably, the ATM gene was located in the region of gain in 8 of 14 samples, but it is noteworthy that one lymphoma (case 1, MPI-626) occurred in a patient with ataxia telangiectasia associated with a homozygous germline deletion of exons 9 and 10 of the ATM gene (c.663_1065del403; EX9_10del) present in a stretch of CNN-LOH.
Recently, hsa-mir-34b was described to be downregulated in BL cases that were negative for MYC translocation.5 Remarkably, the hsa-mir-34b is located in 11q23.1 (chr11:110 888,873-110 888,956bp, hg18) and thus in the recurrently gained rather than in the lost region. Thus we analyzed the DNA methylation of hsa-mir-34b. Heterogeneous methylation patterns of hsa-mir-34b were found in cases 1 through 7 from cohort 2 (supplemental Figure 3). In addition, we found no significant enrichment of hsa-mir-34b annotated target genes (Molecular Signatures Database, MSigDB) among the genes that were differentially expressed between cases with the 11q-gain/loss pattern and IG-MYC–positive mBL nor between these cases and DLBCL.
Minimal loss/homozygously lost 11q region.
Telomeric losses of 11q affecting 11q24.1-qter were observed in the whole cohort 2. The common region of deletion was chr11:126 977 015-134 445 937bp (hg18). Interestingly, one patient (case 9) displayed a focal homozygous deletion within this region located at chr11:127 322 011-128 846 569bp (hg18) (supplemental Figure 4A; supplemental Table 4). The minimal region of loss defined by this homozygous deletion contains, among others, the genes FLI1 and ETS1, which have been shown to be involved in hematologic neoplasms and to be mutated in lymphomas, respectively.25,26
The 11q-breakpoints between regions of gain and loss were not conserved in the different cases, and breakpoints scattered from 117 939 359 to 126 977 015bp (hg18) from 11pter (supplemental Figure 4B).
Identification of lymphomas with the typical 11q-gain/loss pattern in the MMML series (cohort 3)
Having defined the minimal regions of gain and loss in the retrospective cohort 2, we investigated the available CGH-array data of the MMML for cases with simultaneous gain and loss of the minimal regions in 11q. This search yielded a total of 6 cases. Excluding the 2 initial MYC-negative BL and one case that was also in cohort 2 (case 1, MPI-626), this left us with 3 additional cases. Again, these cases resembled BL in many molecular features. Notably, 2 of these cases were classified as BL by one, but not both, of the GEP algorithms (MPI-078 and MPI-148). Table 3 provides an overview on all 6 cases identified from the MMML cohort in which the typical 11q-gain/loss pattern was observed in the absence of an MYC, BCL2, or BCL6 translocation. A comparison of pathological and molecular features contrasting these 6 cases with mBL and DLBCL from the MMML cohort is provided as supplemental Table 6. These results once more indicate that based on CD10 positivity, BCL2 negativity, and high Ki67 expression, the identified cases resembled more BL than DLBCL. Moreover, the cases had an excellent survival rate. Specifically, global survival analysis of the 16 cases from cohort 2 and cohort 3 compared with MYC-positive BL patients younger than 40 years (n = 51 from the MMML series) showed that this subgroup of cases has no significantly different prognosis than typical BL of this age range (3-year overall survival: 92% vs 76%, P = .12) (supplemental Figure 5). In terms of genetic complexity and in line with the results of cohort 2, cohort 3 showed recurrent losses of chromosome 6q as the second most frequent genetic alteration in these cases (4/6 cases) (supplemental Table 4). Combining both data sets, the minimal region of loss was in 6q14.1-q21 and included the EPHA7 and PRDM1 tumor suppressor genes.27,28 Overall, the cases with the typical 11q-gain/loss pattern showed a significantly higher chromosomal complexity based on CGH-array compared with mBL and DLBCL (12.2 vs 4.2 and 7.2 CN alterations, respectively; P < .0001).
Morphologic, genetic, immunohistochemical, and clinical data of the 6 MMML cases carrying 11q-gain/loss pattern selected for GEP analysis (cohort 3)
| Case no. . | Age, sex . | Diagnosis . | Location . | FISH . | . | Immunohistochemistry . | Mutations . | . | . | . | . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MYC break . | BCL6 break . | t(14;18) . | MYC . | CD20 . | CD10 . | CD5 . | BCL2 . | BCL6 . | MUM1 . | Ki67 . | EBER . | IGHV . | BCL6 . | MYC . | GEP ABC/GCB . | Molecular diag . | PAP diag‡ . | Outcome, follow-up . | ||||
| MPI-078 | 8, male | Centroblastic, DLBCL | LN(c) | – | – | – | + | + | – | – | + | + | 95 | – | mut | mut | wt | GCB | mBL | mind-L | In complete remission, alive, 119 mo | |
| MPI-086¶ | 14, male | High grade B-cell lymphoma | LN(c) | – | – | – | + | + | + | – | – | + | – | 90 | – | mut | mut | wt | GCB | mBL | BL-PAP | In complete remission, alive, 161 mo |
| MPI-148* | 26, male | Atypical BL | extranodal | – | – | – | – | + | – | – | + | – | 90 | – | mut | mut | wt | GCB | intermediate | BL-PAP | In complete remission, alive, 163 mo | |
| MPI-315 | 49, female | Centroblastic, DLBCL | intestine | – | – | – | + | – | – | + | + | + | 90 | – | mut | na | na | GCB | intermediate | PAP-1 | In complete remission, alive, 106 mo | |
| MPI-382¶ | 24, male | Centroblastic, DLBCL | LN | – | – | – | + | + | na | na | + | na | 90 | – | mut | na | na | GCB | mBL | BL-PAP | In complete remission, alive, 91 mo | |
| MPI-626† | 7, male | Malignant B-cell lymphoma | LN(c) | – | – | – | + | + | – | – | + | – | 90 | – | mut | na | wt | GCB | intermediate | mind-L | In remission, alive, 62 mo | |
| Case no. . | Age, sex . | Diagnosis . | Location . | FISH . | . | Immunohistochemistry . | Mutations . | . | . | . | . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MYC break . | BCL6 break . | t(14;18) . | MYC . | CD20 . | CD10 . | CD5 . | BCL2 . | BCL6 . | MUM1 . | Ki67 . | EBER . | IGHV . | BCL6 . | MYC . | GEP ABC/GCB . | Molecular diag . | PAP diag‡ . | Outcome, follow-up . | ||||
| MPI-078 | 8, male | Centroblastic, DLBCL | LN(c) | – | – | – | + | + | – | – | + | + | 95 | – | mut | mut | wt | GCB | mBL | mind-L | In complete remission, alive, 119 mo | |
| MPI-086¶ | 14, male | High grade B-cell lymphoma | LN(c) | – | – | – | + | + | + | – | – | + | – | 90 | – | mut | mut | wt | GCB | mBL | BL-PAP | In complete remission, alive, 161 mo |
| MPI-148* | 26, male | Atypical BL | extranodal | – | – | – | – | + | – | – | + | – | 90 | – | mut | mut | wt | GCB | intermediate | BL-PAP | In complete remission, alive, 163 mo | |
| MPI-315 | 49, female | Centroblastic, DLBCL | intestine | – | – | – | + | – | – | + | + | + | 90 | – | mut | na | na | GCB | intermediate | PAP-1 | In complete remission, alive, 106 mo | |
| MPI-382¶ | 24, male | Centroblastic, DLBCL | LN | – | – | – | + | + | na | na | + | na | 90 | – | mut | na | na | GCB | mBL | BL-PAP | In complete remission, alive, 91 mo | |
| MPI-626† | 7, male | Malignant B-cell lymphoma | LN(c) | – | – | – | + | + | – | – | + | – | 90 | – | mut | na | wt | GCB | intermediate | mind-L | In remission, alive, 62 mo | |
−, negative; +, positive; ABC, activated B cell; c, cervical; diag, diagnosis; EGER, Epstein-Barr–encoded RNA test; GCB, germinal center B cell; GEP, gene expression profiling; LN, lymph node; mind-L, molecularly individual lymphoma; mut, mutated; na, not available; PAP, pathway activation pattern algorithm; wt, wild-type.
This case presented the following karyotype: 46,XY,t(7;18)(q32;q21),inv dup(11)(q23q13),add(15)(q22),inv(16)(q11q24).
This case corresponds to case 1 from cohort 2 and was investigated using SNP and CGH-array platforms.
These two cases belong to cohort 1. Lymphoma samples were scored positive if >25% of the tumor cells stained positive except for MYC, where the cutoff was established in 40%.
Identification of target genes of the duplicated/deleted 11q regions
To identify candidate genes potentially deregulated through the gains/amplifications in 11q23 and the telomeric losses in 11q24, we compared the GEP of the 6 lymphomas from the MMML with the typical 11q-gain/loss pattern (cohort 3) to IG-MYC mBL (n = 46) and DLBCL (n = 198) (supplemental Tables 7 and 8). Several genes located in the minimal region of gain at 11q23.3 (PAFAH1B2, ZNF259, PCSK7, CEP164, UBE4A, and ATP5L) were significantly overexpressed in 11q-gain/loss pattern cases compared with both mBL and DLBCL (supplemental Figure 6A-B).
To investigate whether the transcriptional upregulation also translates into changes in protein expression, we performed Western blot analyses of the candidate gene PAFAH1B2 in cell lines with and without 11q-gain/loss pattern as well as in BL cell lines (supplemental Figure 6C). Also on the protein level, this gene was overexpressed in HT and SU-DHL-5 compared with cell lines without an 11q-gain/loss pattern.
In the minimal region of loss at 11q24.1-qter, the FLI1, SNX19, NCAPD3, and ACAD8 genes showed significantly lower mRNA expression in cases with an 11q-gain/loss pattern (supplemental Figure 7A-B). Thus sequence analysis of FLI1 was initiated. In addition, the ETS1 gene located in the focal homozygous deletion of case 9 entered into mutational screening. Sequencing of all exons of FLI1 showed no mutations in the 12 analyzed samples with sufficient material available (data not shown). In contrast, 7 point mutations were detected in ETS1 in 4 of 16 cases studied (supplemental Table 9; supplemental Figure 7C). Two mutations were silent and 5 were nonsilent, corresponding to 3 transitions and 2 transversions. In total, 3 mutations were missense, 1 was located in the splice donor site of intron 1, and the last one was an inactivating mutation (p.Y154X).
In addition to the Y→X/del mutation that removes the DNA binding domain in case 8, no clear functional consequences of any mutation could be derived based on protein modeling, because the modified residues lie outside of regions with resolved structures.
To explore recurrence of ATM, ETS1, and FLI1 gene mutations, data from custom exome sequencing of HT and SU-DHL-5 cell lines with regard to genes located at 11q were investigated. However, the cell lines showed no mutation of the genes in the minimal regions of loss including ETS1 and FLI1 (data not shown). ATM was also not mutated in these cell lines.
Differential gene expression in lymphomas with the typical 11q-gain/loss pattern
Finally, we compared the global GEP of cohort 3 with those of BL and DLBCL. There were 1194 and 1826 genes differentially expressed between 11q-gain/loss cases and IG-MYC–positive BL (n = 46) or DLBCL (n = 198), respectively (supplemental Figures 8 and 9). Compared with DLBCL, cases with 11q-gain/loss pattern showed upregulation of genes involved in cell cycle and proliferation and downregulation of major histocompatibility complex/immune-interferon responses, cell death, and signaling, most of them also with features distinguishing BL from DLBCL. Compared with BL, 11q-gain/loss cases showed downregulation of RNA binding activity and metabolic processes and upregulation of RNA degradation.
Discussion
Here we report the clinical, morphologic, and molecular characterization of a subset of MYC-negative, high-grade B-cell lymphomas sharing similarities with but not being BL. The search for MYC-negative lymphomas with a GEP of BL showed that only 2 cases of 59 were concordantly called “BL” by 2 GEP classifiers, stating that these cases are in general rare. In other series, MYC-negative BL accounted for 14% to 22% of all BL. Nevertheless, many of these studies either do not apply a full set of FISH probes necessary to exclude rare variants of the IG-MYC translocation or are not based on gene expression.5,29 In addition, a recent study applying an extensive set of FISH probes also reached a similar low frequency of MYC-negative BL as described herein.30 Of note, 4 of 6 MYC-negative mBL and 2 of 4 MYC-negative BL-PAP were differently classified by the alternative BL classifier, suggesting that in a high percentage (50-66%) of even molecularly defined MYC-negative BL, the diagnosis should be questioned. The significantly lower expression of MYC mRNA in the MYC-translocation–negative mBL/BL-PAPs supports the view that MYC indeed is not genomically activated in these cases, rendering a technical failure of FISH in detection of the translocation unlikely.
Both molecularly defined MYC-negative BL cases shared a recurrent pattern of chromosome 11q abnormalities that was subsequently found in a series of 12 high-grade B-cell lymphomas with features of BL and in 2 cell lines. A similar pattern was identified in a set of 6 MYC-negative cases from MMML series classified as mBL/intermediate by GEP. The comparison of pathological and molecular features between these 6 cases compared with DLBCL and BL showed that these tumors resembled more BL than DLBCL, reinforcing the idea that this group of samples may represent a genetic variant of BL lacking the MYC translocation. Nevertheless, for the sake of diagnostic clarity, it is proposed here to restrict the term BL to IG-MYC–positive cases and call the lymphomas described herein “high-grade B-cell lymphoma with features of BL, MYC-, with 11q-gain/loss pattern.”
Alterations of the 11q chromosomal region are not frequent in typical BL31,32 but gains/amplifications of 11q22-q24 are found in approximately 8% to 15% of DLBCL33,34 and also in GC-derived IRF4-translocation–positive lymphomas.35 Nevertheless, the region of gain in these cases (minimal region at 11q23.2-q23.3) is not coincident with the minimal region observed in the present series. Indeed, in the DLBCL and IRF4-translocation–positive lymphomas, gains in 11q mostly extend to the telomere. Thus in these lymphomas, genes like ETS1 or FLI1, which are included in our minimal region of loss (11q24.1-qter) and are even homozygously deleted in one case, are considered as oncogenes in DLBCL because they cooperate in sustaining DLBCL proliferation and viability and regulate genes involved in germinal center differentiation,34,36 contrasting the findings herein, which rather suggest a tumor-suppressive function.
The incidence of 11q-gain/loss pattern in the total group of 514 samples from MMML with CGH-array data are the following: 0.7% in DLBCL (2/267), 1.6% in IG-MYC mBL (1/63), 16.7% cell lines (2/12), 0% FL (0/67), 5.2% intermediate (4/77), 50% other mBL without IG-MYC, (3/6), and 0% others (0/22).
When we screened the Mitelman (http://cgap.nci.nih.gov/Chromosomes/Mitelman) and Progenetix (www.Progenetix.org) databases (September 2012) for pediatric lymphomas, we found 10 of 31 cases (32%) diagnosed as BL/Atypical without the t(8;14) translocation or variants presented alterations on chromosome 11.
A potential candidate oncogene in the gained region in 11q in the lymphomas described herein is PAFAH1B2, which we showed to be selectively upregulated also on the protein level in cell lines containing the typical 11q pattern (supplemental Figure 6C). The PAFAH1B2 gene has been previously recognized as target of IGH locus translocations in 2 lymphoma cases.37 Such IGH translocation leads typically to oncogene activation and it is therefore consequent to suggest PAFAH1B2 (previously named PAFAH1A2) as an oncogene in lymphomagenesis. Furthermore, it has been found to interact at protein level with ZFP36L1,38 another partner of IGH translocations described in chronic lymphocytic leukemia39,40 and the target of hypermutation in DLBCL.41 It is also intriguing to speculate that, given the recurrent co-amplification of several genes in 11q, there is a cooperative oncogenic function, as has been proposed for 9p24 gains in some subsets of B-cell lymphomas.42,43
In the minimal region of loss, FLI1 and ETS1 were the candidate tumor suppressor genes, both located in the region of focal homozygous loss. The transcription factor FLI1 is known to be involved in lymphoma development in mice,25 and we found it significantly downregulated in the cohort of MYC-negative lymphomas with the 11q aberrations from cohort 3. Nevertheless, this gene displayed no mutations in any of the samples analyzed. The ETS1 gene, however, was found to be homozygously lost in one and mutated in four of the 16 lymphomas investigated. In addition, ETS1 has been identified as an oncogenic fusion partner of the c-Myb proto-oncogene product in the E26 avian leukemia virus44 and has been found to be recurrently mutated in non-Hodgkin lymphomas,26 although the functional consequences of the detected mutations remain to be defined. Our attempts to model the effect of the mutations yielded no striking results because of the low number of mutations found in our series and the fact that none of the point mutations were within resolved 3-dimensional structures. Additional structures or biophysical studies of ETS1 would be required to resolve the possible roles of these positions in protein function. High levels of expression associated with gains of the 11q24.3 (including FLI1 and ETS1) have been also seen in these genes in DLBCL patients,34 although a different mechanism seems to be involved in these cases.
Recently, several studies showed that ID3 mutations and/or TCF3-activating mutations in BL lead to activation of PI3K signaling.21,23,24 Moreover, PI3K pathway activation has been shown to cooperate with MYC activation in the pathogenesis of BL.45 Our series displayed no mutations in ID3. Nevertheless, PI3K signaling activity in the cases described herein are distributed similarly to that of MYC-positive BL, applying the pathway activity quantification approach of Sander et al (supplemental Figure 10).45 These findings could suggest that either alternative mechanisms cause similar pathway deregulations like in MYC-positive BL, or that the cell of origin of the lymphomas with features of BL characterized by the 11q aberration is the same as that of MYC-positive BL.
In contrast to Leucci et al,5 we failed to identify a role of hsa-mir-34b in MYC-negative BL. These results are in agreement with a recent study performed by Lenze et al in which the MYC-negative BL cases did not show downregulation of this microRNA.46 It is remarkable that this microRNA is located in a region of gain in 6 of the 7 cases with 11q aberration studied.
In conclusion, we have identified a particular molecular subset of high-grade B-cell lymphomas that share a recurrent pattern of 11q aberration. Cases are morphologically and phenotypically similar to BL but differ with regard to the MYC translocation, the 11q-gain/loss pattern and the more complex karyotypes. Of note, lymphomas with the typical 11q-gain/loss pattern seem to have more frequently nodal presentation than BL from patients younger than 40 years (82% vs 55%, P = .074). The specific characteristics detected in this group of lymphomas raise the question of whether these lymphomas truly belong to the entity of BL or whether they could be considered as another set of high-grade, mature B-cell lymphomas sharing features of, but not being, BL. We provide evidence that the presence of the typical 11q-gain/loss pattern could be a genetic hallmark that defines this group of cases. The pattern of aberrations along with the results of our molecular analyses suggest that activation of genes included in the region of gain in 11q may co-operate with the functional inactivation of genes in the recurrent region of loss, including FLI1 and ETS1, most likely through transcriptional mechanisms in this newly identified subgroup of lymphomas.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the staff of the involved cytogenetic and molecular cytogenetic laboratories, in particular Dr Lana Harder, Reina Zühlke-Jenisch, Claudia Becher, Margret Ratjen, and Olivera Batic, for their excellent support.
This work is dedicated to the memory of Prof Dr rer nat Dr hc Lore Zech, who died during the preparation of the manuscript, and her outstanding contributions on the characterization of chromosomal aberrations in Burkitt and other lymphomas.
This study was supported by the Deutsche Krebshilfe (70-3173-Tr3) through the Network project “Molecular Mechanisms in Malignant Lymphomas” (from 2003 to 2011); the KinderKrebsInitiative Buchholz/Holm-Seppensen (W.K., R.S.); the International Cancer Genome Consortium MMML-Seq Project funded by the Federal Ministry of Education and Research in Germany within the Program for Medical Genome Research (01KU1002A to 01KU1002J); a fellowship from the Alexander von Humboldt Foundation and the Subprograma Juan de la Cierva “Ministerio de Ciencia e Innovación” (JCI-2011-10232) (I.S.); a grant from the Basque Government (“Ayuda del programa de perfeccionamiento postdoctoral en el extranjero del Departamento de Educación, Universidades e Investigación”) (I.M.-G.); a Christoph-Schubert-Award of the KinderKrebsInitiative Buchholz/Holm-Seppensen (R.W.); and Dr Werner Jackstädt Stiftung through a Junior Excellence Research Group (J.R.).
Authorship
Contribution: I.S. and I.M.-G. performed molecular analysis, analyzed data, and wrote the manuscript; R.W. performed molecular analysis and analyzed data; M.K., C.W.K., M.L., M.R., and R. Spang performed statistical analyses; J.R., B.P.-G., H.G.D., M.H., R.K., R.A.F.M., I.N., R.B.R., D.S., M. Schlesner, R. Scholtysik, C.S., M. Szczepanowski, I.V., and S.W. performed molecular analysis; B.B., A.C., C.D.-W., E.S.J., C.L., J.L., and L.T. provided clinical data and patient samples; P.A., I.O., G.R., and W.K. provided samples and performed pathology review; and R. Siebert designed the project, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: R. Siebert receives speaker’s honorarium and, in the framework of the MMML, probe discounts from the company Abbott/Vysis and has been supported for a test trial of arrays by the company Affymetrix. The remaining authors declare no competing financial interests.
Correspondence: Reiner Siebert, Institute of Human Genetics, University Hospital Schleswig-Holstein, Campus Kiel, Schwanenweg 24, D-24105 Kiel, Germany; e-mail: rsiebert@medgen.uni-kiel.de; or Itziar Salaverria, Institute of Human Genetics, University Hospital Schleswig-Holstein, Campus Kiel, Schwanenweg 24, D-24105 Kiel, Germany; email: isalaverria@medgen.uni-kiel.de.
References
Author notes
I.S., I.M.-G., R.W., M.K., and C.W.K. share first authorship.