Key Points
Endothelial cells are the predominant (and possibly exclusive) source of coagulation factor VIII.
Hepatocytes do not contribute to plasma FVIII production.
Abstract
The cellular source of coagulation factor VIII (FVIII) remains controversial. Like many coagulation proteins, FVIII is produced in the liver, and FVIII synthesis has long been associated with hepatocytes. But extrahepatic synthesis also occurs, and mounting evidence suggests that hepatocytes are not responsible for FVIII production. To determine the tissue that synthesizes FVIII, we developed a Cre/lox-dependent conditional knockout (KO) model in which exons 17 and 18 of the murine factor VIII gene (F8) are flanked by loxP sites, or floxed (F8F). In cells expressing Cre-recombinase, the floxed sequence is deleted, resulting in F8F→KO gene inactivation. When F8F mice were crossed with various tissue-specific Cre strains, we found that hepatocyte-specific F8-KO mice are indistinguishable from controls, whereas efficient endothelial-KO models display a severe hemophilic phenotype with no detectable plasma FVIII activity. A hematopoietic Cre model was more equivocal, so experimental bone marrow transplantation was used to examine hematopoietic FVIII synthesis. FVIIInull mice that received bone marrow transplants from wild-type donors were still devoid of plasma FVIII activity after hematopoietic donor cell engraftment. Our results indicate that endothelial cells are the predominant, and possibly exclusive, source of plasma FVIII.
Introduction
Coagulation factor VIII (FVIII) deficiency results in the bleeding disorder hemophilia A. Transplantation studies have shown that liver is the major, but not exclusive, site of FVIII synthesis.1-3 Other organs such as lung,4,5 spleen,6-8 and lymphatic tissues9 have also been implicated. F8 gene transcription is widespread, with message present in most of the organs throughout the body.10 Increased FVIII levels were reported in one murine hemophilia model following bone marrow transplantation (BMT),11 but BMT has not been shown beneficial in human12 or canine13,14 hemophilia A.
In liver, about half of total cells are hepatocytes. The remainder consists of 3 major non–parenchymal cell types: liver sinusoidal endothelial cells (LSECs), Kupffer cells, and hepatic stellate cells.15 FVIII production has historically been attributed to hepatocytes,16,17 whereas often-contradictory studies have identified LSECs18-20 and/or Kupffer cells10,21 as sites of hepatic FVIII synthesis. Recently, FVIII activity was demonstrated in isolated LSECs, but not hepatocytes.22
Anatomically diverse microvascular endothelial cells (ECs) have been shown to synthesize and secrete FVIII,23,24 as have induced pluripotent stem cell–derived ECs.25,26 Mesenchymal stem cells isolated from a variety of tissues reportedly produce small amounts of FVIII in vitro.27 A number of cultured primary human cell types, including hepatocytes, mesenchymal stem cells, fibroblasts, monocytes, macrophages, and adipocytes (but not umbilical vein ECs), express roughly equivalent levels of F8 message.28 Although extrahepatic synthesis clearly occurs, the physiological significance of apparent F8 expression in some of the aforementioned cell types remains unclear.
Previous studies have relied upon detection of FVIII protein or messenger RNA (mRNA) using immunochemical, in situ, and cell-isolation techniques to make inferences regarding the contribution of specific cell types to steady-state plasma FVIII levels, a challenging task for a trace protein such as FVIII. We used an alternative approach, introducing genetic changes to the X-chromosomal F8 gene that allow us to eliminate production of functional FVIII in specific cell types. We now report that (1) efficient F8 gene disruption in ECs results in a severe hemophilia phenotype, (2) a hepatocyte-specific F8 knockout (KO) model has a normal phenotype, (3) a hematopoietic F8-KO model results in modestly reduced plasma FVIII levels (but specificity and efficiency are both in question), and (4) transplantation of wild-type (WT) C57BL/6 bone marrow into hemophilia A mice did not correct the coagulation defect.
Methods
F8 gene targeting by homologous recombination
A targeting vector in which F8 exons 17 and 18 (17/18) are flanked by a single upstream loxP site and a downstream <FRT/neomycin resistance/FRT/loxP> cassette was constructed using a combination of ligation-mediated and recombineering29 techniques. F8 gene fragments were retrieved from 129S strain bacterial artificial clone #20642 (Genome Systems, St. Louis, MO). A conditional F8-KO mouse was created by homologous recombination in 129S ECs with subsequent implantation into C57BL/6 embryos to produce chimeric male founders in collaboration with genOway (Lyon, France). Crossbreeding with C57BL/6 females established germline transmission of the targeted F8 allele (Figure 1A). Upon further crossbreeding with B6.Cg-Tg(ACTFLPe)9205Dym/J mice (The Jackson Laboratory, Bar Harbor, ME) expressing Flp-recombinase under control of the human β-actin promoter,30 the FRT-flanked neomycin resistance cassette was excised from intron 18 (Figure 1B), resulting in a conditional KO allele in which exons 17/18 are flanked by loxP sites, or floxed (F8F). Mice carrying the F8F allele were maintained on a mixed 129S-C57BL/6 background.
Conditional F8-KO alleles. (A) Targeted F8 gene. (B) F8F allele. Excision of the neomycin resistance cassette by Flp recombinase produces the floxed (F8F) allele, which is expressed normally. (C) F8KO allele. Excision of exons 17/18 from the F8F allele by Cre recombinase produces a F8KO allele. (D) Alternative mRNA splicing of the F8KO allele. The predicted exon 16/19 splice [F8KO(16/19)] and an alternatively spliced transcript [F8KO(Alt)] in which 46 bp of intron 16 is retained are produced in approximately equal amounts. (E) Domain structure of normal FVIII protein. The position of exons 17/18 within the A3 domain is noted. (F) Predicted FVIII polypeptide encoded by F8KO(16/19) mRNA. (G) Predicted FVIII polypeptide encoded by F8KO(Alt) mRNA. Flp recombinase target (FRT) sites are represented as open triangles, and Cre recombinase target (loxP) sites as gray triangles. The positions of duplex genotyping primers P1, P2, and P3 and RT-PCR primers P4 and P5 are shown. aa, amino acid; Ex, exon.
Conditional F8-KO alleles. (A) Targeted F8 gene. (B) F8F allele. Excision of the neomycin resistance cassette by Flp recombinase produces the floxed (F8F) allele, which is expressed normally. (C) F8KO allele. Excision of exons 17/18 from the F8F allele by Cre recombinase produces a F8KO allele. (D) Alternative mRNA splicing of the F8KO allele. The predicted exon 16/19 splice [F8KO(16/19)] and an alternatively spliced transcript [F8KO(Alt)] in which 46 bp of intron 16 is retained are produced in approximately equal amounts. (E) Domain structure of normal FVIII protein. The position of exons 17/18 within the A3 domain is noted. (F) Predicted FVIII polypeptide encoded by F8KO(16/19) mRNA. (G) Predicted FVIII polypeptide encoded by F8KO(Alt) mRNA. Flp recombinase target (FRT) sites are represented as open triangles, and Cre recombinase target (loxP) sites as gray triangles. The positions of duplex genotyping primers P1, P2, and P3 and RT-PCR primers P4 and P5 are shown. aa, amino acid; Ex, exon.
Crossbreeding with Cre-expressing strains to generate experimental and control animals
In Cre-recombinase–expressing cells, exons 17/18 are excised from the F8F allele, resulting in F8F→KO gene conversion (Figure 1C). Mice carrying the F8F allele were crossbred with tissue-specific Cre strains available from The Jackson Laboratory. The mouse models used in this study are summarized in Table 1. Meox2-Cre mice [B6.129S4-Meox2tm1(cre)Sor/J] ubiquitously express Cre in embryonic epiblast-derived tissues31 and were used to develop a new hemophilic F8KO strain. Alb-Cre [B6.Cg-Tg(Alb-cre)21Mgn/J] efficiently induces hepatocyte-specific recombination of floxed genes.32 Vav1-Cre [B6.Cg-Tg(Vav1-cre)A2Kio/J] reportedly induces recombination in virtually all hematopoietic cells.33 Variable off-target recombination in nonhematopoietic tissues has also been observed, up to and including whole-animal effects.34,35 Three endothelial-specific Cre models were used. Each of the 3 models induces recombination in both endothelial and hematopoietic cells, with varying levels of overall efficiency in the target cell population: Cdh5(Mlia)-Cre [B6.Cg-Tg(Cdh5-cre)7Mlia/J]36 is least efficient, Cdh5(Spe)-Cre [B6;129-Tg(Cdh5-cre)1Spe/J]37 is intermediate, and Tek-Cre [B6.Cg-Tg(Tek-cre)1Ywa/J] causes recombination of floxed alleles in virtually 100% of target cells.38
Cre recombinase mouse models
| Abbreviated name . | Full strain name . | Tissue specificity . |
|---|---|---|
| Meox2-Cre | B6.129S4-Meox2tm1(cre)Sor/J | Embryonic |
| Alb-Cre | B6.Cg-Tg(Alb-cre)21Mgn/J | Hepatocytes |
| Vav1-Cre | B6.Cg-Tg(Vav1-cre)A2Kio/J | Hematopoietic (and various other tissues) |
| Cdh5(Mlia)-Cre | B6.Cg-Tg(Cdh5-cre)7Mlia/J | Endothelial and hematopoietic |
| Cdh5(Spe)-Cre | B6;129-Tg(Cdh5-cre)1Spe/J | Endothelial and hematopoietic |
| Tek-Cre | B6.Cg-Tg(Tek-cre)1Ywa/J | Endothelial and hematopoietic |
| Abbreviated name . | Full strain name . | Tissue specificity . |
|---|---|---|
| Meox2-Cre | B6.129S4-Meox2tm1(cre)Sor/J | Embryonic |
| Alb-Cre | B6.Cg-Tg(Alb-cre)21Mgn/J | Hepatocytes |
| Vav1-Cre | B6.Cg-Tg(Vav1-cre)A2Kio/J | Hematopoietic (and various other tissues) |
| Cdh5(Mlia)-Cre | B6.Cg-Tg(Cdh5-cre)7Mlia/J | Endothelial and hematopoietic |
| Cdh5(Spe)-Cre | B6;129-Tg(Cdh5-cre)1Spe/J | Endothelial and hematopoietic |
| Tek-Cre | B6.Cg-Tg(Tek-cre)1Ywa/J | Endothelial and hematopoietic |
To develop each tissue-specific model, Cre-positive F8+/y males (hemizygous for the X-chromosomal F8 gene) were first mated with F8F/F females to generate F8F/y Cre+/− breeders. Matings with F8F/F females then generate approximately equal numbers of experimental Cre+/− and control Cre−/− offspring of both sexes (F8F/y and F8F/F). Except for the Meox2-Cre–derived F8KO strain, F8F→KO allele conversion occurs only in specific cell types as determined by inheritance of a tissue-specific Cre transgene.
Genotypic and phenotypic assessment of mice
Blood and tail-tip sample collection.
Following excision of the tail tip, blood was collected from >8-week-old mice into 1/10 volume of 4% sodium citrate (Jorgensen Laboratories, Loveland, CO). Following centrifugation for 5 minutes at 1200g, plasma was aspirated and recentrifuged for 10 min at 16 000g, and platelet-poor plasma was aliquoted and frozen at −80°C. White blood cell (WBC) pellets were prepared following red blood cell lysis using RBC Lysis Solution (Qiagen, Valencia CA). Tail tips and/or WBC pellets were frozen at −20°C for later genotyping analysis.
Genotyping.
Animals were genotyped by polymerase chain reaction (PCR) analysis of genomic DNA (gDNA) purified from frozen WBC pellets and/or tail tips using the QIAamp DNA Blood Mini Kit and DNeasy Blood & Tissue Kit (Qiagen), respectively. Floxed (F8F) and Cre-inactivated F8-KO (F8KO) alleles were identified using a duplex PCR reaction in which a common forward primer P1 located in exon 16 (5′-CGTCCCTACTCCTTCTATTCTAGCCTCATTTC-3′) amplifies allele-specific products in combination with 2 alternative reverse primers (Figure 1B-C). Reverse primer P2 (5′-TATCTCTTTCCTAGAGTTCCCAGGAGGG-3′), positioned across the intron 16/exon 17 junction, which is absent in the F8KO allele, amplifies a 712-bp F8F allele-specific product. The alternative reverse primer P3 (5′-CAAATCAGTCTTGACAGCTGC-3′), located downstream from the intron 18 loxP insertion, detects a 462 bp F8KO allele-specific amplicon. This duplex PCR provides direct assessment of F8F→KO gene recombination and was thus used for routine genotyping, with one exception. Such analysis is uninformative for Alb-Cre WBC and tail DNA due to hepatocyte-restricted recombination. Instead, inheritance of the Alb-Cre transgene was detected by amplification of a 401-bp product using primers Cre-s350 (5′-CGATGCAACGAGTGATGAGGTTC-3′) and Cre-a751 (5′-TCCATGAGTGAACGAACCTGGTC-3′).
Tissue gDNA and RNA analysis.
Following whole-animal saline perfusion, various tissue samples were harvested and submerged in RNAlater reagent (Qiagen) to stabilize nucleic acids. gDNA and total RNA were purified using the AllPrep DNA/RNA Mini Kit (Qiagen). The allele-specific duplex genotyping PCR described in the previous section was used to analyze tissue gDNA samples. Total RNA was reverse transcribed using Superscript III according to the manufacturer’s protocol (Life Technologies, Grand Island, NY) and then amplified using a PCR reaction spanning the loxP-flanked exon 17/18 deletion cassette. Primers P4 (5′-ATGGCTCCTTTAGCTAGCCCTT-3′) located in exon 15, and P5 (5′-AAAGAGTGCTCATCCCAGCCTGTA-3′) located in exon 19 amplify an 819-bp product from properly spliced F8 mRNA or a 407-bp product following exon 16/19 splicing as predicted for the F8KO allele (Figure 1D).
Plasma FVIII activity assay.
Plasma FVIII activity was measured using a modified chromogenic assay (Coatest SP4 FVIII Kit; Instrumentation Laboratory, Lexington, MA). Mouse plasmas were diluted in Coatest buffer and assayed against a standard curve consisting of known concentrations of B-domain–deleted human FVIII (Xyntha; Wyeth Pharmaceuticals, Collegeville, PA). A total of 25 µL of diluted samples and standards were added to duplicate wells of a blocked 96-well plate, and then a 75-µL mixture of factor IXa, factor X, phospholipid, and CaCl2 kit components was added, followed by incubation at 37°C for 10 minutes. Fifty μL of chromogenic factor Xa substrate mixture S-2765/I-2581 was then added, and Vmax (Absorbance405-490/min) was measured at 37°C using a ThermoMax microplate reader (Molecular Devices, Sunnyvale, CA). The instrument manufacturer’s software converted Vmax to units of FVIII activity.
BMT
To further investigate the contribution of hematopoietic cells to plasma FVIII, bone marrow from WT C57BL/6 mice was transplanted into FVIIInull mice (the exon 17–disrupted strain of Bi et al),39,40 as described in our previous reports.41,42 Briefly, unfractionated donor bone marrow nucleated cells (BMNCs) or bone marrow mononuclear cells (BMMCs) were isolated. For BMNC isolation, red blood cells were eliminated using RBC lysing buffer (Sigma, St. Louis, MO). For BMMC isolation, cells were fractionated using Fico/Lite-LM (mouse) (Atlanta Biologicals, Lawrenceville, GA). FVIIInull recipient mice (on a C57BL/6 background) were conditioned for cellular transplantation with an 1100 cGy lethal total body irradiation dose, and approximately 1 × 107 donor cells were transplanted by intravenous infusion 24 hours later. Blood was collected 4 and 14 weeks later for plasma FVIII assays. At 15 weeks, a tail-clip survival test41 was used to assess phenotypic correction of the FVIIInull coagulation defect.
Statistical analysis
Plasma FVIII activity assays were assessed using a 2-tailed t test, and BMT experiments were evaluated using the Fisher’s exact test. Values are expressed as mean ± standard deviation, and significance was defined as P ≤ .05.
All animal procedures were performed according to guidelines approved by the Institutional Animal Care and Use Committee of the Medical College of Wisconsin.
Results
Genotyping
A phenotypically normal conditional F8-KO mouse model (F8F) was developed. Embryonic Meox2-Cre was used to develop a new whole-animal F8-KO strain (F8KO), whereas various tissue-specific Cre models differentially cause F8F→KO gene inactivation only in specific cell types (Table 1). A duplex F8 PCR of WBCs and/or tail DNA that provides direct assessment of gene rearrangement at the floxed exon 17/18 locus was used for genotyping whenever possible. Because gene inactivation is hepatocyte restricted in the Alb-Cre model (and liver biopsies are impractical for routine genotyping), a Cre-specific PCR had to suffice for identification of experimental (Alb-Cre+/−) and control (Alb-Cre−/−) animals (Figure 2). Postmortem tests confirmed liver-specific F8F→KO gene conversion in Alb-Cre+/− mice.
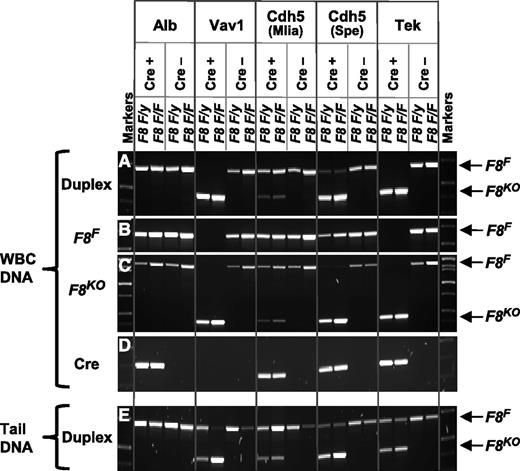
Genotyping PCRs. WBCs or tail DNA PCR was used for routine genotyping. A Cre-positive (experimental) and Cre-negative (control) male (F8 F/y) and female (F8 F/F) are shown for each tissue-specific Cre model. (A) Duplex PCR of WBC DNA using a common forward primer, P1, in combination with 2 alternative reverse primers, P2 and P3. As depicted in Figure 1B-C, a 712-bp product is amplified from the F8F allele and a 462,bp product from the F8KO allele. (B) Singleplex PCR of WBC DNA using primers P1 and P2 amplifies only the 712-bp F8F allele-specific product. (C) Singleplex PCR of WBC DNA using primers P1 and P3 amplifies a 462 bp F8KO allele-specific product but can also amplify a 1683-bp product from the F8F allele. (D) A Cre-recombinase coding sequence–specific PCR of WBC DNA identifies Cre+ mice. No signal is seen for the Vav1-Cre transgene because it contains a codon-optimized iCre sequence, which is not recognized by the native Cre primers we used. (E) Duplex PCR analysis of tail DNA using primers P1, P2, and P3 detects both F8F and F8KO alleles, especially important for Vav1- and Tek-Cre mice, in which WBC DNA undergoes complete F8F→KO gene conversion.
Genotyping PCRs. WBCs or tail DNA PCR was used for routine genotyping. A Cre-positive (experimental) and Cre-negative (control) male (F8 F/y) and female (F8 F/F) are shown for each tissue-specific Cre model. (A) Duplex PCR of WBC DNA using a common forward primer, P1, in combination with 2 alternative reverse primers, P2 and P3. As depicted in Figure 1B-C, a 712-bp product is amplified from the F8F allele and a 462,bp product from the F8KO allele. (B) Singleplex PCR of WBC DNA using primers P1 and P2 amplifies only the 712-bp F8F allele-specific product. (C) Singleplex PCR of WBC DNA using primers P1 and P3 amplifies a 462 bp F8KO allele-specific product but can also amplify a 1683-bp product from the F8F allele. (D) A Cre-recombinase coding sequence–specific PCR of WBC DNA identifies Cre+ mice. No signal is seen for the Vav1-Cre transgene because it contains a codon-optimized iCre sequence, which is not recognized by the native Cre primers we used. (E) Duplex PCR analysis of tail DNA using primers P1, P2, and P3 detects both F8F and F8KO alleles, especially important for Vav1- and Tek-Cre mice, in which WBC DNA undergoes complete F8F→KO gene conversion.
Both F8F and F8KO alleles can be detected in WBCs from Cdh5(Mlia)-Cre+/− and most Cdh5(Spe)-Cre+/− mice. When only the F8KO allele was observed in WBC DNA, as expected for the Vav1-Cre and Tek-Cre models, tail-tip DNA was routinely analyzed to insure against unintended germline transmission and as an indicator of excessive, non–tissue-specific Cre activity.33-35,43 Unexpectedly, variable amounts of residual F8F DNA were present in some Vav1-Cre WBC samples (data not shown), indicative of less than the expected 100% efficiency in the target hematopoietic cell population. Conversely, some Vav1-Cre tail samples contained only F8KO DNA, an indicator of off-target nonhematopoietic effects.
Individual (rather than duplex) PCR provides higher sensitivity, allowing confirmation that both F8F and F8KO alleles are present in a sample, such as when duplex reactions yield only a F8KO product [eg, WBCs from some Cdh5(Spe)-Cre+/− animals]. In addition, although duplex PCR of WBC DNA clearly shows that Cdh5(Spe)-Cre is more efficient than Cdh5(Mlia)-Cre, the relative efficiency of Tek-Cre is less clear. It becomes obvious from singleplex F8F-specific PCR results that Tek-Cre is in fact virtually 100% efficient in WBCs, whereas a significant amount of unrecombined F8F DNA remains for the less efficient Cdh5(Spe)-Cre (Figure 2).
Plasma FVIII levels
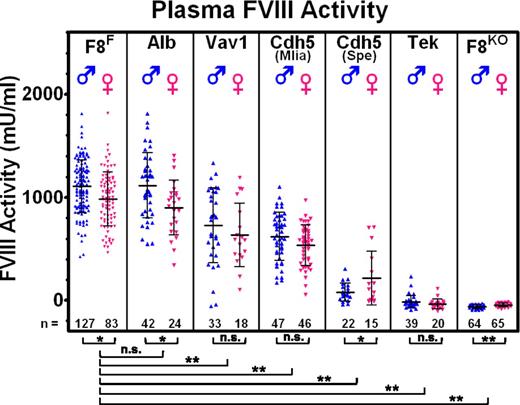
As shown in Figure 3 and as previously reported,44 within a given genotype, plasma FVIII levels tend to be higher in males. FVIII:C levels in F8F Cre−/− controls are 1106 ± 254 mU/mL for males (M) and 983 ± 264 mU/mL for females (F), essentially identical to F8F mice maintained without Cre crossbreeding (not shown in Figure 3), which have 1153 ± 323 mU/mL (M) and 1004 ± 288 mU/mL (F). Alb-Cre mice are also indistinguishable from controls with FVIII:C at 1116 ± 314 mU/mL (M) and 900 ± 266 mU/mL (F). A moderate but significant reduction of FVIII:C is caused by Vav1-Cre, at 728 ± 362 mU/mL (M) and 636 ± 305 mU/mL (F), with a few animals displaying atypically low FVIII levels. Similar modestly reduced levels were observed for Cdh5(Mlia)-Cre, with 621 ± 233 mU/mL (M) and 537 ± 196 mU/mL (F). Cdh5(Spe)-Cre causes a more severe reduction in FVIII levels with 79 ± 87 mU/mL (M) and 216 ± 265 mU/mL (F), which includes a few conspicuously higher-FVIII females. Tek-Cre typically produces a severe hemophilic phenotype, and only rare individual animals have (barely) detectable plasma FVIII activity, whereas F8KO mice uniformly lack detectable FVIII.
Plasma FVIII activity. A chromogenic assay was used to measure FVIII activity in plasma from tail bleeds. Males and females are shown separately, revealing an apparent sex difference. F8F represents Cre−/− control mice that result from breeding F8F/y Cre+/− males of the various Cre stains with F8F/F females. Alb, Vav1, Cdh5(Mlia), Cdh5(Spe), and Tek represent experimental Cre+/− littermates. F8KO is the stably inherited exon 17/18-deleted strain. Hepatocyte-specific Alb-Cre has no effect on FVIII levels, whereas all endothelial Cre models result in reduced plasma FVIII, culminating with a severe hemophilic phenotype in the most efficient Tek-Cre model. *P ≤ .05, **P ≤ .001. n.s., nonsignificant.
Plasma FVIII activity. A chromogenic assay was used to measure FVIII activity in plasma from tail bleeds. Males and females are shown separately, revealing an apparent sex difference. F8F represents Cre−/− control mice that result from breeding F8F/y Cre+/− males of the various Cre stains with F8F/F females. Alb, Vav1, Cdh5(Mlia), Cdh5(Spe), and Tek represent experimental Cre+/− littermates. F8KO is the stably inherited exon 17/18-deleted strain. Hepatocyte-specific Alb-Cre has no effect on FVIII levels, whereas all endothelial Cre models result in reduced plasma FVIII, culminating with a severe hemophilic phenotype in the most efficient Tek-Cre model. *P ≤ .05, **P ≤ .001. n.s., nonsignificant.
Tissue DNA and RNA analysis
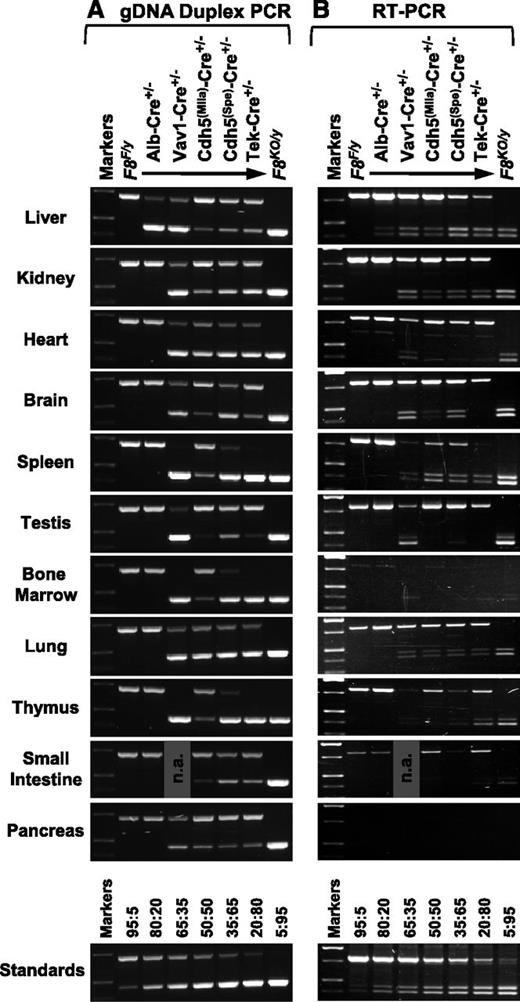
DNA and RNA from various tissues were analyzed by PCR (Figure 4). Only the F8F allele was present in F8F controls and exclusively the F8KO allele in F8KO mice. Reverse-transcription polymerase chain reaction (RT-PCR) analysis revealed widespread distribution of F8 mRNA similar to previous reports,10 with detectable message in all tissues tested except pancreas and only an extremely faint signal in bone marrow. For F8F, only the expected 819-bp RT-PCR product was present, representing normally spliced F8 message. Exon 16/19 mRNA splicing of the F8KO allele (F8KO(16/19)) yields the predicted 407-bp amplicon, but a 453-bp product of similar intensity is also amplified. Sequence analysis showed that this represents an alternatively spliced transcript (F8KO(Alt)), with retention of 46 bases of intron 16 sequence utilizing the same cryptic splice site observed in the hemophilic exon 17–disrupted FVIIInull model of Bi et al.40
Analysis of tissue gDNA and mRNA. Tissue samples were collected from males following whole-animal saline perfusion. DNA and total RNA were isolated from the same tissue sample and analyzed by allele-specific PCR. Except for the F8KO/y mouse, all animals inherited a F8F/y genotype, either without Cre or with coinheritance of a Cre transgene as indicated. (A) A total of 25 ng of gDNA was amplified using a duplex PCR that yields a 712-bp F8F allele-specific product and a 462-bp F8KO allele-specific product. Primer P1, P2, and P3 binding sites are shown in Figure 1B-C. (B) For RT-PCR, 100 ng of total RNA was reverse transcribed and then analyzed by PCR using primers P4 and P5, which span the exon 17/18 deletion cassette (as shown in Figure 1D). An 819-bp product is amplified from the F8F allele, representing normally spliced F8 mRNA. For the F8KO allele, in addition to the 407-bp product predicted for exon 16/19 mRNA splicing (F8KO(16/19)), a 453-bp product is also present due to alternative splicing (F8KO(Alt)). The “Standards” panels consist of PCR products amplified from defined mixtures of F8F/y and F8KO/y liver gDNA (panel A) or total RNA (panel B) at various ratios as indicated (F8F:F8KO). n.a. indicates a sample that was not analyzed.
Analysis of tissue gDNA and mRNA. Tissue samples were collected from males following whole-animal saline perfusion. DNA and total RNA were isolated from the same tissue sample and analyzed by allele-specific PCR. Except for the F8KO/y mouse, all animals inherited a F8F/y genotype, either without Cre or with coinheritance of a Cre transgene as indicated. (A) A total of 25 ng of gDNA was amplified using a duplex PCR that yields a 712-bp F8F allele-specific product and a 462-bp F8KO allele-specific product. Primer P1, P2, and P3 binding sites are shown in Figure 1B-C. (B) For RT-PCR, 100 ng of total RNA was reverse transcribed and then analyzed by PCR using primers P4 and P5, which span the exon 17/18 deletion cassette (as shown in Figure 1D). An 819-bp product is amplified from the F8F allele, representing normally spliced F8 mRNA. For the F8KO allele, in addition to the 407-bp product predicted for exon 16/19 mRNA splicing (F8KO(16/19)), a 453-bp product is also present due to alternative splicing (F8KO(Alt)). The “Standards” panels consist of PCR products amplified from defined mixtures of F8F/y and F8KO/y liver gDNA (panel A) or total RNA (panel B) at various ratios as indicated (F8F:F8KO). n.a. indicates a sample that was not analyzed.
Total F8KO allele-derived message appears to be reduced relative to the F8F allele, possibly indicating transcriptional disregulation and/or accelerated decay of defective transcripts.45 F8KO(16/19) mRNA potentially encodes a FVIII polypeptide truncated within the A3 domain (Figure 1F), whereas the intronic sequence incorporated into alternatively spliced F8KO(Alt) message is predicted to result in 15 miscoded amino acids at the exon 17/18 deletion site, followed by restoration of the normal F8 reading frame at the exon 19 splice junction (Figure 1G). We attempted to detect these truncated polypeptides using commercially available antibodies but encountered specificity issues. We had previously found that some “anti-FVIII” antibodies instead recognize von Willebrand factor (previously known as FVIII-related antigen),46 and our recent experience indicates that caution is still warranted.47 Thus, due to a paucity of reliable anti-mouse FVIII antibodies, we currently have no data regarding translation of F8KO gene products, but it is clear from plasma FVIII assays that no functional FVIII is produced.
As expected for the hepatocyte-specific Alb-Cre model, the F8KO allele was detected only in liver DNA. In spite of efficient F8F→KO gene conversion in hepatocytes, almost exclusively normal F8 mRNA is present, indicating that the Cre-negative nonhepatocyte cell fraction continues to express F8 normally, which is consistent with the normal plasma FVIII levels observed in these mice.
For the other Cre models, gene conversion was more widely distributed. For many Vav1-Cre mice, virtually complete hematopoietic gene inactivation was observed in WBCs as expected, but variable amounts of F8F DNA remained in some animals. Near-complete F8F→KO conversion in tail tips and other nonhematopoietic organs suggests that significant loss of hematopoietic specificity can also occur, as previously reported.33-35 In spite of potentially exaggerated effects due to off-target Cre activity, plasma FVIII levels in Vav1-Cre mice are only modestly reduced.
For the 3 endothelial models, increasing Cre efficiency is correlated with decreasing plasma FVIII levels, indicating that FVIII-producing cells are being successfully targeted. For Cdh5(Mlia)-Cre, some level of gene conversion occurred in all sampled tissues, but normal F8 mRNA predominates. Cdh5(Spe)-Cre drives near-complete F8F→KO conversion in hematopoietic tissues (eg, WBCs, spleen, bone marrow, and thymus), where Tek-Cre appears to be 100% efficient, and both models generate a similar mixture of F8F and F8KO alleles in other tissues, including liver (Figures 2 and 4A). The gradations of efficiency among endothelial models are accompanied by shifting proportions of F8KO vs F8F mRNA, consistent with observed plasma FVIII levels. It is intriguing that normal F8 message persists in numerous tissues of functionally hemophilic Tek-Cre mice that are devoid of plasma FVIII activity.
WT to FVIIInull BMT
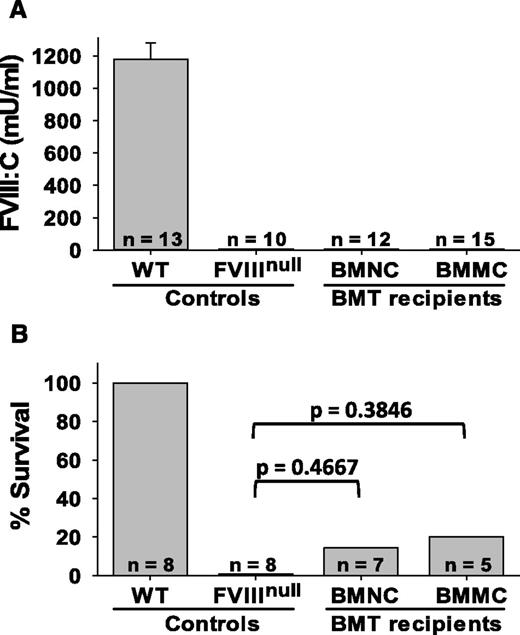
As an additional test of hematopoietic contribution to FVIII synthesis, bone marrow was transplanted from WT mice into FVIIInull recipients. Plasma FVIII was present at 1179 ± 102 mU/mL in WT controls but was undetectable in FVIIInull mice and in all BMT recipients (Figure 5A). While 1 of 7 BMNC recipients and 1 of 5 BMMC recipients survived tail-clipping, neither group is significantly different from FVIIInull controls (Figure 5B).
Transplantation of WT bone marrow into FVIIInull mice. Unfractionated C57BL/6 BMNCs or BMMCs were transplanted into lethally irradiated FVIIInull recipients. (A) Plasma FVIII levels in BMT recipients. Tail blood samples were collected, and FVIII levels were determined by chromogenic assay. (B) Tail-clip survival test. The tail clipping test was performed at 15 weeks after BMT.
Transplantation of WT bone marrow into FVIIInull mice. Unfractionated C57BL/6 BMNCs or BMMCs were transplanted into lethally irradiated FVIIInull recipients. (A) Plasma FVIII levels in BMT recipients. Tail blood samples were collected, and FVIII levels were determined by chromogenic assay. (B) Tail-clip survival test. The tail clipping test was performed at 15 weeks after BMT.
Discussion
While a number of different technologies have been enlisted to identify the specific cells responsible for FVIII synthesis both within the liver and elsewhere, the issue has yet to be definitively resolved. Rather than retrospectively attempting to identify and localize expressed FVIII, we developed a conditional F8-KO model allowing us to preempt FVIII expression in specific cell types. F8F mice express normal levels of plasma FVIII, but when crossbred with Cre-recombinase–expressing mouse strains, F8F→KO gene inactivation occurs in a tissue-specific pattern dependent upon the promoter driving Cre expression. If Cre disrupts the F8 gene in cells responsible for FVIII production, then plasma FVIII levels will be decreased, and if all FVIII-producing cells are affected, plasma FVIII will be absent, resulting in a severe hemophilia phenotype. Conversely, knocking out F8 in cells that do not normally contribute to steady-state plasma FVIII will have no effect.
The pattern of conditional gene inactivation by Cre is largely dependent on the tissue specificity of the model used and its efficiency within the target cell population. Even generally efficient Cre models sometimes produce variegated gene disruption in their target tissues. For example, mosaic F8F/KO/y and F8+/F/KO offspring were not unusual during development of the F8KO strain, indicating that even “ubiquitous” models such as Meox2-Cre sometimes miss cells. Incomplete target-tissue–specific gene conversion in individual animals is thus not unexpected in conditional KO experiments. Conversely, some Cre models induce off-target gene conversion, causing artifactually inflated effects if cells producing the protein of interest are inappropriately affected. These effects can vary significantly, even among littermates, necessitating careful interpretation of conditional KO experiments. The Jax Cre Resource (http://cre.jax.org/data.html) is an expanding repository of information regarding the characteristics of Cre models,35 including some that were used in this study.
Alb-Cre reportedly has the highest fidelity of the tissue-specific models used,32,35 and the pattern of liver-restricted F8F→KO gene conversion we observed is consistent with efficient hepatocyte-specific recombination. If hepatocytes were responsible for FVIII production, then a major effect on plasma levels would be virtually assured, but instead Alb-Cre mice are indistinguishable from controls. Plasma FVIII levels are completely normal, and virtually all F8 message in the liver (and elsewhere) is normal in Alb-Cre mice, derived from Cre-negative (nonhepatocyte) cell types expressing the unrecombined F8F allele. The absence of any phenotypic effect in Alb-Cre mice shows that hepatocytes do not synthesize FVIII and that some other cell type bears that responsibility.
In contrast, each of the endothelial-specific Cre models results in reduced plasma FVIII levels, a clear indication of gene inactivation in FVIII-producing cells. These models are believed to operate with similar efficiency in both endothelial and hematopoietic cells, presumably due to differentiation of both lineages from a shared progenitor.48 There are differences in performance among endothelial models, some of which may be due to onset of Cre expression during subtly different time frames approximating the embryonic endothelial/hematopoietic transition.49 Overall efficiency increases in the order Cdh5(Mlia)-Cre < < Cdh5(Spe)-Cre < Tek-Cre, which parallels observed F8-inactivating activity; plasma FVIII activity is moderately reduced in Cdh5(Mlia)-Cre mice, severely reduced in Cdh5(Spe)-Cre mice, and undetectable in most Tek-Cre mice.
The detectable plasma FVIII observed in a small minority of Tek-Cre mice is presumably due to failed recombination in a small population of target cells in those individual animals. As discussed above, many Cre models are imperfect. The 4 female Cdh5(Spe)-Cre outliers with markedly higher-than-average FVIII levels may be a more extreme example of variegated Cre effects within the target cell population in individual mice. That only females are affected suggests a target-gene dosage effect, since 2 recombination events are required for complete F8F→KO/ F→KO gene conversion in female cells, whereas only a single hit is needed to induce F8F→KO/y gene neutralization in males. The involvement of such effects is purely hypothetical at this point, but these observations provide potential avenues for future investigation.
Because each of the “endothelial” models employed actually produces endothelial plus hematopoietic effects, Vav1-Cre, an ostensibly hematopoietic-specific model, was used as a control. As expected, in most animals, Vav1-Cre causes F8F→KO gene conversion in virtually 100% of WBCs. If FVIII synthesis were a hematopoietic function, then that capacity should be effectively eliminated, but plasma FVIII levels were only modestly reduced in Vav1-Cre mice. Interpretation is complicated by the presence of off-target Vav1-Cre effects, evidenced by higher-than-expected levels of gene recombination in tail tips and other nonhematopoietic tissues. Despite anticipation that this would skew results toward overestimation of hematopoietic effects, the decrease in FVIII was unremarkable. The presence of a few, predominantly male, low-FVIII Vav1-Cre outliers again hints at a target-gene dosage effect, since 1 off-target event is sufficient to inappropriately neutralize a FVIII-producing F8F/y cell, whereas a singly hit F8F→KO /F female cell will continue producing FVIII.
As an alternative approach to the question of hematopoietic involvement in FVIII production, we performed a series of BMT experiments, transferring bone marrow cells from WT mice into FVIIInull recipients. No plasma FVIII was detected in recipients, and the typical FVIIInull lethality following tail clipping was statistically unchanged, indicating that hematopoietic cells do not contribute to the production of plasma FVIII. This further supports the idea that the modest reduction in plasma FVIII levels observed in Vav1-Cre mice is caused by variable off-target effects in FVIII-producing ECs, rather than being due to the generally quite efficient hematopoietic gene disruption that occurs in these mice.
Our conclusion that ECs, which are widely distributed throughout the body, are responsible for FVIII synthesis is fully compatible with comprehensive organ surveys that find F8 mRNA in almost every tissue tested.10 It is interesting that a significant amount of normal F8 mRNA persists in our Tek-Cre mice, which lack detectable plasma FVIII activity. This implies that transcriptional “expression” of F8 does not always culminate in production of plasma FVIII. Possible explanations include failed recombination in a subset of ECs that either continue to produce FVIII at extremely low levels or that produce, but fail to secrete FVIII. Given the high efficiency of the Tek-Cre model, it seems more likely that nonendothelial tissues are involved. Unproductive or “leaky” transcription has been demonstrated in several cell types that are presumed to have no genuine involvement in FVIII protein production,50 providing an alternative explanation. Whatever the source of the remaining normal but apparently unconsummated F8 mRNA, the phenomenon almost certainly occurs in normal animals, becoming apparent only after the production of plasma FVIII is eliminated by Tek-Cre. In the future, we are hopeful that we can determine the source of the residual normal F8 message in hemophilic Tek-Cre mice.
As the body’s main FVIII-producing organ, it is clear that at least one of the cell types present in liver synthesizes FVIII. Previous studies suggest hepatocytes, LSECs, and Kupffer cells as candidates. The Alb-Cre model rules out synthesis by hepatocytes, whereas all endothelial Cre models cause reduced FVIII levels, with a severe hemophilic phenotype in the most efficient model. Our results also indicate that hematopoietic cells do not contribute to the production of plasma FVIII.
Given their importance to the structural and hemostatic maintenance of vascular integrity, a widely distributed network of FVIII-equipped ECs has obvious logistical and mechanistic appeal. Endothelial-restricted expression is compatible with observations of both hepatic and anatomically widespread extrahepatic FVIII expression. It is well established that von Willebrand factor, the protective carrier of FVIII in plasma, is produced and stored in ECs, and coexpression within the same cell provides a compelling explanation for the concurrent release of both proteins that occurs in vivo upon agonist stimulation.3,51,52 We have shown that FVIII synthesis is primarily, and potentially exclusively, an endothelial function. Further characterization of this new conditional F8-KO model is ongoing, directed toward a better understanding of various cell types’ role in FVIII biosynthesis.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Nancy Speck kindly arranged for the transfer of Cdh5(Spe)-Cre mice to our laboratory prior to their availability through The Jackson Laboratory. The authors thank Lesley Everett, Audrey Cleuren, Rami Khoriaty, and David Ginsburg for their helpful critique of this manuscript and for sharing the results of their complementary studies prior to publication.
This work was supported by National Institutes of Health, National Heart, Lung and Blood Institute grants 5P01HL044612 (R.R.M.), 5P01HL081588 (R.R.M.), and R01HL102035 (Q.S.).
Authorship
Contribution: S.A.F. designed and performed research and drafted the manuscript; M.T.H. performed research and assisted in editing the manuscript; Q.S. designed and performed research and assisted in editing the manuscript; H.W. assisted in the design of experiments; R.R.M. designed and supervised research and assisted in writing and editing the manuscript; and all authors participated in critical analysis of research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Robert R. Montgomery, Blood Research Institute, BloodCenter of Wisconsin, PO Box 2178, Milwaukee, WI 53201-2178; e-mail: bob.montgomery@bcw.edu.

![Figure 1. Conditional F8-KO alleles. (A) Targeted F8 gene. (B) F8F allele. Excision of the neomycin resistance cassette by Flp recombinase produces the floxed (F8F) allele, which is expressed normally. (C) F8KO allele. Excision of exons 17/18 from the F8F allele by Cre recombinase produces a F8KO allele. (D) Alternative mRNA splicing of the F8KO allele. The predicted exon 16/19 splice [F8KO(16/19)] and an alternatively spliced transcript [F8KO(Alt)] in which 46 bp of intron 16 is retained are produced in approximately equal amounts. (E) Domain structure of normal FVIII protein. The position of exons 17/18 within the A3 domain is noted. (F) Predicted FVIII polypeptide encoded by F8KO(16/19) mRNA. (G) Predicted FVIII polypeptide encoded by F8KO(Alt) mRNA. Flp recombinase target (FRT) sites are represented as open triangles, and Cre recombinase target (loxP) sites as gray triangles. The positions of duplex genotyping primers P1, P2, and P3 and RT-PCR primers P4 and P5 are shown. aa, amino acid; Ex, exon.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/24/10.1182_blood-2014-02-555151/4/m_3706f1.jpeg?Expires=1769080600&Signature=hMnMDqlc1OTa6BU8qVBzKRyc5hQDjgx~wXmw1VEaxnQoAt6hWMe1dFzKaXThUy~HKiDtxjhNAPFxzUIp83O3YwRGBGp91kcXz5jF7jiTrdkrB967soLF6~q1Y1FG38tZ5q2LIXBTOfpZLkAyCRmIxDRAyWmY96mDCOI6M3NBGJXq7IXcS2BcSlMahGZRQsqFtbRXJCFXg7~MpvobcwqzPf513zTYTJpGZvTWsXCynayN9RdlBARXGPw27qzN7zr4hmq62utZGCTz5ak34XbN9vmbPQc~6latLD5RNGVDGRP1F1hU7aNjhVVYy6uFgQp3vOxegTyMF1J0vSZ8exzvBw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)