Key Points
The level of Sf3b1 expression is critical for the proliferative capacity of hematopoietic stem cells.
Haploinsufficiency for Sf3b1 is not sufficient to induce a RARS-like phenotype in mice.
Abstract
Numerous studies have recently reported mutations involving multiple components of the messenger RNA (mRNA) splicing machinery in patients with myelodysplastic syndrome (MDS). SF3B1 is mutated in 70% to 85% of refractory anemia with ringed sideroblasts (RARS) patients and is highly associated with the presence of RARS, although the pathological role of SF3B1 mutations in MDS-RARS has not been elucidated yet. Here, we analyzed the function of pre-mRNA splicing factor Sf3b1 in hematopoiesis. Sf3b1+/− mice maintained almost normal hematopoiesis and did not develop hematological malignancies during a long observation period. However, Sf3b1+/− cells had a significantly impaired capacity to reconstitute hematopoiesis in a competitive setting and exhibited some enhancement of apoptosis, but they did not show any obvious defects in differentiation. Additional depletion of Sf3b1 with shRNA in Sf3b1+/− hematopoietic stem cells (HSCs) severely compromised their proliferative capacity both in vitro and in vivo. Finally, we unexpectedly found no changes in the frequencies of sideroblasts in either Sf3b1+/− erythroblasts or cultured Sf3b1+/− erythroblasts expressing shRNA against Sf3b1. Our findings indicate that the level of Sf3b1 expression is critical for the proliferative capacity of HSCs, but the haploinsufficiency for Sf3b1 is not sufficient to induce a RARS-like phenotype.
Introduction
Alternative pre-messenger RNA (mRNA) splicing is a key regulator of biological diversity and the normal gene expression in higher eukaryotes.1 More than 90% of multiple-exon genes undergo multiple alternative splicing events within the same transcript.2,3 Nuclear pre-mRNA splicing is catalyzed by the spliceosome, a multi-megadalton ribonucleoprotein (RNP) complex comprised of 5 small nuclear (sn)RNPs and numerous proteins. SF3B1 is a core component of the splicing machinery, forming the U2 snRNP together with the SF3A complex and the U2 small nuclear RNA. The complete U2 snRNP recognizes the 3′ splice site at intron-exon junctions.4,5 Of note, recent next-generation sequencing studies have identified several mutations involving multiple components of the mRNA splicing machinery, including SF3B1, SRSF2, U2AF1, ZRSR2, PRPF40B, U2AF65, and SF1 in myelodysplastic syndrome (MDS) patients.6-8 Among these MDS patients, SF3B1 is one of the most frequently mutated genes, and mutations in SF3B1 have been found in ∼80% of patients with refractory anemia with ringed sideroblasts (RARS) and RARS with thrombocytosis.6 In addition, SF3B1 mutations have also been identified in 15% of patients with chronic lymphocytic leukemia9 and at lower frequencies in patients with solid tumors such as melanoma and breast cancer.10,11
MDS is a heterogeneous group of myeloid malignancies characterized by impaired hematopoiesis with dysplastic blood cells and a predisposition to the development of acute myeloid leukemia.12,13 Various mutations have recently been identified in patients with MDS.14,15 Among these, MDS patients harboring SF3B1 mutations tend to have mild cytopenia and longer survival than patients without the SF3B1 mutation.15 However, it remains unknown how deregulation of SF3B1 is involved in the development of RARS. SF3B1 mutations are confined to a small region corresponding to exons 14 and 15, where no homozygous mutations have been reported. The presence of hot spots and the absence of nonsense or frameshift changes suggest that SF3B1 mutations are associated with some neomorphic function rather than loss of function.6,8 Therefore, it is assumed that depletion of Sf3b1 alone does not recapitulate the pathophysiological consequences of SF3B1 mutations in MDS cells. To address this question, we analyzed the function of Sf3b1 in hematopoiesis utilizing Sf3b1 heterozygous mice, because Sf3b1 null homozygote mice die at around the 16- to 32-cell stage during preimplantation development.16 Sf3b1 heterozygous mice develop normally except for mild skeletal homeotic changes due to deregulated Hox gene expression.16
We found that haploinsufficiency for Sf3b1 significantly impairs the repopulating capacity of hematopoietic stem cells (HSCs) in vivo, but does not induce MDS-like phenotypes such as the appearance of RARs. Additional depletion of Sf3b1 by RNA interference further compromised the proliferative capacity of Sf3b1+/− HSCs both in vitro and in vivo. Our findings show that the level of Sf3b1 expression is critical for the proliferative capacity of HSCs and that the partial loss of Sf3b1 is not sufficient to develop MDS in vivo.
Materials and Methods
Mice
Sf3b1 heterozygous mice were generated as previously described.16 C57BL/6 mice congenic for the Ly5 locus (CD45.1) were purchased from Sankyo-Laboratory Service. All experiments using mice were performed in accordance with our institutional guidelines for the use of laboratory animals and approved by the Review Board for Animal Experiments of Chiba University (approval ID: 25-104).
Flow cytometry and antibodies
Monoclonal antibodies (mAbs) recognizing the following antigens were used in flow cytometry and cell sorting: CD45.2 (104), CD45.1 (A20), Gr-1 (RB6-8C5), CD11b/Mac-1 (M1/70), Ter-119, CD127/IL-7Rα (A7R34), B220 (RA3-6B2), CD4 (L3T4), CD8α (53-6.7), CD43 (S7), IgM (1B4B1), CD117/c-Kit (2B8), Sca-1 (D7), and CD16/32/FcγRII-III (93). The mAbs were purchased from BD BioSciences (San Jose, CA), eBioScience (San Diego, CA), or BioLegend (San Diego, CA). Dead cells were eliminated by staining with propidium iodide (1 µg/mL, Sigma). All flow cytometric analyses and cell sorting were performed on a JSAN (Bay Bioscience, Kobe, Japan), FACSAria II, or FACSCanto II (BD Biosciences).
Purification of hematopoietic cells
Hematopoietic cells were harvested from long bones that were triturated and passed through 40-µm nylon mesh to obtain a single cell suspension. Mononuclear cells (MNCs) were isolated on Ficoll-Paque PLUS (GE Healthcare, Buckinghamshire, UK) and incubated with a mixture of biotin-conjugated mAbs against lineage markers, including Gr-1, Mac-1, Ter-119, B220, IL-7Rα, CD4, and CD8α. The cells were further stained with allophycocyanin (APC)-Cy7-conjugated streptavidin and a combination of mAbs, including fluorescein isothiocyanate-conjugated anti-CD34, phycoerythrin (PE)- or PE-Cy7-Sca-1, PE-FcγR, and APC-c-Kit. Pacific Blue-CD45.2 mAb was used as an additional marker for donor-derived cells in the bone marrow (BM) of B6-CD45.1 recipient mice. HSC (Lin–CD34–/lowc-Kit+Sca-1+), multipotent progenitor (MPP) (Lin–CD34+c-Kit+Sca-1+), common myeloid progenitor (Lin–Sca-1–c-Kit+CD34+FcγRlow), granulocyte/macrophage progenitor (Lin–Sca-1–c-Kit+CD34+FcγRhi), and megakaryocyte/erythroid progenitor (MEP) (Lin–Sca-1–c-Kit+CD34–FcγRlow) fractions were defined as previously described.17,18
BM transplantation
BM cells (2 × 106) from CD45.2 mutant mice (test cells) were transplanted intravenously into 8-week-old CD45.1 recipients irradiated at a dose of 9.5 Gy with or without the same number of BM cells from 8-week-old CD45.1 congenic mice (competitor cells). At 3 months after transplantation, serial transplants were carried out by transferring 2 × 106 BM cells to secondary recipients. The chimerism of donor-derived hematopoiesis was monitored monthly by flow cytometry. Peripheral blood (PB) cells were stained with a mixture of mAbs that included PE-anti-Gr-1, PE-anti-Mac-1, APC-anti-B220, APC-Cy7-anti-CD4, APC-Cy7-anti-CD8α, fluorescein isothiocyanate-CD45.2, and PE-Cy7-anti-CD45.1. The proportion of donor cells was evaluated by dividing the number of CD45.2-single positive cells by the total number of CD45-positive cells (CD45.1 + CD45.2).
Homing assay
Wild-type (WT) and Sf3b1+/− BM MNCs (106 cells) were labeled with 5 μM carboxyfluorescein diacetate, succinimidyl ester (CFSE, Molecular Plobes) in phosphate-buffered saline at 37°C for 10 minutes. The cells were then transplanted via tail vein. After 8 hours, recipient BM cells were isolated and analyzed for CFSE+ cells with a FACSCanto II (Beckton Dickinson).
Knockdown of Sf3b1
For knockdown of Sf3b1, pCS-H1-shRNA-EF-1-EGFP was used.19 Target sequences were as follows; Sh-Sf3b1#525; GAGCTAAAGCTGGAGAACTA and Sh-Sf3b1#1282; GCTCGAAAGCTGACAGCAA. CD34-KSL HSCs were transduced with the indicated lentivirus as previously described with minor modifications.20 CD34-KSL HSCs were sorted into 96-well microtiter plates coated with the recombinant human fibronectin fragment CH-296 (RetroNectin; Takara Shuzo) at 100 cells per well and then incubated in α- minimum essential medium supplemented with 1% fetal bovine serum (FBS), 1% l-glutamine, penicillin, streptomycin solution (GPS; Sigma-Aldrich), 50 μM 2-mercaptoethanol (2-ME), 100 ng/mL mouse stem cell factor (SCF; PeproTech), and 100 ng/mL human thrombopoietin (TPO; PeproTech) for 24 hours. Next, cells were transduced with the indicated virus at a multiplicity of infection of 1500 in the presence of 1 μg/mL RetroNectin and 10 μg/mL protamine sulfate (Sigma-Aldrich) for 24 hours. After transduction, cells were further incubated in S-Clone SF-O3 (Sanko Junyaku) supplemented with 0.2% bovine serum albumin, 50 μM 2-ME, 1% GPS, 50 ng/mL SCF, and 50 ng/mL TPO for 2.5 days, and then subjected to competitive repopulation assays. For in vitro proliferation assay, infected cells were further incubated in S-Clone SF-O3 supplemented with 0.2% bovine serum albumin, 50 μM 2-ME, 1% GPS, 50 ng/mL SCF, and 50 ng/mL TPO for 14 days. The transduction efficiency was 60% to 90%, as judged from green fluorescent protein (GFP) expression. To induce knockdown in erythroblasts, c-Kit+ progenitors preincubated in Iscove modified Dulbecco medium supplemented with 10% FBS and 50 ng/mL of SCF, TPO, and FP6 (kindly provided by Kyowa Hakko Kirin Pharma, Inc.) and 40 ng/mL interleukin-3 (IL-3; PeproTech) were similarly transduced with the indicated virus at a multiplicity of infection of 20. At 24 hours posttransduction, cells were cultured for 3 days in Iscove modified Dulbecco medium supplemented with 10% FBS and 50 units/mL of human erythropoietin (EPO; kindly provided by Kyowa Hakko Kirin) then subjected to cell sorting to recover the GFP+CD71+ transduced erythroblasts.
Microarray analysis
Total RNA was extracted from pooled BM lineage−Sca-1+c-Kit+ (LSK) cells (isolated from 4-5 mice per each genotype) by using an RNeasy Plus Mini Kit (Qiagen). Microarray analysis using a SurePrint G3 Mouse GE Microarray 8x60K kit (Agilent) was carried out as previously described.21 The raw data were deposited in Gene Expression Omnibus under the accession number GSE51038.
Real-time quantitative RT-PCR
Total RNA was isolated using TRIZOL LS solution (Invitrogen) and reverse-transcribed by the ThermoScript RT-PCR system (Invitrogen) with an oligo-dT primer. Real-time quantitative reverse-transcription polymerase chain reaction (RT-PCR) was performed with an ABI Prism 7300 Thermal Cycler (Applied Biosystems) using FastStart Universal Probe Master (Roche) and the indicated combinations of Universal Probe Library (Roche). Primer sequences are as follows:
Gene/Forward/Reverse primer (5′ to 3′)/probe
Sf3b1 agcaagctcgtggatgatg/ccacaggttattatgaccaagaaa/#31
Hprt1 tcttcctcagaccgctttt/cctggttcatcatcgctaatc/#95
Mycn tgtgtctgttccagctactgc/ ttcctcctcgtcatcctcat/#68
Gdf15 gagctacggggtcgcttc/ gagtcctctcggctctggt/#62
Epx tcagcaagtgagaaggatcg/ agcgtctccaggcattgtat/#97
Ear2 gggagccacaaagcagac/ ttgatatgctggatggcaaa/#48
Cd83 tggttctgaaggtgacagga/ caaccagagagaagagcaacac/#29
Bcl2a1d tttccagttttgtggcagaat/ tcaaacttctttatgaagccatctt/#2
Blnk tcccatttaattcgacgtttg/ agcataccagggcttaccg/#75
Skil agtgtcttacccagacgtctcac/ agcagactcagatttctttctacttgt/#68
Statistical analysis
Statistical significance was analyzed with Student t test. The level of significance was set at 0.05.
Results
Minimal defects of hematopoiesis in Sf3b1 heterozygous mice
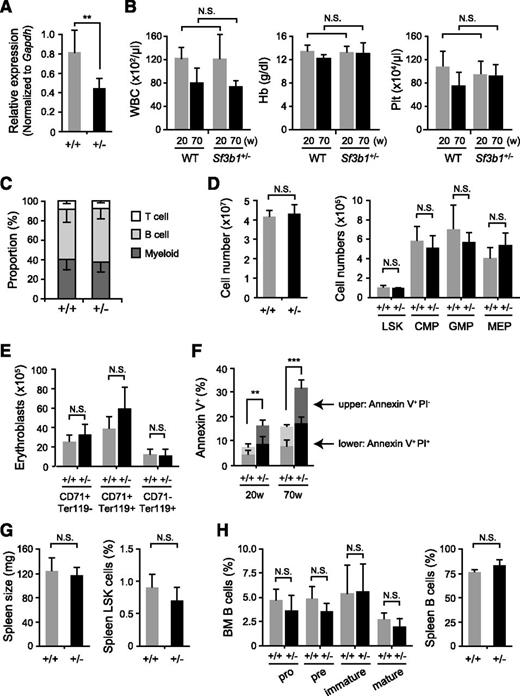
Given that Sf3b1-deficient mice die at a very early embryonic stage,16 we first examined hematopoiesis in Sf3b1 heterozygous (Sf3b1+/−) mice that show almost normal development compared with the WT littermates (data not shown). We confirmed that Sf3b1+/− mice exhibited a 40% reduction in the expression of Sf3b1 in LSK hematopoietic stem/progenitor cells (HSPCs) (Figure 1A). Sf3b1+/− mice did not show any significant changes in white blood cell counts, hemoglobin levels, or platelet counts in their PB at any time points up to 70 weeks old (Figure 1B). Sf3b1+/− mice had a lineage composition of PB MNCs comparable with that of WT mice (Figure 1C). In addition, Sf3b1+/− mice had total BM cell counts comparable with those of WT mice and did not show any significant changes in the numbers of LSKs, common myeloid progenitors, granulocyte/macrophage progenitors, and megakaryocyte/erythroid progenitors (Figure 1D). Consistent with the absence of anemia in Sf3b1+/− mice, Sf3b1+/− mice showed almost normal differentiation of erythroid lineage cells in the BM (Figure 1E). Furthermore, they did not develop any hematological disorders, including MDS by 70 weeks of age (data not shown). Of note, however, Sf3b1+/− mice showed a mild but significant increase in apoptosis in LSK cells compared with WT mice (Figure 1F), though the number of LSK cells did not change in Sf3b1+/− mice. Thus, Sf3b1+/− mice do not show obvious phenotypic changes in hematopoiesis except for mildly enhanced apoptosis.
Minimal defects of hematopoiesis in Sf3b1 heterozygous mice. (A) Quantitative RT-PCR analysis of the expression of Sf3b1 in LSK cells from WT and Sf3b1+/− mice. Gapdh was used to normalize the amount of input RNA. Data are shown as the mean ± standard deviation (SD) for triplicate analyses. (B) PB cell counts in 20- and 70-week-old WT and Sf3b1+/− mice. White blood cell (WBC), hemoglobin (Hb), and platelet (Plt) counts are presented as mean ± SD (WT n = 4; Sf3b1+/− n = 5). (C) The proportion of each cell lineage in PB from 70-week-old WT and Sf3b1+/− mice shown as mean ± SD (WT n = 4; Sf3b1+/− n = 5). (D) Absolute numbers of total MNCs, LSK cells, and myeloid progenitors in a unilateral femur and tibia from 70-week-old WT and Sf3b1+/− mice. Data are shown as the mean ± SD (WT n = 4; Sf3b1+/− n = 5). (E) Absolute numbers of CD71+Ter119−, CD71+Ter119+, and CD71−Ter119+ erythroblasts in a unilateral femur and tibia from WT and 70-week-old Sf3b1+/− mice. Data are shown as the mean ± SD (WT n = 4; Sf3b1+/− n = 5). (F) Percentage of AnnexinV+PI− and AnnexinV+PI+ apoptotic cells in BM LSK cells from 70-week-old WT and Sf3b1+/− mice. Data are shown as the mean ± SD (WT n = 4; Sf3b1+/− n = 5). (G) Size of the spleen and the proportion of LSK cells in the spleen from 32-week-old WT and Sf3b1+/− mice. Data are shown as the mean ± SD (WT n = 5; Sf3b1+/− n = 5). (H) B-cell differentiation in the BM and proportion of B cells in the spleen from 32-week-old WT and Sf3b1+/− mice. The proportion of Pro-B (B220+CD43+IgM−), Pre-B (B220+CD43−IgM−), and immature B (B220+CD43−IgM+) and mature B (B220hiCD43−IgM+) cells in BM is shown as the mean ± SD (WT n = 5; Sf3b1+/− n = 5). *P < .05; **P < .01. N.S., not significant.
Minimal defects of hematopoiesis in Sf3b1 heterozygous mice. (A) Quantitative RT-PCR analysis of the expression of Sf3b1 in LSK cells from WT and Sf3b1+/− mice. Gapdh was used to normalize the amount of input RNA. Data are shown as the mean ± standard deviation (SD) for triplicate analyses. (B) PB cell counts in 20- and 70-week-old WT and Sf3b1+/− mice. White blood cell (WBC), hemoglobin (Hb), and platelet (Plt) counts are presented as mean ± SD (WT n = 4; Sf3b1+/− n = 5). (C) The proportion of each cell lineage in PB from 70-week-old WT and Sf3b1+/− mice shown as mean ± SD (WT n = 4; Sf3b1+/− n = 5). (D) Absolute numbers of total MNCs, LSK cells, and myeloid progenitors in a unilateral femur and tibia from 70-week-old WT and Sf3b1+/− mice. Data are shown as the mean ± SD (WT n = 4; Sf3b1+/− n = 5). (E) Absolute numbers of CD71+Ter119−, CD71+Ter119+, and CD71−Ter119+ erythroblasts in a unilateral femur and tibia from WT and 70-week-old Sf3b1+/− mice. Data are shown as the mean ± SD (WT n = 4; Sf3b1+/− n = 5). (F) Percentage of AnnexinV+PI− and AnnexinV+PI+ apoptotic cells in BM LSK cells from 70-week-old WT and Sf3b1+/− mice. Data are shown as the mean ± SD (WT n = 4; Sf3b1+/− n = 5). (G) Size of the spleen and the proportion of LSK cells in the spleen from 32-week-old WT and Sf3b1+/− mice. Data are shown as the mean ± SD (WT n = 5; Sf3b1+/− n = 5). (H) B-cell differentiation in the BM and proportion of B cells in the spleen from 32-week-old WT and Sf3b1+/− mice. The proportion of Pro-B (B220+CD43+IgM−), Pre-B (B220+CD43−IgM−), and immature B (B220+CD43−IgM+) and mature B (B220hiCD43−IgM+) cells in BM is shown as the mean ± SD (WT n = 5; Sf3b1+/− n = 5). *P < .05; **P < .01. N.S., not significant.
Sf3b1+/− mice showed no sign of extramedullary hematopoiesis as judged by the normal spleen size and no increase in LSK cells in the spleen (Figure 1G). In addition, Sf3b1+/− mice showed almost normal differentiation and expansion of B cells in the BM and spleen (Figure 1H). These findings indicate that Sf3b1 haploinsufficiency is not sufficient to induce MDS or clonal expansion of B lymphoid cells in vivo.
Sf3b1 haploinsufficiency results in impaired competitive reconstitution capacity of HSCs
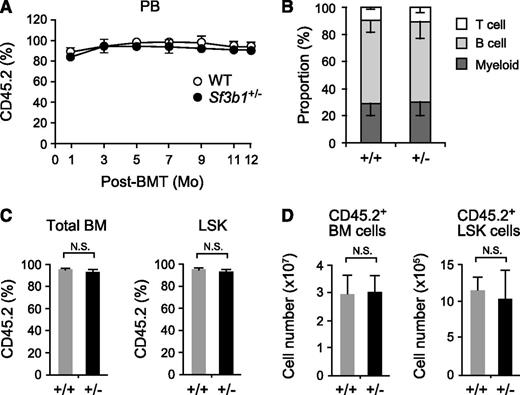
To exclude any effects of reduced Sf3b1 expression in nonhematopoietic cells in vivo, we next transplanted 5 × 106 WT and Sf3b1+/− BM cells (CD45.2+) into lethally irradiated recipient (CD45.1+) mice. Sf3b1+/− hematopoietic cells efficiently reconstituted hematopoiesis of recipient mice at least up to 12 months posttransplantation (Figure 2A) and contributed to myeloid and lymphoid cells in PB in a manner similar to WT hematopoietic cells (Figure 2B). Sf3b1+/− hematopoietic cells also reconstituted BM cells, including LSK cells to levels comparable with WT cells in proportion (Figure 2C) as well as in absolute number (Figure 2D). Reconstitution of myeloid progenitors and erythroid lineage cells in BM was also comparable between WT and Sf3b1+/− mice (data not shown). These findings confirm that haploinsufficiency of Sf3b1 in hematopoietic cells does not largely impair hematopoiesis. Once again, we did not find any hematological features of MDS in mice reconstituted with Sf3b1+/− BM cells up to 1 year postransplantation.
Sf3b1+/− BM cells normally reconstitute hematopoiesis in vivo. (A) Contribution of donor cells to PB hematopoiesis. WT and Sf3b1+/− BM cells (5 × 106 cells, CD45.2+) were transplanted into lethally irradiated recipient (CD45.1+) mice. Chimerism of donor-derived CD45.2+ cells in the PB is shown as mean ± SD (n = 7). BMT, BM transplant. (B) Lineage contribution of WT and Sf3b1+/− donor cells in PB. The proportion of each cell lineage in donor-derived CD45.2+ cells in the PB at 12 months posttransplantation. (C-D) Contribution of donor cells to BM hematopoiesis. Chimerism of donor-derived CD45.2+ cells in total BM cells and LSK cells is shown as mean ± SD (n = 7) in C. Absolute numbers of donor-derived CD45.2+ BM cells and LSK cells are shown as mean ± SD (n = 7) in D. N.S., not significant.
Sf3b1+/− BM cells normally reconstitute hematopoiesis in vivo. (A) Contribution of donor cells to PB hematopoiesis. WT and Sf3b1+/− BM cells (5 × 106 cells, CD45.2+) were transplanted into lethally irradiated recipient (CD45.1+) mice. Chimerism of donor-derived CD45.2+ cells in the PB is shown as mean ± SD (n = 7). BMT, BM transplant. (B) Lineage contribution of WT and Sf3b1+/− donor cells in PB. The proportion of each cell lineage in donor-derived CD45.2+ cells in the PB at 12 months posttransplantation. (C-D) Contribution of donor cells to BM hematopoiesis. Chimerism of donor-derived CD45.2+ cells in total BM cells and LSK cells is shown as mean ± SD (n = 7) in C. Absolute numbers of donor-derived CD45.2+ BM cells and LSK cells are shown as mean ± SD (n = 7) in D. N.S., not significant.
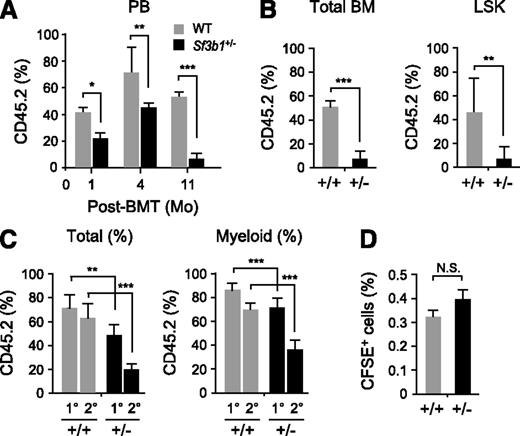
We next performed competitive reconstitution assays utilizing Sf3b1+/− (CD45.2+) BM cells together with an equal number of CD45.1+ competitor BM cells to test whether functional defects in Sf3b1+/− HSCs become obvious under stressful conditions. Sf3b1+/− cells showed significantly less contribution to the PB of recipient mice at all time points compared with WT cells, and their contribution gradually declined over time (Figure 3A). Eventually, chimerism of Sf3b1+/− cells declined to very low levels in both PB and BM hematopoietic cells and in BM LSK cells (Figure 3A-B).
Sf3b1 haploinsufficiency causes impaired competitive repopulating capacity of HSCs. (A) Contribution of donor cells to PB hematopoiesis. WT and Sf3b1+/− BM cells (2 × 106 cells, CD45.2+) together with an equal number of CD45.1+ competitor BM cells were transplanted into lethally irradiated recipient (CD45.1+) mice. Chimerism of donor-derived CD45.2+ cells in the PB is shown as mean ± SD (n = 5). (B) Chimerism of donor-derived CD45.2+ cells in total BM cells and LSK cells at 11 months posttransplantation shown as mean ± SD (n = 5). (C) Serial transplantation assays. WT and Sf3b1+/− BM cells (2 × 106 cells, CD45.2+) together with an equal number of CD45.1+ competitor BM cells were transplanted into lethally irradiated recipient (CD45.1+) mice. At 4 months posttransplantation, 2 × 106 whole BM cells from the primary recipient mice were transplanted into the secondary recipient mice. Chimerism of donor-derived CD45.2+ cells was detected in total MNCs and myeloid cells in the PB at 4 months postprimary (1°) and -secondary (2°) transplantation and is shown as mean ± SD (n = 9). (D) Homing assays. CFSE-labeled WT and Sf3b1+/− BM MNCs (106 cells) were transplanted into lethally irradiated mice. Frequencies of CFSE+ cells detected in BM at 8 hours posttransplantation are shown as mean ± SD (n = 5). *P < .05; **P < .01; ***P < .001. N.S., not significant.
Sf3b1 haploinsufficiency causes impaired competitive repopulating capacity of HSCs. (A) Contribution of donor cells to PB hematopoiesis. WT and Sf3b1+/− BM cells (2 × 106 cells, CD45.2+) together with an equal number of CD45.1+ competitor BM cells were transplanted into lethally irradiated recipient (CD45.1+) mice. Chimerism of donor-derived CD45.2+ cells in the PB is shown as mean ± SD (n = 5). (B) Chimerism of donor-derived CD45.2+ cells in total BM cells and LSK cells at 11 months posttransplantation shown as mean ± SD (n = 5). (C) Serial transplantation assays. WT and Sf3b1+/− BM cells (2 × 106 cells, CD45.2+) together with an equal number of CD45.1+ competitor BM cells were transplanted into lethally irradiated recipient (CD45.1+) mice. At 4 months posttransplantation, 2 × 106 whole BM cells from the primary recipient mice were transplanted into the secondary recipient mice. Chimerism of donor-derived CD45.2+ cells was detected in total MNCs and myeloid cells in the PB at 4 months postprimary (1°) and -secondary (2°) transplantation and is shown as mean ± SD (n = 9). (D) Homing assays. CFSE-labeled WT and Sf3b1+/− BM MNCs (106 cells) were transplanted into lethally irradiated mice. Frequencies of CFSE+ cells detected in BM at 8 hours posttransplantation are shown as mean ± SD (n = 5). *P < .05; **P < .01; ***P < .001. N.S., not significant.
To evaluate the reconstitution capacity of Sf3b1+/− HSCs more precisely, we performed serial transplantation assays using 2 × 106 whole BM cells from the primary recipient mice. As in Figure 3A, chimerism of Sf3b1+/− cells in total MNCs and myeloid cells in PB was significantly less than WT even in the primary recipients (Figure 3A) and further declined in the secondary recipients (Figure 3C). However, Sf3b1+/− cells did not show obvious defects in differentiation, because there were no significant changes in the proportion of myeloid and lymphoid lineage cells within CD45.2+Sf3b1+/− cells compared with WT cells (data not shown). To explore whether compromised homing capability may account for the impaired reconstitution capacity of Sf3b1+/− hematopoietic cells, we performed in vivo homing assay utilizing CFSE-labeled WT and Sf3b1+/− BM MNCs. There were no significant differences in the proportion of CFSE-positive cells in the BM at 8 hours postinjection (Figure 3D), indicating that Sf3b1+/− cells retain normal homing capability. Taken together, these findings indicate that Sf3b1 haploinsufficiency significantly impairs the competitive reconstitution capacity of HSCs.
Level of Sf3b1 is critical for the proliferative capacity of HSCs
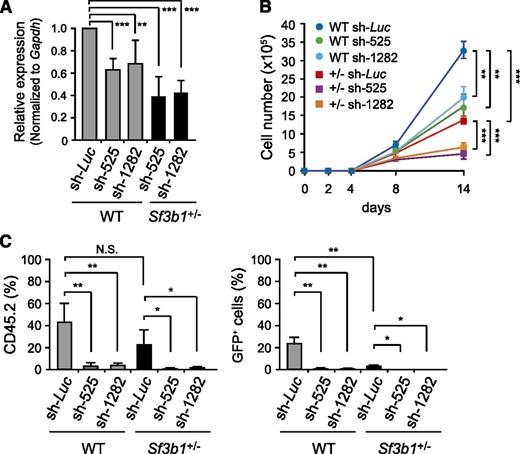
We next examined the effects of further down-regulation of Sf3b1 on HSCs. To do so, we prepared shRNAs directed to Sf3b1 (525 and 1282). Transduction efficiency was ∼80% as detected by GFP expression on flow cytometry, and no significant difference was observed in transduction efficiencies among samples. Quantitative RT-PCR confirmed that Sf3b1 levels were reduced by 40% and 60% in WT and Sf3b1+/− LSK cells transduced with shRNA against Sf3b1, respectively (Figure 4A). We then transduced CD34− LSK HSCs with these viruses. We first evaluated the growth of HSCs in liquid culture supplemented only with SCF and TPO, a condition that supports the growth of immature HSPCs rather than their differentiation. Under these conditions, Sf3b1+/− HSCs showed significant growth retardation compared with WT HSCs, and depletion of Sf3b1 by shRNAs further attenuated their growth compared with the controls expressing shRNA directed to Luciferase (Figure 4B). We next transplanted WT and Sf3b1+/− HSCs transduced with shRNAs against Sf3b1 together with radio-protective CD45.1+ BM cells into lethally irradiated CD45.1+ recipient mice. Depletion of Sf3b1 severely impaired reconstitution capacity of both WT and Sf3b1+/− HSCs (Figure 4C). The effect of Sf3b1 knockdown was more obvious in Sf3b1+/− HSCs compared with WT HSCs, showing a clear dependency of reconstitution capacity of HSCs on Sf3b1 levels. In contrast, we did not see any obvious defects in the differentiation of HSCs, including erythroblasts in BM upon depletion of Sf3b1 (data not shown). These findings imply that Sf3b1 is required for the self-renewal capacity of HSCs but is not critical for the differentiation capabilities of HSCs.
Knockdown of Sf3b1 compromises proliferative capacity of HSCs. (A) Knockdown efficiencies of shRNAs against Sf3b1. WT and Sf3b1+/− LSK cells were transduced with the indicated shRNAs and cultured in the presence of SCF and TPO. At day 3 postinfection, GFP+Lin−c-Kit+ cells were purified by cell sorting, and the levels of Sf3b1 were analyzed by quantitative RT-PCR analysis. mRNA levels were normalized to Gapdh expression. Expression levels relative to that in the control cells transduced with an shRNA against Luciferase are shown as the mean ± SD for triplicate analyses. (B) Growth of WT and Sf3b1+/− CD34−LSK HSCs upon knockdown of Sf3b1 in culture. One hundred WT and Sf3b1+/− HSCs were transduced with the indicated shRNAs and cultured in the presence of SCF and TPO. Data are shown as mean ± SD (n = 5). The transduction efficiency was ∼80% as detected by GFP expression on flow cytometry. (C) Reconstitution capacity of WT and Sf3b1+/− CD34−LSK HSCs upon knockdown of Sf3b1 in vivo. WT and Sf3b1+/− HSCs were transduced with the indicated shRNAs as in (B), and 100 transduced HSCs were transplanted into lethally irradiated recipient (CD45.1+) mice together with 2 × 105 CD45.1+ competitor BM cells. Data are shown as mean ± SD (n = 5). Chimerism of donor-derived CD45.2+ cells (left panel) and CD45.2+GFP+ transduced cells (right panel) in the PB at 4 months posttransplantation are shown as mean ± SD (n = 5). *P < .02; **P < .01; ***P < .001.
Knockdown of Sf3b1 compromises proliferative capacity of HSCs. (A) Knockdown efficiencies of shRNAs against Sf3b1. WT and Sf3b1+/− LSK cells were transduced with the indicated shRNAs and cultured in the presence of SCF and TPO. At day 3 postinfection, GFP+Lin−c-Kit+ cells were purified by cell sorting, and the levels of Sf3b1 were analyzed by quantitative RT-PCR analysis. mRNA levels were normalized to Gapdh expression. Expression levels relative to that in the control cells transduced with an shRNA against Luciferase are shown as the mean ± SD for triplicate analyses. (B) Growth of WT and Sf3b1+/− CD34−LSK HSCs upon knockdown of Sf3b1 in culture. One hundred WT and Sf3b1+/− HSCs were transduced with the indicated shRNAs and cultured in the presence of SCF and TPO. Data are shown as mean ± SD (n = 5). The transduction efficiency was ∼80% as detected by GFP expression on flow cytometry. (C) Reconstitution capacity of WT and Sf3b1+/− CD34−LSK HSCs upon knockdown of Sf3b1 in vivo. WT and Sf3b1+/− HSCs were transduced with the indicated shRNAs as in (B), and 100 transduced HSCs were transplanted into lethally irradiated recipient (CD45.1+) mice together with 2 × 105 CD45.1+ competitor BM cells. Data are shown as mean ± SD (n = 5). Chimerism of donor-derived CD45.2+ cells (left panel) and CD45.2+GFP+ transduced cells (right panel) in the PB at 4 months posttransplantation are shown as mean ± SD (n = 5). *P < .02; **P < .01; ***P < .001.
Sf3b1 haploinsufficiency does not activate the non-sense–mediated mRNA decay pathway
To understand the molecular mechanism through which Sf3b1 haploinsufficiency affects the function of HSCs, we performed gene expression analysis using LSK HSPCs isolated from 11-month-old WT and Sf3b1+/− mice. We also harvested LSK cells from recipient mice reconstituted with WT and Sf3b1+/− cells at 9 months posttransplantation. Microarray analysis revealed that more genes were altered by >2-fold in Sf3b1+/− LSK cells from primary recipient mice compared with those from 11-month-old mice (Figure 5A; supplemental Table 1, available on the Blood Web site). Among these, 12 and 27 genes were up- and down-regulated, respectively, in both analyses. Expression of a portion of genes was validated by quantitative RT-PCR (supplemental Figure 1). Unexpectedly, however, these genes did not include those known to be involved in the regulation of HSC functions.
Sf3b1 haploinsufficiency does not activate the NMD pathway. (A) A scatter diagram of microarray analysis. WT and Sf3b1+/− LSK cells were isolated from 11-month-old WT and Sf3b1+/− mice and recipient mice reconstituted with WT and Sf3b1+/− BM cells at 9 months posttransplantation and then analyzed by microarray-based expression analysis. The average signal levels of Sf3b1+/− cells compared with those of WT cells are plotted. The red and green lines represent the borderline for 2-fold increase and 2-fold decrease, respectively, and the numbers of genes altered by >2-fold are indicated. (B) The gene set enrichment analysis demonstrating a significant negative enrichment of the NMD pathway genes in Sf3b1+/− LSK cells relative to WT LSK cells. Normalized enrichment scores (NSE), nominal P values, and false discovery rates (FDRs) are indicated. Red and blue represent positive (up-regulated in the given genotype relative to WT) and negative (up-regulated in WT relative to the given genotype) enrichment, respectively. Heat maps showing the gene expression profiles of 30 randomly selected genes are indicated below the plot.
Sf3b1 haploinsufficiency does not activate the NMD pathway. (A) A scatter diagram of microarray analysis. WT and Sf3b1+/− LSK cells were isolated from 11-month-old WT and Sf3b1+/− mice and recipient mice reconstituted with WT and Sf3b1+/− BM cells at 9 months posttransplantation and then analyzed by microarray-based expression analysis. The average signal levels of Sf3b1+/− cells compared with those of WT cells are plotted. The red and green lines represent the borderline for 2-fold increase and 2-fold decrease, respectively, and the numbers of genes altered by >2-fold are indicated. (B) The gene set enrichment analysis demonstrating a significant negative enrichment of the NMD pathway genes in Sf3b1+/− LSK cells relative to WT LSK cells. Normalized enrichment scores (NSE), nominal P values, and false discovery rates (FDRs) are indicated. Red and blue represent positive (up-regulated in the given genotype relative to WT) and negative (up-regulated in WT relative to the given genotype) enrichment, respectively. Heat maps showing the gene expression profiles of 30 randomly selected genes are indicated below the plot.
In contrast, gene set enrichment analysis22 revealed a number of differences in canonical pathways in gene expression profiles between WT and Sf3b1+/− LSK cells. Many gene sets were commonly enriched in Sf3b1+/− LSK cells isolated from 11-month-old mice and recipient mice at 9 months posttransplantation. Those included gene sets related to transcription, translation, cell cycle and metabolism (supplemental Table 2). Among these, the gene set for the non-sense–mediated mRNA decay (NMD) pathway was negatively enriched in Sf3b1+/− LSK cells, particularly those from primary recipient mice (Figure 5B; supplemental Table 2). NMD is a surveillance pathway that surveys and degrades abnormal transcripts that contain premature stop codons.23,24 Because of aberrant splicing, the genes in this pathway, such as MAGOH, SMG6, SMG7, SMG9, and UPF3B, are activated in HeLa cells overexpressing an U2AF35 mutant identified in MDS patients.6 Conversely, these NMD pathway genes were expressed at levels comparable with WT LSK cells or mildly down-regulated in Sf3b1+/− LSK cells (Figure 5B), and this trend was validated by quantitative RT-PCR (data not shown). These findings indicate that Sf3b1 haploinsufficiency is not sufficient for activating NMD pathway.
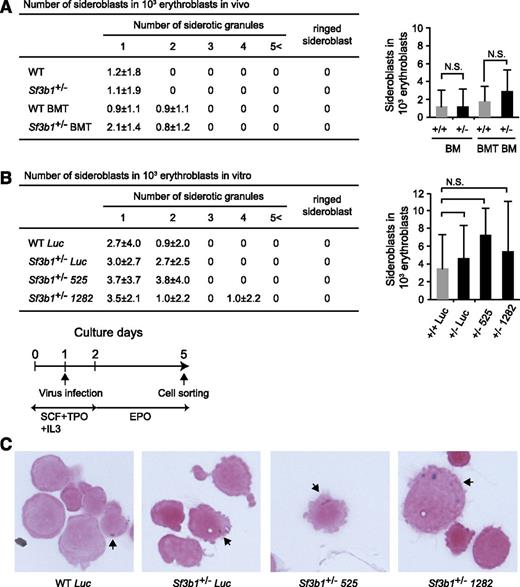
Reduced Sf3b1 expression is not sufficient to induce RARs
Given that SF3B1 is very frequently mutated in RARS patients, we finally examined the frequencies of sideroblasts in CD71+ BM erythroblasts from 11-month-old WT and Sf3b1+/− mice. Sideroblasts are erythroblasts with granules of iron accumulated in perinuclear mitochondria and are classified according to the WHO International Working Group on Morphology of MDS25 : type 1 sideroblasts with fewer than 5 siderotic granules in the cytoplasm, Type 2 sideroblasts with 5 or more siderotic granules but not in a perinuclear distribution, and Type 3 or RARs with 5 or more granules in a perinuclear position surrounding the nucleus or encompassing at least one-third of the nuclear circumference. Although a recent study has shown that Sf3b1+/− BM cells have a significantly increased number of RARs,26 we did not detect any RARs in our mice. We found only type 1 sideroblasts at frequencies comparable between WT and Sf3b1+/− cells (Figure 6A). To examine whether the level of Sf3b1 expression is critical for the induction of RARs, we transduced WT and Sf3b1+/− c-Kit+ progenitor cells with shRNAs against Sf3b1 and then cultured them in the presence of EPO to induce erythroblasts (Figure 6B). Again, depletion of Sf3b1 in erythroblasts in culture did not increase the number of CD71+ sideroblasts compared with the control erythroblasts expressing shRNA against Luciferase or induce RARs (Figure 6B-C). Taken together, these findings indicate that the reduction in the levels of Sf3b1 does not cause appearance of RARs that is the definitive dysplastic feature of RARS.
Reduced Sf3b1 expression is not sufficient to induce RARs. (A) Frequencies of sideroblastic erythroblasts in Sf3b1+/− erythroblasts. CD71+ erythroblasts in BM of WT and Sf3b1+/− mice and recipients repopulated with WT and Sf3b1+/− BM cells at 11 months posttransplantation (WT BMT and Sf3b1+/− BMT, respectively) were isolated by cell sorting. The cells were cytospun onto slide glasses, subjected to Prussian Blue staining, and then examined under a light microscope. Numbers of cells with siderotic granules per 1000 erythroblasts are shown in a table and a graph. Data are shown as mean ± SD. Counts of 1000 cells were independently performed 3 times. (B) Frequencies of sideroblastic erythroblasts in Sf3b1+/− erythroblasts upon knockdown of Sf3b1. c-Kit+ progenitors transduced with shRNA against Sf3b1 were cultured in the presence of EPO to induce erythroblasts (bottom). GFP+CD71+-transduced erythroblasts were isolated by cell sorting, then evaluated for the presence of sideroblasts in a manner similar to (A). (C) Representative sideroblastic erythroblasts observed in (B). Arrows indicate siderotic granules.
Reduced Sf3b1 expression is not sufficient to induce RARs. (A) Frequencies of sideroblastic erythroblasts in Sf3b1+/− erythroblasts. CD71+ erythroblasts in BM of WT and Sf3b1+/− mice and recipients repopulated with WT and Sf3b1+/− BM cells at 11 months posttransplantation (WT BMT and Sf3b1+/− BMT, respectively) were isolated by cell sorting. The cells were cytospun onto slide glasses, subjected to Prussian Blue staining, and then examined under a light microscope. Numbers of cells with siderotic granules per 1000 erythroblasts are shown in a table and a graph. Data are shown as mean ± SD. Counts of 1000 cells were independently performed 3 times. (B) Frequencies of sideroblastic erythroblasts in Sf3b1+/− erythroblasts upon knockdown of Sf3b1. c-Kit+ progenitors transduced with shRNA against Sf3b1 were cultured in the presence of EPO to induce erythroblasts (bottom). GFP+CD71+-transduced erythroblasts were isolated by cell sorting, then evaluated for the presence of sideroblasts in a manner similar to (A). (C) Representative sideroblastic erythroblasts observed in (B). Arrows indicate siderotic granules.
Discussion
Our study demonstrated that heterozygosity for Sf3b1 does not significantly perturb hematopoiesis except for mildly enhancing apoptosis in HSPCs. It also fails to induce any hematological malignancies, including MDS, within an observation period up to 70 weeks. Nevertheless, Sf3b1+/− HSCs showed significantly impaired reconstitution capacity in a competitive setting, and further reduction in Sf3b1 levels by RNA interference severely compromised the reconstitution capacity of HSCs, though it did not affect their differentiation potential. These findings are consistent with the effect of U2AF35 mutants in HSCs.6 Exogenous U2AF35 mutants compromised the reconstitution capacity of WT HSCs, suggesting a dominant-negative action of U2AF35 mutants relative to the WT U2AF35 protein. Indeed, there are reported mutational hot spots in U2AF35 in patients with MDS,6,27 none of which are homozygous, nonsense, or frameshift mutations. All of these findings suggest that mutations in U2AF35 are associated with some gain of function rather than simple loss of function. SF3B1 mutations are assumed to behave in a fashion similar to U2AF35 mutations because of its similar mutational characteristics. Therefore, we are not sure whether the depletion of Sf3b1 that we used in this study precisely reflects the pathophysiological consequences of SF3B1 mutations in MDS cells. However, it is intriguing that both U2AF35 mutants and depletion of Sf3b1 compromised the proliferative capacity of HSCs in a similar fashion. These findings suggest that SF3B1 mutations do not confer a growth advantage to MDS clones over normal hematopoietic cells, thus highlighting the requirement of cooperation with other gene mutations.
As expected from its function in pre-mRNA splicing, RNA sequencing analysis of BM cells from a patient with RARS with an SF3B1 mutation showed alternative splicing in 350 of 9069 genes, including 4 relevant genes to the pathology of MDS such as ASXL1, CBL, EZH1, and RUNX3.26 In addition, the NMD pathway, which surveys and degrades abnormal transcripts, was activated in HeLa cells overexpressing an U2AF35 mutant identified in MDS patients. In contrast, the NMD pathway was suppressed in Sf3b1+/− LSK cells in this study, even in LSK cells reconstituted from donor BM cells in recipient mice, suggesting that Sf3b1 haploinsufficiency does not cause abnormal RNA splicing sufficient for activating the NMD pathway. Nonetheless, Sf3b1+/− HSCs showed impaired reconstitution capacity in vivo and growth retardation in culture. It is possible that Sf3b1 haploinsufficiency induces subclinical levels of splicing abnormalities that cause impairment in HSC functions only under stressful conditions such as transplant and in vitro culture and that mild but significant levels of altered gene expression detected in microarray analyses could be attributed to the splicing defect.
Notably, we did not detect any changes in the frequencies of sideroblasts and did not detect RARs in Sf3b1+/− BM cells and Sf3b1+/− erythroblasts depleted of Sf3b1. Down-regulation of ABCB7 and overexpression of MITOCHONDRIAL FERRITIN (FTMT) in immature red cells are typical features of RARS.28 We analyzed the expression of Abcb7 and Ftmt in Sf3b1+/− CD71+Ter119+ erythroblasts and observed no significant changes in Abcb7 and Ftmt expression (data not shown). These results indicate that simple reduction in the levels of Sf3b1 does not cause abnormal iron metabolism in mitochondria and present a striking contrast with the recent report showing an increased frequency of RARs in the same Sf3b1+/− BM cells.26 The difference in the results of the detection of RARs in the same mouse strain using a conventional staining method is obscure but could be partly due to the different conditions under which the mice were maintained. Nevertheless, this point requires further investigation in comparison with the effects of SF3B1 mutants in the induction of RARs. As suggested from the mutational properties of the SF3B1 gene in MDS, SF3B1 mutants might acquire some aberrant functions, like IDH1/2 mutants in cancer, that lead to dysregulated iron metabolism.29
In conclusion, our findings indicate that the level of Sf3b1 gene expression is critical in the maintenance of proliferative capacity of HSCs. However, heterozygosity for Sf3b1 alone does not induce apparent RNA splicing abnormalities or lead to the development of RARS. Further analysis of mice expressing SF3B1 mutants specifically in hematopoietic cells would promote precise understanding of the pathological function of SF3B1 mutations in the development of MDS.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs K. Matsushita and K. Kitamura for Prussian Blue staining and members of the Iwama Laboratory for discussion during the preparation of this manuscript, particularly George R. Wendt for a critical reading of the manuscript.
This work was supported in part by Grants-in-Aid for Scientific Research (24249054 and 25130702) and Scientific Research on Innovative Areas “Cell Fate” (22118004) from MEXT, Japan, a Grant-in-Aid for Core Research for Evolutional Science and Technology from the Japan Science and Technology Corporation, and grants from the Uehara Memorial Foundation and the Takeda Science Foundation.
Authorship
Contribution: C.W. performed the experiments, analyzed results, made the figures, and actively wrote the manuscript; G.S., A.S., R.I., S.K., and M.O. assisted with the experiments, including the hematopoietic analyses; K.I. and H.K. provided mice; and A.I. conceived of and directed the project, secured funding, and actively wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Atsushi Iwama, 1-8-1 Inohana, Chuo-ku, Chiba, 260-8670 Japan; e-mail: aiwama@faculty.chiba-u.jp.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal