Key Points
Compared with ubiquitously expressed PI3K p110α, genetic inhibition of PI3K p110δ uniquely normalizes mutant Shp2-induced GM-CSF hypersensitivity.
Potent pharmacologic inhibitors of PI3K p110δ cooperate with MEK inhibition to reduce mutant Shp2-induced hyperproliferation.
Abstract
Although hyperactivation of the Ras-Erk signaling pathway is known to underlie the pathogenesis of juvenile myelomonocytic leukemia (JMML), a fatal childhood disease, the PI3K-Akt signaling pathway is also dysregulated in this disease. Using genetic models, we demonstrate that inactivation of phosphatidylinositol-3-kinase (PI3K) catalytic subunit p110δ, but not PI3K p110α, corrects gain-of-function (GOF) Shp2-induced granulocyte macrophage–colony-stimulating factor (GM-CSF) hypersensitivity, Akt and Erk hyperactivation, and skewed hematopoietic progenitor distribution. Likewise, potent p110δ-specific inhibitors curtail the proliferation of GOF Shp2-expressing hematopoietic cells and cooperate with mitogen-activated or extracellular signal-regulated protein kinase kinase (MEK) inhibition to reduce proliferation further and maximally block Erk and Akt activation. Furthermore, the PI3K p110δ-specific inhibitor, idelalisib, also demonstrates activity against primary leukemia cells from individuals with JMML. These findings suggest that selective inhibition of the PI3K catalytic subunit p110δ could provide an innovative approach for treatment of JMML, with the potential for limiting toxicity resulting from the hematopoietic-restricted expression of p110δ.
Introduction
Class IA phosphatidylinositol-3-kinase (PI3K) activity is composed of catalytic subunits (p110α, p110β, or p110δ) and regulatory subunits (p85α, p55α, p50α, or p85β) and is commonly upregulated in human malignancies as a result of somatic mutations in the genes encoding p110α and p85α.1,2 Although mutations in the gene encoding p110δ have not been found, the p110δ protein is commonly overexpressed in myeloid leukemia3,4 and is unique among the class IA catalytic subunits in its capacity to induce malignant transformation independent of Ras.5
Juvenile myelomonocytic leukemia (JMML) is an aggressive childhood myeloproliferative neoplasm characterized as being Ras-driven because of mutations in NF1, CBL, KRAS, NRAS, or PTPN11.6 Accordingly, inhibition of the Ras-Erk signaling pathway using mitogen-activated or extracellular signal-regulated protein kinase kinase (MEK) inhibitors effectively normalizes loss-of-function (LOF) Nf1-induced and gain-of-function (GOF) Kras-induced disease in murine models.7,8 These findings are relevant, as MEK inhibitors have demonstrated success in BRAF and NRAS mutant melanoma in humans.9,10 However, MEK inhibition is accompanied by undesirable and dose-limiting adverse effects.9,11,12 Furthermore, resistance to MEK inhibitors commonly develops via multiple resistance mechanisms, including aberrant upregulation of the PI3K-Akt pathway.13
Previous studies demonstrate that PTEN expression, a negative regulator of PI3K-Akt signaling, frequently is reduced in primary JMML samples.14 Likewise, we recently found that genetic disruption of the PI3K regulatory subunit, p85α, normalizes GOF PTPN11-induced hypersensitivity to granulocyte macrophage–colony-stimulating factor (GM-CSF).15 Thus, we hypothesized that PI3K hyperactivation promotes myeloid cell growth in JMML beyond the customary task of a hyperactivated Ras effector and that the PI3K catalytic subunit, p110δ, uniquely promotes JMML because of its Ras-independent oncogenic properties.
Methods
Animal husbandry
Mice bearing a conditional GOF Ptpn11 allele (LSL-Shp2D61Y/+) have been described.16 Shp2D61Y/+;Mx1Cre+ animals were crossed with Pik3caflox/flox and Pik3cdD910A/D910A animals17,18 to genetically inactivate p110α and p110δ, respectively. Eight weeks after polyI:polyC treatment (300 µg × 3 intraperitoneal injections), animals were euthanized for functional and biochemical analyses. This study was approved by the Institutional Animal Care and Use Committee of the Indiana University School of Medicine.
Phenotypic and biochemical analyses
Murine bone marrow low-density mononuclear cells (LDMNCs) were plated into methylcellulose-based colony and [3H]-thymidine incorporation assays in the presence or absence of pharmacologic inhibitors, as described.15,19,20 Cell lysate preparation and immunoblotting also have been described.15,19,20 Phenotypically defined progenitor populations from mouse bone marrow were measured using flow cytometry, as described.21 Idelalisib (formerly known as GS-1101)22 and GS-982023 were provided by Gilead Sciences, Inc, and PD0325901 was purchased from Selleck. Ficoll-purified LDMNCs from JMML patients (meeting World Health Organization diagnostic criteria) or control (Lonza) were plated into methylcellulose-based assays with increasing concentrations of idelalisib. Samples were obtained with informed consent at the University of California, San Francisco, under a locally approved institutional review board protocol and were used in assays under approval from the institutional review board at the Indiana University School of Medicine. This study was conducted in accordance with the Declaration of Helsinki.
Results and discussion
Shp2D61Y/+;Mx1Cre+ mice16 were crossed with mice bearing a conditional knockout allele of p110α (Pik3caflox/flox).18 Although p110α protein expression was ablated without compensatory upregulation of p110β or p110δ, spleen sizes from Shp2D61Y/+;Pik3caflox/flox;Mx1Cre+ mice were not reduced substantially compared with those of Shp2D61Y/+;Pik3ca+/+;Mx1Cre+ mice (Figure 1A,C). Genetic disruption of p110α also failed to normalize GOF Shp2-induced GM-CSF hypersensitivity in methylcellulose-based progenitor or 3H-thymidine incorporation assays (Figure 1D,E) and did not reduce basal or GM-CSF-stimulated hyperphosphorylation of Erk or Akt (Figure 1F, compare lanes 7 and 8 with lane 6).
Genetic inhibition of PI3K catalytic subunit p110δ, but not p110α, normalizes GOF Shp2-induced hypersensitivity to GM-CSF. (A) and (B) Representative spleens from polyI:polyC-treated Shp2D61Y/+;Mx1Cre− mice (D61Y;Cre−, negative controls), Shp2D61Y/+;Mx1Cre+ mice (D61Y;Cre+, positive controls), and Shp2D61Y/+;Pik3caflox/flox;Mx1Cre+ (D61Y;Cre+;p110αFl/Fl) or Shp2D61Y/+;Pik3cdD910A/D910A;Mx1Cre+ (D61Y;Cre+;p110δD910A/D910A) mice. (C) Quantification of spleen weight:body weight (n = 6 to 9 mice per group). *P = .02 for D61Y;Cre− vs D61Y;Cre+; **P = .04 for D61Y;Cre+;p110δD910A/D910A vs D61Y;Cre+, statistics by unpaired, 2-tailed student’s t test. (D) Bone marrow LDMNCs from 6 to 8 mice per genotype were plated in methylcellulose, and colony-forming unit-GM assays were carried out in duplicate in 3 to 4 independent experiments. Data are represented as percentage maximal colony formation, calculated by dividing the number of colonies at each GM-CSF concentration by the average number of colonies at GM-CSF 10 ng/mL. *P = .0005 for D61Y;Cre+;p110δD910A/D910A vs D61Y;Cre+. (E) Bone marrow LDMNCs from 6 to 8 mice per genotype were subjected to [3H]-thymidine incorporation assays in replicates of 6 in 3 to 4 independent experiments. ^P < .0001 for D61Y;Cre+;p110δD910A/D910A vs D61Y;Cre+. For colony-forming unit-GM and [3H]-thymidine incorporation assays, data were analyzed using mixed-effects models with random intercept, using GM-CSF concentration as a categorical variable. (F) Immunoblot demonstrating Erk and Akt hyperphosphorylation in D61Y;Cre+ mice compared with D61Y;Cre− mice, without normalization on successful knockout of p110α protein expression in 2 independent D61Y;Cre+;p110αFl/Fl mice. (G) Immunoblot demonstrating normalization of Erk and Akt hyperphosphorylation in 2 independent D61Y;Cre+;p110δD910A/D910A mice compared with 2 independent D61Y;Cre+ mice. (H) Representative flow cytometry analysis demonstrating increased megakaryocyte erythroid progenitor and decreased granulocyte macrophage progenitor in D61Y;Cre+ mice compared with D61Y;Cre− mice, as described previously.16 (I) Quantification of phenotypically defined bone marrow myeloid precursor distribution. *P = .02, with the D61Y;Cre+;p110δD910A/D910A group being significantly closer to D61Y;Cre− than the D61Y;Cre+;p110αFl/Fl group to D61Y;Cre−. Progenitor distribution between genotypes was analyzed by comparing Euclidian distance between mean vectors to quantify the similarity among group mean levels, with the P value determined using the bootstrapping resampling method.
Genetic inhibition of PI3K catalytic subunit p110δ, but not p110α, normalizes GOF Shp2-induced hypersensitivity to GM-CSF. (A) and (B) Representative spleens from polyI:polyC-treated Shp2D61Y/+;Mx1Cre− mice (D61Y;Cre−, negative controls), Shp2D61Y/+;Mx1Cre+ mice (D61Y;Cre+, positive controls), and Shp2D61Y/+;Pik3caflox/flox;Mx1Cre+ (D61Y;Cre+;p110αFl/Fl) or Shp2D61Y/+;Pik3cdD910A/D910A;Mx1Cre+ (D61Y;Cre+;p110δD910A/D910A) mice. (C) Quantification of spleen weight:body weight (n = 6 to 9 mice per group). *P = .02 for D61Y;Cre− vs D61Y;Cre+; **P = .04 for D61Y;Cre+;p110δD910A/D910A vs D61Y;Cre+, statistics by unpaired, 2-tailed student’s t test. (D) Bone marrow LDMNCs from 6 to 8 mice per genotype were plated in methylcellulose, and colony-forming unit-GM assays were carried out in duplicate in 3 to 4 independent experiments. Data are represented as percentage maximal colony formation, calculated by dividing the number of colonies at each GM-CSF concentration by the average number of colonies at GM-CSF 10 ng/mL. *P = .0005 for D61Y;Cre+;p110δD910A/D910A vs D61Y;Cre+. (E) Bone marrow LDMNCs from 6 to 8 mice per genotype were subjected to [3H]-thymidine incorporation assays in replicates of 6 in 3 to 4 independent experiments. ^P < .0001 for D61Y;Cre+;p110δD910A/D910A vs D61Y;Cre+. For colony-forming unit-GM and [3H]-thymidine incorporation assays, data were analyzed using mixed-effects models with random intercept, using GM-CSF concentration as a categorical variable. (F) Immunoblot demonstrating Erk and Akt hyperphosphorylation in D61Y;Cre+ mice compared with D61Y;Cre− mice, without normalization on successful knockout of p110α protein expression in 2 independent D61Y;Cre+;p110αFl/Fl mice. (G) Immunoblot demonstrating normalization of Erk and Akt hyperphosphorylation in 2 independent D61Y;Cre+;p110δD910A/D910A mice compared with 2 independent D61Y;Cre+ mice. (H) Representative flow cytometry analysis demonstrating increased megakaryocyte erythroid progenitor and decreased granulocyte macrophage progenitor in D61Y;Cre+ mice compared with D61Y;Cre− mice, as described previously.16 (I) Quantification of phenotypically defined bone marrow myeloid precursor distribution. *P = .02, with the D61Y;Cre+;p110δD910A/D910A group being significantly closer to D61Y;Cre− than the D61Y;Cre+;p110αFl/Fl group to D61Y;Cre−. Progenitor distribution between genotypes was analyzed by comparing Euclidian distance between mean vectors to quantify the similarity among group mean levels, with the P value determined using the bootstrapping resampling method.
Given the lack of effect of p110α loss, we hypothesized that PI3K catalytic subunit p110δ might specifically contribute to GOF Shp2-induced GM-CSF hypersensitivity. Indeed, Shp2D61Y/+;Mx1Cre+ mice homozygous for kinase-dead p110δ (Pik3cdD910A/D910A)17 demonstrated reduced spleen size (Figure 1B,C), normalized hypersensitivity to GM-CSF (Figure 1D,E), and reduced Akt hyperactivation (Figure 1G, compare lanes 7 and 8 with lanes 5 and 6). Although PI3K canonically activates Akt, genetic disruption of p110δ kinase activity also reduced GOF Shp2-induced Erk activation (Figure 1G, compare lanes 7 and 8 with 5 and 6), indicating that p110δ inhibition reduces positive crosstalk to the Ras-Erk pathway. Genetic inhibition of p110δ also normalized the skewed hematopoietic progenitor distribution (increased megakaryocyte erythroid progenitor, decreased granulocyte macrophage progenitor reported in Shp2D61Y/+;Mx1Cre+ mice,16 whereas genetic disruption of p110α failed to do so (Figure 1H,I). These findings reveal that p110δ, a highly expressed hematopoietic class IA PI3K isoform, is fundamental in GOF Shp2-induced myeloproliferative neoplasm.
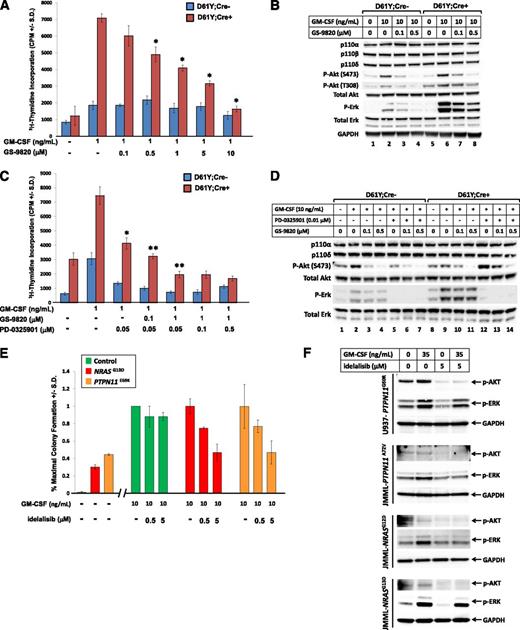
We next examined proliferation of GOF Shp2-expressing cells in response to the potent p110δ-specific inhibitors, GS-9820 and idelalisib, which are currently under investigation in clinical trials for hematological malignancies.22 Bone marrow LDMNCs from Shp2D61Y/+;Mx1Cre− and Shp2D61Y/+;Mx1Cre+ mice were treated with increasing concentrations of GS-9820, and GOF Shp2-expressing cells demonstrated a dose-dependent reduction in proliferation, whereas the wild-type (WT) Shp2-expressing cells were less sensitive (Figure 2A). As expected, GS-9820 decreased Akt phosphorylation and, importantly, also reduced Erk phosphorylation (Figure 2B), similar to the effects of genetic inhibition of p110δ (Figure 1G).
Pharmacologic inhibition of PI3K catalytic subunit p110δ reduces proliferation of GOF-Shp2-expressing cells and JMML primary cells. (A) Proliferation of bone marrow LDMNCs from polyI:polyC-treated Shp2D61Y/+;Mx1Cre− (D61Y;Cre−) and Shp2D61Y/+;Mx1Cre+ (D61Y;Cre+) mice in response to GM-CSF 1 ng/mL in the presence of increasing concentrations of the p110δ-specific inhibitor, GS-9820; n = 4. *P < .05 for D61Y;Cre+ cells treated with GS-9820 compared with no drug; statistics performed using unpaired, 2-tailed student’s t test. (B) Immunoblot demonstrating reduced phospho-Akt and phospho-Erk in response to increasing concentrations of GS-9820. (C) Proliferation of bone marrow LDMNCs from polyI:polyC-treated Shp2D61Y/+;Mx1Cre− (D61Y;Cre−) and Shp2D61Y/+;Mx1Cre+ (D61Y;Cre+) mice in response to GM-CSF 1 ng/mL in the presence of the MEK inhibitor, PD-0325901, and increasing concentrations of the p110δ-specific inhibitor, GS-9820; n = 4. *P = .0002 for D61Y;Cre+ cells treated with 0.05 μM PD0325901 vs no PD0325901. **P = .01 or **P = .0002 for D61Y;Cre+ cells treated with either 0.05 μM PD0325901 + 0.1 μM GS-9820 or 0.05 μM PD0325901 + 1 μM GS-9820, respectively, compared with 0.05 μM PD-0325901, statistics performed using unpaired, 2-tailed student’s t test. (D) Immunoblot demonstrating maximal inhibition of Erk and Akt activation in the presence of both PD0325901 and GS-9820. (E) Control human bone marrow LDMNCs or primary JMML LDMNCs (2 independent patient samples) were plated in duplicate in methylcellulose-based progenitor assays in the absence or presence of human GM-CSF 10 ng/mL and increasing concentrations of the PI3K p110δ-specific inhibitor, idelalisib. Data are represented as percentage maximal colony formation, calculated by dividing the number of colonies in each condition by the average number of colonies in the presence of 10 ng/mL GM-CSF. (F) Myelomonocytic cell line U937 or primary JMML LDMNCs (3 independent samples) were treated with GM-CSF, idelalisib alone, or GM-CSF plus idelalisib, followed by examination of AKT (S473) and ERK activation by immunoblot.
Pharmacologic inhibition of PI3K catalytic subunit p110δ reduces proliferation of GOF-Shp2-expressing cells and JMML primary cells. (A) Proliferation of bone marrow LDMNCs from polyI:polyC-treated Shp2D61Y/+;Mx1Cre− (D61Y;Cre−) and Shp2D61Y/+;Mx1Cre+ (D61Y;Cre+) mice in response to GM-CSF 1 ng/mL in the presence of increasing concentrations of the p110δ-specific inhibitor, GS-9820; n = 4. *P < .05 for D61Y;Cre+ cells treated with GS-9820 compared with no drug; statistics performed using unpaired, 2-tailed student’s t test. (B) Immunoblot demonstrating reduced phospho-Akt and phospho-Erk in response to increasing concentrations of GS-9820. (C) Proliferation of bone marrow LDMNCs from polyI:polyC-treated Shp2D61Y/+;Mx1Cre− (D61Y;Cre−) and Shp2D61Y/+;Mx1Cre+ (D61Y;Cre+) mice in response to GM-CSF 1 ng/mL in the presence of the MEK inhibitor, PD-0325901, and increasing concentrations of the p110δ-specific inhibitor, GS-9820; n = 4. *P = .0002 for D61Y;Cre+ cells treated with 0.05 μM PD0325901 vs no PD0325901. **P = .01 or **P = .0002 for D61Y;Cre+ cells treated with either 0.05 μM PD0325901 + 0.1 μM GS-9820 or 0.05 μM PD0325901 + 1 μM GS-9820, respectively, compared with 0.05 μM PD-0325901, statistics performed using unpaired, 2-tailed student’s t test. (D) Immunoblot demonstrating maximal inhibition of Erk and Akt activation in the presence of both PD0325901 and GS-9820. (E) Control human bone marrow LDMNCs or primary JMML LDMNCs (2 independent patient samples) were plated in duplicate in methylcellulose-based progenitor assays in the absence or presence of human GM-CSF 10 ng/mL and increasing concentrations of the PI3K p110δ-specific inhibitor, idelalisib. Data are represented as percentage maximal colony formation, calculated by dividing the number of colonies in each condition by the average number of colonies in the presence of 10 ng/mL GM-CSF. (F) Myelomonocytic cell line U937 or primary JMML LDMNCs (3 independent samples) were treated with GM-CSF, idelalisib alone, or GM-CSF plus idelalisib, followed by examination of AKT (S473) and ERK activation by immunoblot.
Abundant data indicate that MEK inhibition ameliorates GOF Kras-induced and LOF Nf1-induced disease.7,8 As both genetic and pharmacologic p110δ-specific inhibition reduces Ras-Erk signaling, we evaluated whether p110δ cooperates with the Ras-Erk pathway by examining whether inhibition of p110δ adds to or is redundant with MEK inhibition. Treatment with PD0325901 reduced proliferation of GOF Shp2-expressing cells, and addition of the p110δ-specific inhibitor, GS-9820, further reduced proliferation in a dose-dependent manner (Figure 2C). Addition of higher PD0325901 concentrations (0.1 and 0.5 μM) to 1 μM GS-9820 did not further reduce proliferation, indicating that relatively low doses of a MEK inhibitor, in combination with a p110δ inhibitor, achieve optimal reduction in proliferation. This is significant, as MEK inhibitors have significant toxicity.9,11,12
Biochemically, although GS-9820 reduced Akt phosphorylation and PD0325901 reduced Erk phosphorylation, PD0325901 induced a compensatory increase in Akt activation in cells expressing GOF Shp2 (Figure 2D, compare lane 12 with lane 9, observed in multiple experiments). Notably, this upregulation of Akt phosphorylation was not observed in WT Shp2-expressing cells (Figure 2D, compare lane 5 with lane 2). Addition of GS-9820 to PD0325901 reduced Akt phosphorylation levels to that observed with GS-9820 treatment alone (compare lane 14 with lane 11). These findings suggest that MEK inhibition reduces proliferation by inhibiting Erk activation, and p110δ inhibition further reduces proliferation because of effective inhibition of residual (and potentially upregulated) Akt activation in GOF Shp2-expressing cells. Furthermore, the differential effect of MEK inhibition on Akt activation in the GOF Shp2 vs WT Shp2-expressing cells could provide a therapeutic window permitting selective inhibition of disease vs normal cells.
Finally, we examined the effect of idelalisib22 on primary JMML cell-derived colony growth. Although a single JMML sample failed to demonstrate a response (up to 5 μM idelalisib, data not shown), 2 independent samples demonstrated a dose-dependent reduction in colony formation, whereas control bone marrow cells demonstrated only a modest response (Figure 2E). Activation of AKT and ERK was also reduced by idelalisib in the myelomonocytic cell line, U937, and in primary JMML patient samples (Figure 2F). Notably, 5 μM is an achievable concentration of idelalisib in humans.24,25
Collectively, our findings demonstrate that PI3K catalytic subunit p110δ works jointly with the Ras-Erk signaling pathway to promote GOF Shp2-induced hypersensitivity to GM-CSF. These studies support further investigation into the putative selective role of p110δ in JMML, as well as into the use of p110δ-specific inhibitors, alone or in combination with MEK inhibitors, as a novel therapeutic strategy for JMML.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors gratefully acknowledge the administrative assistance of Marilyn L. Wales.
This work was supported by grants from the National Institutes of Health (HL104867 [to C.B.G.]; HL092524 and CA134777 [to R.J.C.]; R01HL077177, R01HL075816 and R01CA134777 [to R.K.]; and R37 CA49432 [to B.G.N.]), the Riley Children’s Foundation, the Indiana University-Purdue University Indianapolis Office of the Vice Chancellor for Research, and Gilead Sciences (for providing p110δ inhibitors). B.G.N. is a Canada Research Chair, Tier 1, and is partially supported by the Princess Margaret Foundation and the Ontario Ministry of Health and Long Term Care.
Authorship
Contribution: C.B.G. and X.J.L. directed and performed research and analyzed results; R.S.M. designed and performed research; G.C., M.K., B.V., and B.G.N. provided reagent; Z.L. performed statistical analysis; M.L.L. provided primary JMML samples; B.J.L. and R.K. designed research; and R.J.C. directed research, analyzed results, and wrote the manuscript.
Conflict-of-interest disclosure: B.V. is a consultant to Karus Therapeutics and Activiomics.
Correspondence: Rebecca J. Chan, associate professor, Indiana University School of Medicine, Department of Pediatrics, Herman B Wells Center for Pediatric Research; e-mail: rchan@iu.edu.

![Figure 1. Genetic inhibition of PI3K catalytic subunit p110δ, but not p110α, normalizes GOF Shp2-induced hypersensitivity to GM-CSF. (A) and (B) Representative spleens from polyI:polyC-treated Shp2D61Y/+;Mx1Cre− mice (D61Y;Cre−, negative controls), Shp2D61Y/+;Mx1Cre+ mice (D61Y;Cre+, positive controls), and Shp2D61Y/+;Pik3caflox/flox;Mx1Cre+ (D61Y;Cre+;p110αFl/Fl) or Shp2D61Y/+;Pik3cdD910A/D910A;Mx1Cre+ (D61Y;Cre+;p110δD910A/D910A) mice. (C) Quantification of spleen weight:body weight (n = 6 to 9 mice per group). *P = .02 for D61Y;Cre− vs D61Y;Cre+; **P = .04 for D61Y;Cre+;p110δD910A/D910A vs D61Y;Cre+, statistics by unpaired, 2-tailed student’s t test. (D) Bone marrow LDMNCs from 6 to 8 mice per genotype were plated in methylcellulose, and colony-forming unit-GM assays were carried out in duplicate in 3 to 4 independent experiments. Data are represented as percentage maximal colony formation, calculated by dividing the number of colonies at each GM-CSF concentration by the average number of colonies at GM-CSF 10 ng/mL. *P = .0005 for D61Y;Cre+;p110δD910A/D910A vs D61Y;Cre+. (E) Bone marrow LDMNCs from 6 to 8 mice per genotype were subjected to [3H]-thymidine incorporation assays in replicates of 6 in 3 to 4 independent experiments. ^P < .0001 for D61Y;Cre+;p110δD910A/D910A vs D61Y;Cre+. For colony-forming unit-GM and [3H]-thymidine incorporation assays, data were analyzed using mixed-effects models with random intercept, using GM-CSF concentration as a categorical variable. (F) Immunoblot demonstrating Erk and Akt hyperphosphorylation in D61Y;Cre+ mice compared with D61Y;Cre− mice, without normalization on successful knockout of p110α protein expression in 2 independent D61Y;Cre+;p110αFl/Fl mice. (G) Immunoblot demonstrating normalization of Erk and Akt hyperphosphorylation in 2 independent D61Y;Cre+;p110δD910A/D910A mice compared with 2 independent D61Y;Cre+ mice. (H) Representative flow cytometry analysis demonstrating increased megakaryocyte erythroid progenitor and decreased granulocyte macrophage progenitor in D61Y;Cre+ mice compared with D61Y;Cre− mice, as described previously.16 (I) Quantification of phenotypically defined bone marrow myeloid precursor distribution. *P = .02, with the D61Y;Cre+;p110δD910A/D910A group being significantly closer to D61Y;Cre− than the D61Y;Cre+;p110αFl/Fl group to D61Y;Cre−. Progenitor distribution between genotypes was analyzed by comparing Euclidian distance between mean vectors to quantify the similarity among group mean levels, with the P value determined using the bootstrapping resampling method.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/18/10.1182_blood-2013-10-535104/4/m_2838f1.jpeg?Expires=1769140321&Signature=QiAHVvaRddB72056CZw3WBExJbLL2JGC8Rc~3Pt~rBdxMiFxM9s1Hu4XtRJ3gzKJFw4SllclFa-x7cKd3Ibdy5i9M-7dgaxOQ2PdfzRCjNNASScO8Kt2v5myIQTnTRp7Efj1lFYdnxt8h~SumPZ4T-UtqrWlJ9XsPKjRGXwt5nMLW4zg44leW7Kqbemt68f~IgfQq9odFa50OM1n6mEHXDn3yYO~nzOmkDJgA9fQ~9pEDoc62TxQE8PXS58QwtxRvGKjc6KKsT9-qevPYnJxCjD8pv4L484KwAsaqU~Fz6jZ~~SWQL5Y~2VMl~9S7tJpt-UFD1e6NqgiBjC32Oejng__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)