In this issue of Blood, Goodwin et al investigate the pathogenesis of juvenile myelomonocytic leukemia (JMML), demonstrating that mutant Shp2 induces granulocyte macrophage–colony-stimulating factor (GM-CSF) hypersensitivity and that the p110δ subunit of phosphatidylinositol 3-kinase (PI3K) further promotes this dysregulation.1
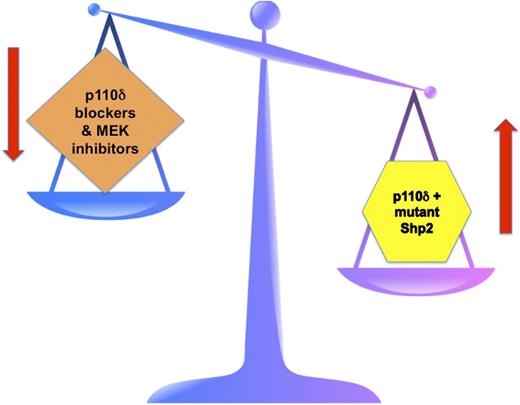
The right side of the scale depicts JMML pathogenesis with Shp2 mutants favoring uncontrolled leukemic cell growth. This is further promoted by the p110δ subunit of PI3K. The left side of the scale depicts the goal of targeted therapy in JMML. By blocking p110δ PI3K and by inhibiting MEK, hopefully the “scales of cell growth” will once again balance out.
The right side of the scale depicts JMML pathogenesis with Shp2 mutants favoring uncontrolled leukemic cell growth. This is further promoted by the p110δ subunit of PI3K. The left side of the scale depicts the goal of targeted therapy in JMML. By blocking p110δ PI3K and by inhibiting MEK, hopefully the “scales of cell growth” will once again balance out.
JMML has been classified by the World Health Organization as a mixed myeloproliferative neoplasm/myelodysplastic disorder. Many years ago, we demonstrated that a hallmark of JMML was that JMML progenitors displayed an in vitro cellular biology characteristic of a selective, hypersensitive growth pattern to GM-CSF while demonstrating a normal sensitivity to interleukin-3 (IL-3).2 Subsequent investigations over the ensuing years by several groups have delineated mutations in NF1, RAS, PTPN11, and CBL in nearly 85% of JMML patients. These mutations are almost always exclusive of each other, and all of them hyperactivate and dysregulate the Ras signaling pathway, with an end result being the hallmark selective GM-CSF hypersensitivity. No one has yet been able to explain why there is selective hypersensitivity to GM-CSF with IL-3 sensitivity being spared, even though they share a common β subunit to their cell-surface receptor. More recently, additional but secondary mutations have been described in SETBP1 and JAK3.3 Given this knowledge of one specific pathway appearing to account for the disease phenotype, many have attempted to disrupt JMML signaling via Ras. But, just like other investigators in other cancers, efforts to normalize Ras signaling have fallen dismally short. In 2013, National Cancer Institute Director Harold Varmus initiated a new push to formulate agents capable of blocking Ras.
JMML is not the only proliferative disease of the marrow to show cytokine hypersensitivity. As far back as 1974, Prchal and Axelrad first described hypersensitivity of polycythemia vera (PV) progenitor cells to erythropoietin (Epo).4 Abnormal erythroid colony growth in vitro was a hallmark of PV long before the JAK2 V617F mutation, observed in ∼98% of PV patients, took center stage. Epo hypersensitivity has also been noted in primary and familial congenital polycythemia.5 In at least some of these families, this Epo hypersensitivity is the result of Epo receptor mutations.6 Additionally, a recent paper shows that the Cbl protein (encoded by the CBL gene) interacts with JAK2 and with the p85 regulatory subunit of PI3K.7 An essential negative regulatory mechanism to turn off Epo signaling occurs when Epo induces Cbl to ubiquitinate the p85 subunit of PI3K, which ultimately leads to Epo receptor endocytosis, which terminates Epo signaling. Mutated Epo receptors, and/or Cbl deficiencies, result in Epo hypersensitivity.
The contrary situation is found in JMML, where despite diligent searching, no mutations of either the α or the β subunit of the GM-CSF receptor have ever been found in any JMML patient. Rather, the mutations occur downstream in cytoplasmic signaling components. Several groups, including Goodwin, Chan, and colleagues, previously demonstrated that mutations in PTPN11, which encodes Shp2, induces GM-CSF hypersensitivity.8 In this current paper, they convincingly demonstrate that the p110δ subunit of PI3K helps to promote this dysregulation.1 Such crosstalk between the PI3K and Ras signaling pathways is not novel.9 But importantly, demonstrating such crosstalk and further promotion of a dysregulated effect leading to more leukemogenic growth has significant implications in JMML, especially when we are thinking about targeted therapies. Goodwin et al hypothesized that blocking p110δ, along with blocking mitogen-activated protein kinase kinase (MEK) downstream in the Ras pathway, may prove effective in JMML. They present in vitro evidence in Figure 2 in their paper that this might work in a clinical setting. Further, the level of MEK blockade might be able to be reduced, thus dampening the untoward side effects of MEK inhibitors, which have been a recurring theme in clinical trials of various MEK inhibitors in various cancers. The concept for therapy in JMML is one of “balancing the scales” (see figure). PTPN11 mutations in JMML lead to mutant Shp2 and subsequent hyperactivation of the Ras signaling pathway. In similar fashion, mutations in NF1, RAS, and CBL have similar effects in JMML. Thus, mutant Shp2 tips the scales in JMML in favor of uncontrolled growth that cannot be turned off. The PI3K pathway gets into the act by the p110δ subunit further promoting the effects of Shp2. So, the scales are even more tipped in favor of uncontrolled growth. The hope is that by specifically blocking the p110δ subunit of PI3K and by inhibiting MEK downstream of Ras, we may coordinately be able to in effect rebalance the scales and once again achieve a normal growth pattern.
Combining the specific p110δ inhibitor, idelalisib (which is not yet US Food and Drug Administration approved), with other agents has been recently reported wherein idelalisib was combined with rituximab in chronic lymphocytic leukemia.10 Currently, the only known effective therapy for JMML is allogeneic stem cell transplantation. Other forms of chemotherapy, even induction-type therapy, have not been consistently effective in JMML. Because we understand so much regarding JMML pathogenesis and dysregulated signaling pathways, certainly targeted therapy in this disease makes sense. Thus, the horizon appears brighter for effective therapeutics in JMML more so than it probably ever has been. But the cautionary note is that blocking 2 common signaling pathways will require careful study in clinical trials. In our efforts to balance the scales, we do not want to create new problems.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal