Key Points
Overall, our results suggest that NOTCH2 and FLT3 aberrant splicing is a common event in AML that correlates with disease status and may correlate with disease outcomes.
Selected variants of NOTCH2 and FLT3 transcripts were detected in a significant number of AML patients and could be useful as disease markers.
Abstract
Our previous studies revealed an increase in alternative splicing of multiple RNAs in cells from patients with acute myeloid leukemia (AML) compared with CD34+ bone marrow cells from normal donors. Aberrantly spliced genes included a number of oncogenes, tumor suppressor genes, and genes involved in regulation of apoptosis, cell cycle, and cell differentiation. Among the most commonly mis-spliced genes (>70% of AML patients) were 2, NOTCH2 and FLT3, that encode myeloid cell surface proteins. The splice variants of NOTCH2 and FLT3 resulted from complete or partial exon skipping and utilization of cryptic splice sites. Longitudinal analyses suggested that NOTCH2 and FLT3 aberrant splicing correlated with disease status. Correlation analyses between splice variants of these genes and clinical features of patients showed an association between NOTCH2-Va splice variant and overall survival of patients. Our results suggest that NOTCH2 and FLT3 mis-splicing is a common characteristic of AML and has the potential to generate transcripts encoding proteins with altered function. Thus, splice variants of these genes might provide disease markers and targets for novel therapeutics.
Introduction
In acute myeloid leukemia (AML), the leukemic blasts are arrested at various stages of granulocytic and monocytic differentiation.1 Most patients with this incurable disease have chromosome translocations or mutations that promote self-renewal of leukemic stem cells, block differentiation, enhance growth, and block apoptosis.2,3 In addition, epigenetic abnormalities are very common in AML. Recently discovered gene mutations in AML, such as in TET2, DNMT3A and EZH2, can potentially alter global gene expression by modulating epigenetic regulatory mechanisms.4,5 Understanding the epigenetic abnormalities in AML is of great interest, particularly given their potential role as targets for therapy.
Alternative splicing (AS) is a normal epigenetic phenomenon and a key regulator of gene expression. AS produces multiple transcripts and consequently multiple proteins from a single gene, thereby increasing the use of genetic information in any given cell.6,7 Even though AS is a normal process, mistakes are inevitable. Aberrant splicing events have been linked to malignant transformation, particularly in genes associated with susceptibility and/or progression of cancer.6,8-16 For some cancers, these splicing alterations create functionally significant biomarkers.9,12,14,17-21
Using exome sequencing, frequent mutations in genes involved in regulating splicing were identified in myelodysplasia (MDS) and in some AML patients.22 Recently, using a single nucleotide polymorphism (SNP) array and targeted sequencing of 1000 genes, novel somatic mutations of splicing factor SFPQ and splicing regulator CTCF were identified in 10% of AML patients.23,24 Graubert et al reported that mutations in the U2AF1 gene contribute to progression from MDS to secondary AML.25 Prior studies of AS identified several genes mis-spliced in AML patients, including AML1, Gfi1b, CD96, survivin-2B, and GSK3beta.8,15,21,26-29 In our previous study, we used exon arrays to compare genome-wide gene-splicing events in AML vs normal CD34+ cells and observed widespread differences.30 Among the most frequently mis-spliced genes in AML patients were NOTCH2 and FLT3, and here we describe these events in detail.
Materials and methods
Tissue and cell preparation
Diagnostic samples from 193 AML patients were obtained after institutional review board–approved informed consent. This study was conducted in accordance with the Declaration of Helsinki. Supplemental Table 1 on the Blood Web site describes the demographic and clinical characteristics of the patients with AML that were included in our studies. Details of the sample preparations are described in supplemental Methods.
Reverse-transcriptase polymerase chain reaction
cDNA for the reverse transcriptase-polymerase chain reaction (RT-PCR) and PCR reactions was prepared as described previously.31 The NOTCH2 and FLT3 gene primer sequences are provided in supplemental Table 2. Obtained PCR products were detected using gel electrophoresis or capillary electrophoresis and DNA fragment analysis on the ABI31/30XL DNA genetic analyzer (Life Technologies). Capillary electrophoresis and DNA fragment analyses were carried out as described previously.31
Cloning and sequencing experiments
cDNAs for cloning and sequencing were prepared from 1 μg RNA isolated from the blasts of 8 AML patients all expressing NOTCH2 and FLT3 splice variants. TOPO-TA cloning and PCR reactions for sequencing were performed as described previously.31 Positive colonies containing NOTCH2 and FLT3 plasmids were identified through testing individual bacterial colonies by colony PCR using gene-specific primer sets (supplemental Table 2). Sequencing was performed at the Dana-Farber Cancer Institute (DFCI)-Molecular Biology Laboratory facility on the ABI3730 DNA Genetic Analyzer.
Real-time quantitative RT-PCR
cDNAs for quantitative RT-PCR (qRT-PCR) were prepared as above. qRT-PCR was performed using inventoried Taq-man assays for HES1, DTX1, and HEY1 with the TaqMan Universal PCR Master Mix from Life Technologies. All samples were run in triplicate on a 7500Real-Time PCR system. Results were analyzed using the relative standard curve method. In AML patients, the expression level of each gene was determined by comparing it to the gene level in CD34+ cells obtained from 3 normal donor (ND) bone marrow (BM) aspirates (AllCells).
Confocal laser scanning microscopy
A NOTCH2-FL open reading frame was obtained from OreGene Inc. NOTCH2-Va and NOTCH2-Vb transcripts were engineered according to the splicing modes detected by cloning and sequencing analysis of 8 AML patient samples. NOTCH2 splice variants were subcloned into a pCMV6-AN-(GFP plasmid (OriGene). FLT3-FL and splice variants were generated by a gene synthesis approach (GeneArt Services; Life Technologies) followed by cloning them into a pDONR221 vector and subcloning into a pcDNA6.2_C-EmGFP destination vector according to the manufacturer’s recommendations (Life Technologies). Details of confocal microscopy experiments are described in supplemental Methods.
Western blotting
HEK293T cells expressing Notch2-FL-GFP, Notch2-Va-GFP, and Notch2-Vb-GFP were cocultured 24 hours with 3T3 cells or 3T3 cells expressing Jagged2 previously generated in our laboratory. After incubation, cells were scraped and washed, and GFP-positive cells were sorted on the flow sorter. From sorted cells, whole cell lysates, and nuclear fractions were obtained using the Nuclear Extract kit (Active Motif) according to the manufacturer’s recommendations. In other experiments, HEK293T cells expressing FLT3-FL, FLT3-Va, and FLT3-Vb splice variants were serum deprived, and 24 hours later, cells were stimulated for 10 minutes with Flt3L. After stimulation, whole cell lysates were obtained using the RIPA lyses buffer from Cell Signaling Technology. Cell lysates were separated by gradient (4-12%) sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to PVDF membranes. Membranes were probed with the appropriate antibodies and visualized using standard techniques. As loading controls, we used anti-actin or anti-TBP (TATA box binding protein) antibodies. For western blotting, anti-Notch2 anti-Hes1, anti-actin, and anti-TBP were purchased from Cell Signaling Technology, anti-pTyr antibody (Clone 4G10) was from Millipore, and anti-Flt3 antibodies were from eBioscience.
Statistical analysis
Statistical analysis is described in supplemental Methods. Concordance index, confidence interval (CI), and corresponding P values were computed with use of the survcomp package (version 1.6.0) of the R statistical software (version 2.15.2).32,33 Associations between splice variant expression and overall survival of AML patients were assessed using univariate Cox regression analyses, followed by the Kaplan-Meier analyses with the R survival package (version 2.36-14). All P values were 2-sided, and if necessary, nominal P values were corrected for multiple testing by estimating the false discovery rate (FDR).34
Results
Identification of splicing patterns in NOTCH2 and FLT3 in patients with AML
In preliminary studies, we looked for splicing abnormalities in 66 AML patients and 10 NDs using exon arrays.30 In addition, RNA-seq analysis was performed in 12 of these AML patients and 3 of the NDs (unpublished data) to validate exon array studies. We identified widespread RNA splicing abnormalities in AML patients compared with NDs (supplemental Table 3A). For the current study, we focused on genes encoding myeloid cell surface proteins Notch2 and Flt3 that were most frequently spliced in AML patients.
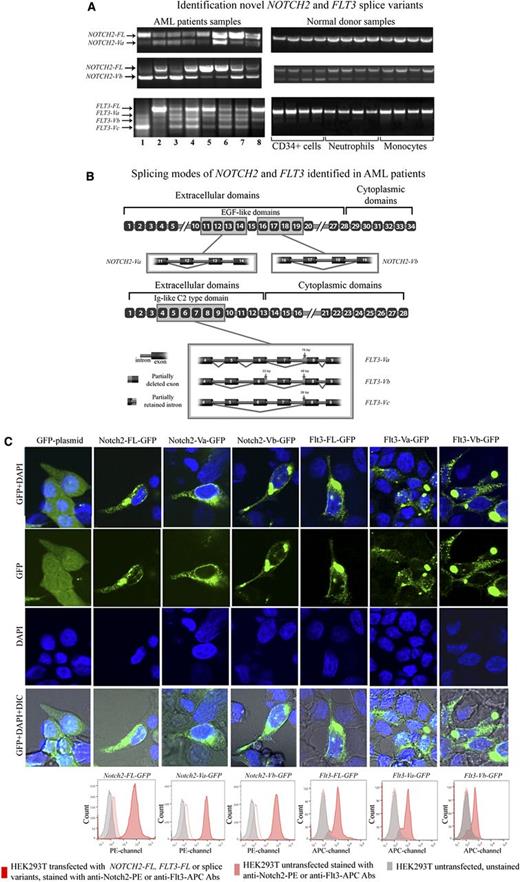
To determine the precise splicing pattern for NOTCH2 and FLT3, we performed RT-PCR followed by cloning and sequencing of the cDNAs of these transcripts from the blasts of 8 AML patients and 12 NDs (Figure 1A). Two novel variants of NOTCH2 and 3 novel variants of FLT3, referred to as NOTCH2-Va and NOTCH2-Vb, and FLT3-Va, FLT3-Vb, and FLT3-Vc. respectively, were identified (Figure 1A). Except for NOTCH2-Vb, NOTCH2, and FLT3 splice variants were absent from NDs. The levels of NOTCH2-Vb were elevated in some AML samples compared with the NDs (Figure 1A).
Identification novel NOTCH2 and FLT3 splice variants. (A) This figure displays NOTCH2 and FLT3 RT-PCR agarose gel electrophoresis results. (B) This figure displays AML-associated splicing patterns of NOTCH2 and FLT3 splice variants. After cloning and sequencing experiments, novel splice variant sequences were identified through alignment with published sequences for human NOTCH2 and FLT3 mRNA (gi:317008612 and gi:178535 NCBI, respectively). NOTCH2-Va and NOTCH2-Vb are the result of complete deletions of exon 12 (111 bp) or exons 17 (120 bp) and 18 (153 bp), respectively. These splicing aberrations did not cause any frame shifts. FLT3-Va is a result of skipping exon 7 (140 bp) and a 76-bp deletion of exon 8 at the 5′ end, whereas FLT3-Vb was due to skipping exons 5 (130 bp) and 7 (140 bp), with partial deletions of exons 6 (33-bp deletion at the 3′ end of exon 6) and 8 (48-bp deletion at the 5′ end of exon 8). FLT3-Vc appears to be most severely affected by splicing events compared with other FLT3 splice variants. FLT3-Vc is a result of skipping of exons 5 (130 bp), 6 (128 bp), and 7 (140 bp) and a 26-bp deletion of exon 8 at the 5′ end. As a result of these aberrations 240 bp are spliced out from FLT3-Va, 351 bp from FLT3-Vb, and 398 bp from FLT3-Vc. Thus, these aberrations did not cause a frame shift on FLT3-Va and FLT3-Vb transcripts, whereas FLT3-Vc transcripts were subjected to a frame shift. For both NOTCH2 and FLT3, the splice variant transcripts retained their original start codons and conserved signal sequences. Also, bioinformatics and alignment analysis showed that exons affected by aberrant splicing events mapped to extracellular domains of the Notch2 and Flt3 protein sequences. As a result of the splicing alterations, 3 EGF-like domains were entirely deleted from the Notch2 protein and 2 were partially affected. Splicing alterations caused deletion of the entire Ig-like C2 type domain on the Flt3 protein. In the figure, we zoomed out gene segments of NOTCH2 and FLT3 that we cloned and sequenced. In the figure, yellow-boxed exons are those that are affected by the splicing events. (C) HEK293T cells were transiently transfected with NOTCH2-FL-GFP, NOTCH2-Va-GFP, NOTCH2-Vb-GFP, FLT3-FL-GFP, FLT3-Va-GFP, FLT3-Vb-GFP, or GFP backbone plasmids. Seventy-two hours after transfection cells were stained using the live cell nuclear staining reagent (Life Technologies) that includes DAPI. Cells were visualized under the Zeiss 710 confocal laser-scanning microscope. On the figure, GFP signal shown in green, and DAPI signal in blue. HEK293T cells transfected with NOTCH2 and FLT3 splice-variants were stained with anti-Notch2-PE (16F11, eBioscience) and anti-Flt3-APC (BV10A4H2, eBioscience) antibodies, then membrane expression of these variants was evaluated by flow cytometry. On the flow cytometry histograms, gray peaks represent HEK293T cells untransfected and unstained, pink peaks represent HEK293T cells untransfected and stained with anti-Notch2-PE or anti-Flt3-APC antibodies, red peaks represent HEK293T cells transfected with NOTCH2-FL, FLT3-FL, or splice variants tagged with GFP. These cells were stained with anti-Notch2-PE or anti-Flt3-APC antibodies and staining was determined in GFP gated cells. IgG stained controls are provided in Supplemental Figure 1A.
Identification novel NOTCH2 and FLT3 splice variants. (A) This figure displays NOTCH2 and FLT3 RT-PCR agarose gel electrophoresis results. (B) This figure displays AML-associated splicing patterns of NOTCH2 and FLT3 splice variants. After cloning and sequencing experiments, novel splice variant sequences were identified through alignment with published sequences for human NOTCH2 and FLT3 mRNA (gi:317008612 and gi:178535 NCBI, respectively). NOTCH2-Va and NOTCH2-Vb are the result of complete deletions of exon 12 (111 bp) or exons 17 (120 bp) and 18 (153 bp), respectively. These splicing aberrations did not cause any frame shifts. FLT3-Va is a result of skipping exon 7 (140 bp) and a 76-bp deletion of exon 8 at the 5′ end, whereas FLT3-Vb was due to skipping exons 5 (130 bp) and 7 (140 bp), with partial deletions of exons 6 (33-bp deletion at the 3′ end of exon 6) and 8 (48-bp deletion at the 5′ end of exon 8). FLT3-Vc appears to be most severely affected by splicing events compared with other FLT3 splice variants. FLT3-Vc is a result of skipping of exons 5 (130 bp), 6 (128 bp), and 7 (140 bp) and a 26-bp deletion of exon 8 at the 5′ end. As a result of these aberrations 240 bp are spliced out from FLT3-Va, 351 bp from FLT3-Vb, and 398 bp from FLT3-Vc. Thus, these aberrations did not cause a frame shift on FLT3-Va and FLT3-Vb transcripts, whereas FLT3-Vc transcripts were subjected to a frame shift. For both NOTCH2 and FLT3, the splice variant transcripts retained their original start codons and conserved signal sequences. Also, bioinformatics and alignment analysis showed that exons affected by aberrant splicing events mapped to extracellular domains of the Notch2 and Flt3 protein sequences. As a result of the splicing alterations, 3 EGF-like domains were entirely deleted from the Notch2 protein and 2 were partially affected. Splicing alterations caused deletion of the entire Ig-like C2 type domain on the Flt3 protein. In the figure, we zoomed out gene segments of NOTCH2 and FLT3 that we cloned and sequenced. In the figure, yellow-boxed exons are those that are affected by the splicing events. (C) HEK293T cells were transiently transfected with NOTCH2-FL-GFP, NOTCH2-Va-GFP, NOTCH2-Vb-GFP, FLT3-FL-GFP, FLT3-Va-GFP, FLT3-Vb-GFP, or GFP backbone plasmids. Seventy-two hours after transfection cells were stained using the live cell nuclear staining reagent (Life Technologies) that includes DAPI. Cells were visualized under the Zeiss 710 confocal laser-scanning microscope. On the figure, GFP signal shown in green, and DAPI signal in blue. HEK293T cells transfected with NOTCH2 and FLT3 splice-variants were stained with anti-Notch2-PE (16F11, eBioscience) and anti-Flt3-APC (BV10A4H2, eBioscience) antibodies, then membrane expression of these variants was evaluated by flow cytometry. On the flow cytometry histograms, gray peaks represent HEK293T cells untransfected and unstained, pink peaks represent HEK293T cells untransfected and stained with anti-Notch2-PE or anti-Flt3-APC antibodies, red peaks represent HEK293T cells transfected with NOTCH2-FL, FLT3-FL, or splice variants tagged with GFP. These cells were stained with anti-Notch2-PE or anti-Flt3-APC antibodies and staining was determined in GFP gated cells. IgG stained controls are provided in Supplemental Figure 1A.
Sequence alignment indicated that splice variants of NOTCH2 and FLT3 derived from exon skipping and/or partial deletions of some exons (Figure 1B). Alignment followed by bioinformatic analysis showed that some of these splicing events avoided frame shifts (NOTCH2-Va, NOTCH2-Vb, FLT3-Va, and FLT3-Vb). whereas other combinations of AS led to frame-shifted transcripts (eg, for FLT3-Vc; Figure 1B). To determine whether splice variants of NOTCH2 and FLT3 encode proteins, we generated NOTCH2 and FLT3 splice variant plasmids tagged with GFP and expressed them in HEK293T cells. Transfected cells were harvested and examined by laser-scanning confocal microscopy and by flow cytomerty (Figure 1C; supplemental Figure 1A). These analyses demonstrated that Notch2-Va-GFP, Notch2-Vb-GFP, Flt3-Va-GFP, and Flt3-Vb-GFP proteins were readily d in HEK293T cells. Further, flow cytometric analyses with anti-Notch2 conjugated to phycoerythrin (PE) and anti-Flt3 conjugated to allophycocyanin (APC) showed that a significant amount of Notch2 and Flt3 splice variant proteins were expressed on the cell surface. Because the predicted difference between Notch2-FL or Flt3-FL and their splice variants is small, our ability to detect splice variant proteins in AML patients or cell lines using western blotting was felt to be unreliable. Antibody production against the novel Notch2 and Flt3 splice variant proteins is underway.
Correlation of NOTCH2 splice variants with NOTCH2 target gene expression
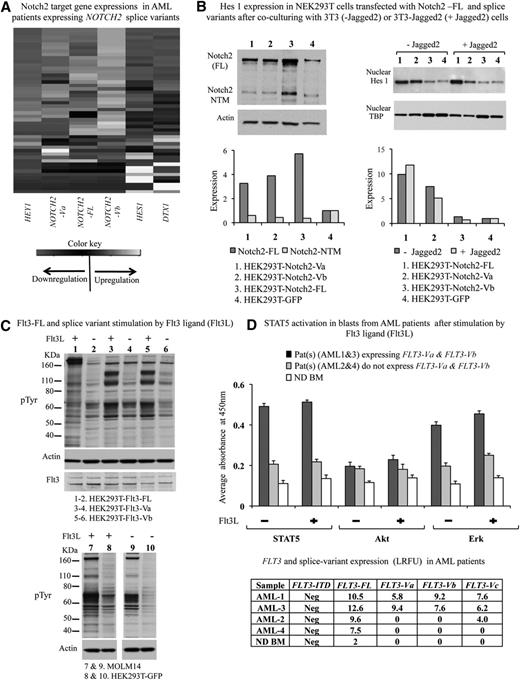
We evaluated the expression of 3 known Notch target genes, HES1, HEY1, and DTX1, in 49 AML patients expressing the NOTCH2 splice variant using Taqman gene expression assays. As a control, CD34+ BM cells from 3 NDs were used. Expression levels of HES1, HEY1, and DTX1 in AML patients were compared with expression levels of these genes in NDs. We observed down-regulation of HES1 and DTX1 transcripts (on average, 30-fold) in patients expressing both NOTCH2 splice variants in combination with NOTCH2-FL or expressing NOTCH2-Vb and NOTCH2-FL transcripts compared with expression levels in normal CD34+ cells (Figure 2A). We observed low expression of the HEY1 gene in all samples regardless of expression levels of NOTCH2-FL or splice variants (Figure 2A).
Functional effects of NOTCH2 and FLT3 splice variants. (A) This figure displays unsupervised clustering analyses results obtained from the TaqMan gene expression assays of Notch2 target genes HES1, DXT1 and HEY1 carried out in 49 AML patients expressing NOTCH2-FL, FLT3-FL, and their splice variants. Expression levels of NOTCH2-FL, FLT3-FL, and their splice variants were determined by RT-PCR and DNA fragment analysis described in this paper. In these studies, relative transcript expressions are calculated compared with the expression levels of the corresponding transcripts detected in NDs. (B) This figure is a summary of coculture experiments performed 3 times. HEK293T cells expressing NOTCH2-FL-GFP, NOTCH2-Va-GFP, and NOTCH2-Vb-GFP were cocultured 24 hours with 3T3 cells (−Jagged2) or 3T3 cells expressing Jagged2 (+Jagged2). After incubation, cells were scraped, and GFP-positive cells were sorted. Western blotting analysis for Notch2 and Hes 1 was performed as described in “Materials and methods”. Western blots were quantified using the ImageJ software (http://rsb.info.nih.gov/ij). Densitometry measurements were normalized to loading control amount. (C) FLT3-FL, FLT3-Va, and FLT3Vb splice variants were stably expressed in HEK293T cells. Serum-deprived, transfected cells were stimulated for 10 minutes with FLT3L. Cellular tyrosine phosphorylation was analyzed by immunoblotting with a pTyr antibody (Clone 4G10 from Millipore). Elevation of tyrosine phosphorylation of bands at approximately 100 kDa was determined compared with FLT3L unstimulated cells. Additionally, in the samples, Flt3 expression was determined by immunoblotting using anti-FLT3 antibodies from eBiosciences. As a loading control, the same membranes were reprobed with anti-actin antibody. As a control, MOLM14 and HEK293T-GFP stimulated and unstimulated cell lysates were used for pTyr immunoblotting analyses. (D) Serum-starved blasts from patients were stimulated for 10 minutes with 100 ng/mL Flt3L and washed, and cell lysates were prepared to measure phosphorylation levels of STAT5, Akt, and Erk using InstantOne enzyme-linked immunosorbent assay kits according to the manufacturer’s suggestions. Absorbance was measured at 450 nm using an automated enzyme-linked immunosorbent assay plate reader. In the figure, results are presented as bar graphs. The x-axis shows the samples analyzed, and the y-axis displays the phosphorylation level as an absorbance. Results obtained from positive and negative control samples are not displayed on the graph. Expression levels of FLT3-FL and its splice variants in patient samples are reported in a table included in this figure and presented as RFU = log2RFU (relative fluorescence units, described in Figure 3).
Functional effects of NOTCH2 and FLT3 splice variants. (A) This figure displays unsupervised clustering analyses results obtained from the TaqMan gene expression assays of Notch2 target genes HES1, DXT1 and HEY1 carried out in 49 AML patients expressing NOTCH2-FL, FLT3-FL, and their splice variants. Expression levels of NOTCH2-FL, FLT3-FL, and their splice variants were determined by RT-PCR and DNA fragment analysis described in this paper. In these studies, relative transcript expressions are calculated compared with the expression levels of the corresponding transcripts detected in NDs. (B) This figure is a summary of coculture experiments performed 3 times. HEK293T cells expressing NOTCH2-FL-GFP, NOTCH2-Va-GFP, and NOTCH2-Vb-GFP were cocultured 24 hours with 3T3 cells (−Jagged2) or 3T3 cells expressing Jagged2 (+Jagged2). After incubation, cells were scraped, and GFP-positive cells were sorted. Western blotting analysis for Notch2 and Hes 1 was performed as described in “Materials and methods”. Western blots were quantified using the ImageJ software (http://rsb.info.nih.gov/ij). Densitometry measurements were normalized to loading control amount. (C) FLT3-FL, FLT3-Va, and FLT3Vb splice variants were stably expressed in HEK293T cells. Serum-deprived, transfected cells were stimulated for 10 minutes with FLT3L. Cellular tyrosine phosphorylation was analyzed by immunoblotting with a pTyr antibody (Clone 4G10 from Millipore). Elevation of tyrosine phosphorylation of bands at approximately 100 kDa was determined compared with FLT3L unstimulated cells. Additionally, in the samples, Flt3 expression was determined by immunoblotting using anti-FLT3 antibodies from eBiosciences. As a loading control, the same membranes were reprobed with anti-actin antibody. As a control, MOLM14 and HEK293T-GFP stimulated and unstimulated cell lysates were used for pTyr immunoblotting analyses. (D) Serum-starved blasts from patients were stimulated for 10 minutes with 100 ng/mL Flt3L and washed, and cell lysates were prepared to measure phosphorylation levels of STAT5, Akt, and Erk using InstantOne enzyme-linked immunosorbent assay kits according to the manufacturer’s suggestions. Absorbance was measured at 450 nm using an automated enzyme-linked immunosorbent assay plate reader. In the figure, results are presented as bar graphs. The x-axis shows the samples analyzed, and the y-axis displays the phosphorylation level as an absorbance. Results obtained from positive and negative control samples are not displayed on the graph. Expression levels of FLT3-FL and its splice variants in patient samples are reported in a table included in this figure and presented as RFU = log2RFU (relative fluorescence units, described in Figure 3).
In comparing expression levels of target genes across the AML samples, expression levels of HES1, HEY1, and DTX1 genes were generally inversely correlated with expression of NOTCH2 splice variants or NOTCH2-FL and relatively higher in patient samples expressing only NOTCH2-FL or not expressing any NOTCH2 transcripts (Figure 2A). However, these results were not statistically significant (P < .1) as determined by the Spearman rank correlation coefficient test (analysis results not shown).
In an effort to determine whether splice variants of NOTCH2 might be functional, HEK293T cells were transfected with NOTCH2-FL or splice variant transcripts tagged with GFP (Figure 2B) and cocultured overnight with 3T3 cells stably expressing the Jagged2 Notch ligand. Cells expressing GFP were enriched by flow cytometry, seeded into dishes with glass coverslips, fixed-permeabilized, and stained with anti-Hes1 (supplemental Figure 1B). Alternatively, after sorting, whole cell lysates and nuclear fractions were prepared and subjected to western blotting for Hes1 expression. Jagged2 modestly increased expression of Hes1 in cells transfected with NOTCH2-FL, but had no effect on Hes1 expression in cells expressing either NOTCH2-Va or NOTCH2-Vb (Figure 2B).
FLT3 splice variant activation in response to Flt3L
We evaluated the functionality of the FLT3 splice variant transcripts in HEK293T cells. The FLT3-FL and splice variants (FLT3-Va and FLT3-Vb) were stably expressed in HEK293T cells. Cells were serum-starved overnight and stimulated for 10 minutes with 100 ng/mL Flt3 ligand (Flt3L). Flt3L increased the level of tyrosine phosphorylation in cells expressing either FLT3-FL or splice variants (Figure 2C). We next examined the functionality of the FLT3 splice variants in primary blasts from 4 patients by evaluating activation of important downstream targets of the activated FLT3 receptor, such as signal transducer and activator of transcription (STAT)5, mitogen-activated protein kinase (MAPK), and serine/threonine protein kinase AKT as markers for the activation of phosphatidylinositol 3-kinase (PI3K)-dependent pathways. Constitutive activation of STAT5 and MAPK/Erk was detected in AML patient cells expressing Flt3 splice variants regardless of Flt3L activation (Figure 2D). The Flt3 activation signature was not observed in AML patient cells lacking FLT3-Va and FLT3-Vb splice variant expression (Figure 2D). More detailed studies regarding functional effects of splice variants are underway.
NOTCH2 and FLT3 splice variant expression pattern in large cohort of AML patients
We analyzed expression of the NOTCH2 and FLT3 novel variants by the RT-PCR capillary electrophoresis and DNA fragment analysis, in the peripheral blood (PB) or in BM samples from 193 AML patients, in CD34+ cells from BM of 8 NDs, and in PB monocytes and neutrophils from 4 NDs (Figure 3; supplemental Figure 2).
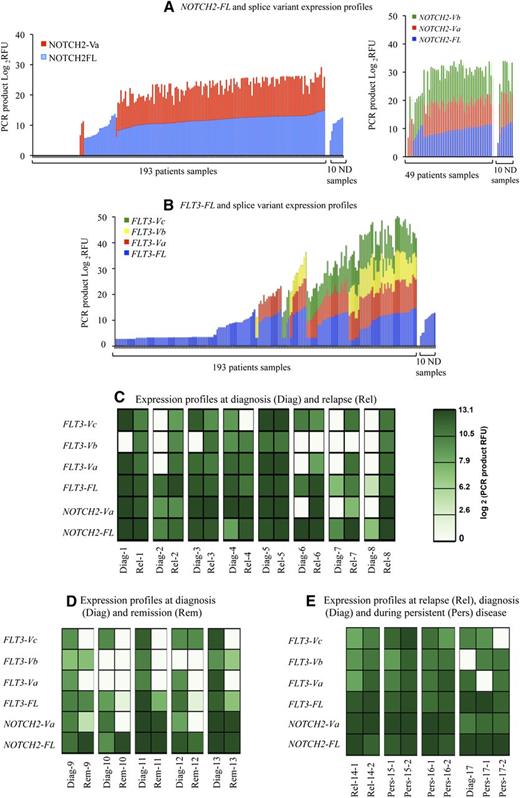
NOTCH2-FL and FLT3-FL and their novel splice variant expression frequencies in AML patients and their association with disease status. (A-B) These figures display overall expression patterns of (A) NOTCH2 and (B) FLT3 FL transcripts and their splice variants. On the figures, the x-axes display patient and normal donor samples, and the y-axes display relative fluorescent units (RFU). PCR product RFU = log2RFU. RFU is a unit of measurement calculated relative to the size standards included in each reaction. For relative level determination, product levels were kept below 3500 RFU, and size standard levels were within 500 to 800 units as recommended by the manufacturer. All calculations and an instrument calibration were done according to the Life Technologies recommendations. We note that in some patients, FLT3-FL expression is lower than the cutoff and not displayed on the graph. (C-E) These figures display overall expression patterns of NOTCH2-FL and FLT3-FL transcripts and their splice variants over the course of the disease in 17 patients. Patients are numbered from 1 to 17; patients samples were taken (C) at diagnosis and relapse (patients 1-8), (D) at diagnosis and remission (patients 9-13), or (E) over the course of refractory disease (patients 14-17). A total of 35 samples were collected including 14 samples obtained at diagnosis: Diag 1 to 13 and Diag 17; 10 samples obtained at relapse: Rel 1 to 8, Rel-14-1, and Rel-14-2 (first and second relapses); 5 samples obtained at remission: Rem 9 to 13; 6 samples obtained during regular visits over the course of refractory disease: Pers 15 to 17 (samples taken at first and second visits are marked as −1 or −2). On the panels, NOTCH2-FL and FLT3-FL and their splice variant transcripts expression levels are presented as log2(PCR product RFU) and shown in a green color-coded scale.
NOTCH2-FL and FLT3-FL and their novel splice variant expression frequencies in AML patients and their association with disease status. (A-B) These figures display overall expression patterns of (A) NOTCH2 and (B) FLT3 FL transcripts and their splice variants. On the figures, the x-axes display patient and normal donor samples, and the y-axes display relative fluorescent units (RFU). PCR product RFU = log2RFU. RFU is a unit of measurement calculated relative to the size standards included in each reaction. For relative level determination, product levels were kept below 3500 RFU, and size standard levels were within 500 to 800 units as recommended by the manufacturer. All calculations and an instrument calibration were done according to the Life Technologies recommendations. We note that in some patients, FLT3-FL expression is lower than the cutoff and not displayed on the graph. (C-E) These figures display overall expression patterns of NOTCH2-FL and FLT3-FL transcripts and their splice variants over the course of the disease in 17 patients. Patients are numbered from 1 to 17; patients samples were taken (C) at diagnosis and relapse (patients 1-8), (D) at diagnosis and remission (patients 9-13), or (E) over the course of refractory disease (patients 14-17). A total of 35 samples were collected including 14 samples obtained at diagnosis: Diag 1 to 13 and Diag 17; 10 samples obtained at relapse: Rel 1 to 8, Rel-14-1, and Rel-14-2 (first and second relapses); 5 samples obtained at remission: Rem 9 to 13; 6 samples obtained during regular visits over the course of refractory disease: Pers 15 to 17 (samples taken at first and second visits are marked as −1 or −2). On the panels, NOTCH2-FL and FLT3-FL and their splice variant transcripts expression levels are presented as log2(PCR product RFU) and shown in a green color-coded scale.
NOTCH2 and FLT3 splice variants were expressed in the majority of AML patients. These variants, with the exception of NOTCH2-Vb, were not detectable in the NDs. DNA fragment analysis also documented overexpression of 2 novel splice variants in AML, NOTCH2-Va and FLT3-Va, respectively, in 73% and 50% of the AML cases (Figure 3A-B; Table 1). Because NOTCH2-Vb expression was seen in NDs (Figure 1A), this variant was excluded from validation studies performed within the 193 AML patient sample set. However, we analyzed an additional 49 AML patient samples and 10 NDs for the expression of a NOTCH2-Vb splice variant (Figure 3A). This analysis demonstrated that the NOTCH2-Vb/FL expression ratio in NDs ranges from 2.1 to −3.7, whereas in AML patients, it ranges from 5.9 to −2.4; thus, NOTCH2-Vb is up-regulated in AML patients compared with NDs (supplemental Figure 2B).
NOTCH2-FL, FLT3-FL, and their splice variant expression patterns examined in 193 patients with AML
| Splice variants . | Patient number . | Patient (%) . |
|---|---|---|
| NOTCH2 | ||
| No expression | 31 | 16 |
| FL | 21 | 11 |
| Va | 3 | 1.5 |
| FL+Va | 138 | 72 |
| FLT3 | ||
| No expression | 65 | 34 |
| Vc | 3 | 2 |
| Vb | 2 | 1 |
| Va+Vc | 7 | 4 |
| Va+Vb | 2 | 1 |
| Va+Vb+Vc | 7 | 4 |
| FL | 25 | 13 |
| FL+Vc | 2 | 1 |
| FL+Va | 14 | 7 |
| FL+Va+Vc | 20 | 10 |
| FL+Va+Vb | 9 | 5 |
| FL+Va+Vb+Vc | 37 | 19 |
| Splice variants . | Patient number . | Patient (%) . |
|---|---|---|
| NOTCH2 | ||
| No expression | 31 | 16 |
| FL | 21 | 11 |
| Va | 3 | 1.5 |
| FL+Va | 138 | 72 |
| FLT3 | ||
| No expression | 65 | 34 |
| Vc | 3 | 2 |
| Vb | 2 | 1 |
| Va+Vc | 7 | 4 |
| Va+Vb | 2 | 1 |
| Va+Vb+Vc | 7 | 4 |
| FL | 25 | 13 |
| FL+Vc | 2 | 1 |
| FL+Va | 14 | 7 |
| FL+Va+Vc | 20 | 10 |
| FL+Va+Vb | 9 | 5 |
| FL+Va+Vb+Vc | 37 | 19 |
RT-PCR DNA fragment analyses show that the majority (141 of 193; 73%) of the AML patients express NOTCH2-Va transcripts, either alone or in combination with NOTCH2-FL (Figure 3A; Table 1). However, 3 patients expressed only the NOTCH2-Va variant, and 31 did not express either transcript. Similar to NOTCH2-Va, splice variants of FLT3 were also expressed in various combinations with FLT3-FL transcripts or with other FLT3 variants. Of 193 patients, 98 expressed ≥1 splice variants in various combinations with other FLT3 variants (16 patients) or FLT3FL (82 patients), whereas 65 patients lacked any FLT3 variants but expressed FLT3-FL at low levels (Figure 3B). Figure 3 shows that expression of ≥2 splice variants in various combinations was the most common in AML patients (Table 1). The most frequently expressed splice variant combinations were FLT3-FL/-Va/-Vb/-Vc and FLT3-FL/-Va/-Vc, expressed in 37 and 20 patients, respectively. We also investigated association between FLT3 gene splicing and the presence of a FLT3 gene mutation. There was only a trend in the associations between certain FLT3 variant expressions with the FLT3-ITD/Asp835 mutations (supplemental Table 3B).
NOTCH2 and FLT3 aberrant splicing reflects disease status
To evaluate whether aberrant splicing reflects disease status, we examined novel splice variant expression over the course of disease in individual AML patients. NOTCH2-FL or FLT3-FL transcripts and their splice variant expression levels were monitored longitudinally in paired samples of 17 AML patients; samples were taken at diagnosis and remission or relapse or over the course of refractory disease. A total of 35 samples were collected.
This analysis showed specific expression patterns and levels of the novel splice variants at different time points during disease development (Figure 3C-E); for example, at diagnosis, >79% of patients expressed 2 or 3 NOTCH2 and FLT3 novel splice variants out of the 4 we have identified; at remission, expression frequencies of these splice variants were markedly decreased (to 0% or to 40%); and at relapse, expression of all splice variants was sharply increased by which point >90% of the patients expressed ≥1 variant in various combinations (Figure 3 and Table 2); Likewise, in a group of patients with persistent AML, novel splice variant expression frequencies remained high, such that >83% of patients expressed all splice variants, alone or in various combinations.
NOTCH2-FL, FLT3-FL, and their splice variant expression profiles reflect disease status
| Samples . | NOTCH2 variants . | FLT3 variants . | ||||
|---|---|---|---|---|---|---|
| -FL [n (%)] . | -Va [n (%)] . | -FL [n (%)] . | -Va [n (%)] . | -Vb [n (%)] . | -Vc [n (%)] . | |
| Diagnosis (n = 14) | 14 (100%) | 11 (79%) | 14 (100%) | 9 (64%) | 5 (36%) | 11 (79%) |
| Remission (n = 5) | 5 (100%) | 3 (60%) | 5 (100%) | 0 (0%) | 2 (40) | 1 (20%) |
| Relapse (n = 10) | 8 (80%) | 8 (80%) | 8 (80%) | 8 (80%) | 6 (60%) | 7 (70%) |
| Persistent AML (n = 6) | 6 (100%) | 6 (100%) | 6 (100%) | 5 (83%) | 6 (100%) | 5 (83%) |
| Samples . | NOTCH2 variants . | FLT3 variants . | ||||
|---|---|---|---|---|---|---|
| -FL [n (%)] . | -Va [n (%)] . | -FL [n (%)] . | -Va [n (%)] . | -Vb [n (%)] . | -Vc [n (%)] . | |
| Diagnosis (n = 14) | 14 (100%) | 11 (79%) | 14 (100%) | 9 (64%) | 5 (36%) | 11 (79%) |
| Remission (n = 5) | 5 (100%) | 3 (60%) | 5 (100%) | 0 (0%) | 2 (40) | 1 (20%) |
| Relapse (n = 10) | 8 (80%) | 8 (80%) | 8 (80%) | 8 (80%) | 6 (60%) | 7 (70%) |
| Persistent AML (n = 6) | 6 (100%) | 6 (100%) | 6 (100%) | 5 (83%) | 6 (100%) | 5 (83%) |
Frequently occurring spliced transcripts may be associated with clinical outcome in AML
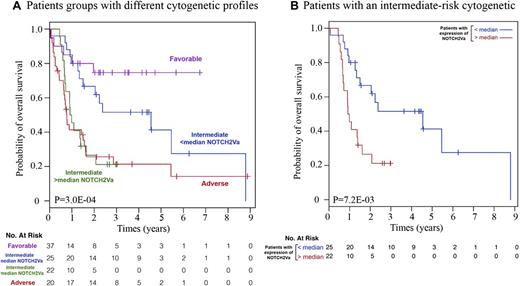
To test whether splicing events are associated with clinical outcomes, we estimated the prognostic value of NOTCH2-FL, FLT3-FL and their splice variant expression levels in the 193 patient sample set; NOTCH2-Va splice variant expression was significantly associated with prognosis (overall survival; concordance index, CINDEX = 0.60; 95% CI, 0.53-0.67; nominal P = .005; FDR = 5.8%; Table 3). To further assess the clinical significance of the presence of NOTCH2 and FLT3 splice variants, patients were classified into 2 equal-sized groups divided by median splice variant transcript expression levels (supplemental Figure 3A), and survival curves were estimated using the Kaplan-Meier method. Among NOTCH2 and FLT3 splice variants, patients expressing higher-than-median levels of the NOTCH2-Va transcripts had significantly inferior survival (hazard ratio [HR] = 1.79; 95% CI, 1.15-2.78; nominal P = .0086; FDR = 9.5%; supplemental Figure 3B). Similar results were obtained in the group of patients <50 years (HR = 3; 95% CI, 1.28-7.09; nominal P = .0084; FDR = 9.2%; supplemental Figure 3B). First we tested the significance of the association between FAB subtypes and expression status of NOTCH2/FLT3 splice variants (low or high expression); we did not find any significant associations (data not shown). To determine whether binary classification based on NOTCH2-Va expression has a prognostic value that is independent of well-established clinical variables, we fitted multivariate Cox models including FLT3-ITD, FLT3-D835 mutations, cytogenetic risk groups, percentage of blasts in BM, percentage of blasts in PB, and patient age (supplemental Table 4). We conclude that NOTCH2-Va binary classification may have prognostic value (significance at P < .05). Furthermore, a multivariate Cox model with selected clinical variables that were significantly associated with survival (ie, cytogenetic risk group of patients and age of patients) revealed that higher-than-median expression levels of NOTCH2-Va transcripts still correlate significantly with overall survival (HR = 1.81; 95% CI, 1.06-3.10; P = .029). The relevance of NOTCH2-Va expression for clinical outcome was further illustrated by use of the Kaplan-Meier estimator in the subgroup of patients within each cytogenetic risk group (Figure 4A). We observed that patients with an intermediate cytogenetic risk profile that express higher-than-median levels of NOTCH2-Va had a trend toward worse survival than patients in the group that express NOTCH2-Va at lower-than-median levels (HR = 2.74; 95% CI, 1.27-5.89; nominal P = 0.0072; FDR = 8%; Figure 4B). Interestingly, these patients exhibited survival similar to patients with an adverse risk cytogenetic profile.
Concordance indexes of each splice variant in the dataset of 193 patients
| Splice variants . | CINDEX . | Lower 95% CI . | Upper 95% CI . | Nominal P value . | FDR . |
|---|---|---|---|---|---|
| NOTCH2-FL | 0.564 | 0.494 | 0.634 | .074 | 0.406 |
| NOTCH2-Va | 0.600 | 0.530 | 0.670 | .005 | 0.058 |
| FLT3-FL | 0.559 | 0.478 | 0.641 | .152 | 0.419 |
| FLT3-Va | 0.538 | 0.460 | 0.615 | .339 | 0.600 |
| FLT3-Vb | 0.482 | 0.378 | 0.587 | .740 | 0.904 |
| FLT3-Vc | 0.545 | 0.459 | 0.632 | .304 | 0.600 |
| Splice variants . | CINDEX . | Lower 95% CI . | Upper 95% CI . | Nominal P value . | FDR . |
|---|---|---|---|---|---|
| NOTCH2-FL | 0.564 | 0.494 | 0.634 | .074 | 0.406 |
| NOTCH2-Va | 0.600 | 0.530 | 0.670 | .005 | 0.058 |
| FLT3-FL | 0.559 | 0.478 | 0.641 | .152 | 0.419 |
| FLT3-Va | 0.538 | 0.460 | 0.615 | .339 | 0.600 |
| FLT3-Vb | 0.482 | 0.378 | 0.587 | .740 | 0.904 |
| FLT3-Vc | 0.545 | 0.459 | 0.632 | .304 | 0.600 |
For each concordance index, the lower and upper bound 95% CI is provided along with the corresponding P value and FDR.
The relevance of NOTCH2-Va expression to the clinical outcome of patients with AML. The relevance of NOTCH2-Va expression to clinical outcome was estimated in the subgroup of patients within each cytogenetic risk group. As A demonstrates, a patient group expressing higher than median levels of NOTCH2-Va has a similar prognosis as the adverse risk group of patients. B shows the overall survival of AML patients with an intermediate risk cytogenetic profile. When dividing patients into 4 categories based on NOTCH2-Va expression (divided by quartile) the resulting classification was borderline significant (P = 5.9 × 10−2) as expected due to the small number of patients in each group. However, separation trend remained the same.
The relevance of NOTCH2-Va expression to the clinical outcome of patients with AML. The relevance of NOTCH2-Va expression to clinical outcome was estimated in the subgroup of patients within each cytogenetic risk group. As A demonstrates, a patient group expressing higher than median levels of NOTCH2-Va has a similar prognosis as the adverse risk group of patients. B shows the overall survival of AML patients with an intermediate risk cytogenetic profile. When dividing patients into 4 categories based on NOTCH2-Va expression (divided by quartile) the resulting classification was borderline significant (P = 5.9 × 10−2) as expected due to the small number of patients in each group. However, separation trend remained the same.
Discussion
A genome-wide AS screen in AML patients identified NOTCH2 and FLT3 as the 2 most frequently mis-spliced transcripts in patients compared with NDs. FLT3 is already known as a frequently mutated oncogene in AML, whereas the role of NOTCH2 in this disease is not well understood, although decreased NOTCH2 expression has been linked to AML in 2 reports.35,36
Cloning and sequencing analysis identified several aberrant splice variants of NOTCH2 and FLT3 that resulted from either skipping of ≥1 exon or activation of cryptic splicing sites. All these splicing alterations created transcripts that encoded proteins, as demonstrated through expression of NOTCH2-Va, NOTCH2-Vb, FLT3-Va, and FLT3-Vb splice variants in HEK293T cells. These results were expected, because splicing alterations do not cause frame shifts on NOTCH2 and FLT3 genes and because they occur on the gene segments that encode proteins in the extracellular domain, (spliced-out exons of NOTCH2 encode extracellular EGF-like domains, whereas FLT3 shows splicing aberrations in exons encoding extracellular Ig-like motifs).
The functionality of Notch2 splice variant proteins was explored using primary blasts from AML patients. We observed an inverse correlation between the expression of NOTCH2 splice variants and Notch2 target genes HES1, TXL1, and HEY1 in AML patients. Because splice variants are expressed in various combinations with NOTCH2-FL transcripts, it is intriguing to speculate that, similar to splice variants of many other genes (including MDM2, CD44, and p53). Notch2 splice variants may act in a dominant-negative manner that compromises the functions of Notch2-FL and are likely to block corresponding signaling pathways in AML cells.7,37,37-39 Based on these discoveries and the findings presented here, we suggest that silencing of the Notch2 signaling pathway could be a due to dominant-negative effects of Notch2 splice variants on Notch2-FL. It is thus possible to speculate that Notch2 pathway reactivation could be achieved by the using of blocking antibodies against Notch2-Va and Notch2-Vb splice variants. Our observations indicate that NOTCH2 splice variants are attractive targets for novel AML therapeutics. Also, based on our preliminary studies on FLT3 splice variants, we speculate that some of these variants may be functional. Additional studies need to be done to determine the functional consequences of these variants.
Expression profiling of NOTCH2-FL, FLT3-FL, and their splice variants in 193 AML patients identified NOTCH2-Va (73%) and FLT3-Va (50%) as the most frequently expressed variants in AML. These splice variants were undetectable in plasma cells from 12 myloma patients and in several lymphoma cell lines (data not shown). More detailed study regarding expression of these splice variants in different cell populations in AML patients, as well as in patients with other hematological malignances, is underway. Based on our studies, we speculate that these splice variants represent potential novel markers to differentiate AML cells from normal stem cells, offering the possibility for selective targeting of AML stem cells.
Recent reports of mutations in splicing factors in many different cancers, including MDS and AML, suggest that splicing events could contribute directly to disease initiation or progression.22,25 Also, aberrant splicing events have been linked to malignant transformation, particularly in genes associated with susceptibility and/or progression of cancer.6,8-16 For some cancers, these splicing alterations create functionally significant biomarkers.9,12,14,17-21 In light of these studies and the results presented here, we believe that NOTCH2 and FLT3 splicing events are associated with pathogenesis in AML and that identification of the causes and consequences of these splicing alterations will provide a better understanding of the biology of this disease. Because it is well known that splicing alterations can result from mutations and/or could be due to modulated expression of splicing factors, we evaluated whether expression of NOTCH2 and FLT3 splice variants correlated with mutations detected on commonly mutated genes such as U2AF1 and SF3B1 in patients with AML (supplemental Figure 4). We did not find any significant correlation from this preliminary study, possibly because of the small size of the cohort. It may also reflect the extensive complexity of the splicing machinery. Evaluation of a larger cohort of patients in these studies is underway. Additionally, to understand causes of altered splicing in AML, we tested if expression levels of splice variants correlate with classical mutations such as NPM1 and IDH1/2. We did not find any significant correlation from this preliminary study (supplemental Figure 4).
In these studies, we investigated the clinical relevance of AS in AML patients. In longitudinal studies, we detected significant differences in the frequency of expression of particular splice variants of NOTCH2 and FLT3 at time points throughout the course of disease progression; for example, certain splicing events were elevated at diagnosis and relapse and were “normalized” during remission. We also noted correlations between splicing events and the clinical outcome; NOTCH2-Va expression was associated with a poor outcome, particularly for patients with an intermediate risk cytogenetic profile. Furthermore, patients who expressed NOTCH2-Va at higher-than-median levels had the same median overall survival as those with cytogenetically determined adverse risk profiles. Thus, NOTCH2-Va expression has potential use for identifying a subgroup of patients within the larger group having cytogenetically determined intermediate risk profiles. Additional studies with larger AML patient cohorts are needed to verify the clinical significance of splicing events of the NOTCH2 gene.
Our results suggest that AS of NOTCH2 and FLT3 is a common event in AML. Because NOTCH2 and FLT3 variants are detected in a significant number of patients with AML, they can be used as novel disease markers and as selective targets for either therapeutic antibodies or for the delivery of cytotoxic drugs, toxins, or radionucleotides to leukemic cells. More extended functional studies are underway to clarify the oncogenic potential of NOTCH2 and FLT3 splice variants in AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Linda M. Pilarski (Cross Cancer Institute and University of Alberta) for editing and providing critical comments on the manuscript.
This study was supported in part by National Cancer Institute grant PO1CA66996-13. S.A. is supported by the American Society of Hematology scholar award for Clinical/Translational Research and supported by the American Association for Cancer Research-Amgen, Inc. Fellowship in Clinical/Translational Cancer Research. J.Q. was supported by National Library of Medicine of the National Institutes of Health grant R01 LM010129-01. B.H.-K. was supported by internal funding at the Institut de Recherches Cliniques de Montréal. P.M.P. is supported by the Alberta Innovates Centre for Machine Learning. C.B. was supported by the German Research Foundation (DFG Fellowship BA 4186/1-1).
Authorship
Contribution: S.A. conceived the idea, executed experiments, coordinated and directed the study, and wrote the manuscript; M.B.-N. collected and processed patients’ clinical data; B.H.-K. performed statistical and bioinformatics analyses and wrote the manuscript; P.M.P. and S.P. performed bioinformatics analyses; C.B. helped with the sequencing experiments; T.C. performed microscopy experiments; H.A.-L. and L.L. provided University Health Network samples and clinical data; S.V. and E.A.F. collected patient samples; I.G., D.P.S., G.M., and D.J.D. provided DFCI patient samples; M.W., S.M., and I.D.-J. collected patient clinical data; D.G.T. and J.Q. provided discussions and advice; R.M.S. provided DFCI patient samples and assisted in writing the manuscript; and J.D.G. directed the study and edited the manuscript.
Conflict-of-interest disclosure: J.D.G. receives research support and is a consultant for Novartis Pharma AG. The remaining authors declare no competing financial interests.
Correspondence: Sophia Adamia, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215; e-mail: sophia_adamia@dfci.harvard.edu; and James D. Griffin, Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215; e-mail: James_Griffin@dfci.harvard.edu.
References
Author notes
M.B.-N., B.H.-K., and P.M.P. contributed equally to this work.