Key Points
FL carries mutations in linker histone H1 B, C, D, and E genes in 27% of cases.
FL carries recurrent mutations in OCT2 (POU2F2), IRF8, and ARID1A.
Abstract
Follicular lymphoma (FL) constitutes the second most common non-Hodgkin lymphoma in the western world. FL carries characteristic recurrent structural genomic aberrations. However, information regarding the coding genome in FL is still evolving. Here, we describe the results of massively parallel exome sequencing and single nucleotide polymorphism 6.0 array genomic profiling of 11 highly purified FL cases, and 1 transformed FL case and the validation of selected mutations in 102 FL cases. We report the identification of 15 novel recurrently mutated genes in FL. These include frequent mutations in the linker histone genes HIST1H1 B-E (27%) and mutations in OCT2 (also known as POU2F2; 8%), IRF8 (6%), and ARID1A (11%). A subset of the mutations in HIST1H1 B-E affected binding to DNMT3B, and mutations in HIST1H1 B-E and in EZH2 or ARID1A were largely mutually exclusive, implicating HIST1H1 B-E in epigenetic deregulation in FL. Mutations in OCT2 (POU2F2) affected its transcriptional and functional properties as measured through luciferase assays, the biological analysis of stably transduced cell lines, and global expression profiling. Finally, multiple novel mutated genes located within regions of acquired uniparental disomy in FL are identified. In aggregate, these data substantially broaden our understanding of the genomic pathogenesis of FL.
Introduction
Follicular lymphoma (FL) is the most common indolent B-cell lymphoma, with an incidence and prevalence of ∼14 000 and ∼100 000 cases in the US, respectively.1 FL has a varied clinical course that is influenced by FL cell-intrinsic and cell-extrinsic aberrations, and FL remains incurable using conventional therapies.2-6 Genetically, FL is characterized by the hallmark balanced chromosomal translocation t(14;18), found in about 85% of FL cases, leading to deregulated expression of BCL2.7-9 A subtype of FL (∼15%) that lacks translocation t(14;18) has also been described.10,11
At the genomic level, FL is characterized by recurrent acquired structural abnormalities which include acquired genomic copy number aberrations (aCNA) and loss-of-heterozygosity (LOH), as well as frequent acquired uniparental disomy (aUPD)/copy neutral LOH (cnLOH). Largely as a result of karyotyping and, more recently, the application of single nucleotide polymorphism (SNP) array profiling and array CGH to FL genome analysis, a relatively complete view of aCNA/LOH and cnLOH in FL has emerged.12-23 The subsequent application of this knowledge to pathogenetic gene discovery has resulted in the identification of novel genes important to the biology of FL.12,15,24 However, compared with other hematologic malignancies, like diffuse large B-cell lymphoma (DLBCL), chronic lymphocytic leukemia, acute lymphoblastic anemia, or multiple myeloma, the current understanding of the incidence of recurrent gene mutations and the interplay of gene mutations and aCNA/LOH and cnLOH in FL is in its early stages and largely incomplete.3,25-35
The discovery of genes mutated in various forms of cancer has further advanced our understanding of the specific pathogenetic mechanisms that operate in cancer cell subtypes. Recently, as a result of the study of DLBCL, 2 novel high-frequency mutated genes (MLL2 and CREBBP) in non-Hodgkin lymphoma (NHL) were identified.31,36 The subsequent expansion of these findings to FL identified very frequent mutations in these genes in FL as well, demonstrating that at the genomic level, FL and DLBCL share common aberrations. Together, these data support a major driver role for MLL2 and CREBBP in NHL pathogenesis. However, to date, an unbiased discovery screen for gene mutations in FL has not been reported, thus limiting our ability to fully assemble and analyze the landscape of genomic changes in FL.
In this study, we describe the results of massively parallel exome sequencing and high-resolution SNP 6.0 array profiling of 11 FL cases and 1 transformed FL (t-FL) case and validation of mutations in various expansion cohorts of a combined 114 FL cases, with the goal of broadening our knowledge base of recurrent gene mutations in FL and their relation to aCNA/LOH and cnLOH. Aided by the use of highly purified sorted lymphomatous B cells and paired normal T cells as the cellular source for DNA used in all genomic analyses, we identify 15 novel recurrently mutated genes in FL, including frequent mutations in the linker histone genes HIST1H1 B-E (27%) and mutations in OCT2 (also known as POU2F2; 8%), IRF8 (6%), and ARID1A (11%). Furthermore, we place novel and known mutated genes in FL within regions of aUPD, thus further substantiating this lesion type as a marker for gene mutations and providing multiple candidate genes for further detailed genomic and biological follow-up studies. In aggregate, this study advances the current knowledge of the genetics of FL and stimulates multiple novel directions for research in this common lymphoma subtype.
Methods
Patient characteristics and study source material
Please see supplemental Methods, available on the Blood Web site. This study received the following approvals: Medical School Institutional Review Boards #HUM00007985 and Medical School Institutional Review Boards #HUM00017055. This study was conducted in accordance with the Declaration of Helsinki.
Solution-based exome capture and HiSeq2000-based massively parallel sequencing and bioinformatic pipeline analysis of next-generation sequence data
Please see supplemental Methods for details.
Results
Massively parallel sequencing of the coding genome of FL samples
To further our understanding of the genetic basis of FL, we used solution exon capture of sheared and processed genomic DNA isolated from flow-sorted immunoglobulin light chain–restricted lymphomatous B cells and paired CD3+ T cells isolated from 11 cases of FL and 1 case of DLBCL transformed from prior FL followed by paired-end, massively parallel sequencing. FL tumor B-cell DNA purity was conservatively estimated based on the B-cell population size expressing κ or λ light chains (supplemental Table 1; mean purity 90%; range, 77%-100%) and separately using the degree of immunoglobulin-locus deletions for the 6 cases with informative rearrangements (supplemental Table 2; conservative mean purity estimate 82%). The sequence data were characterized by a depth of coverage range of 43 to 73, with 90% of bases in the target region covered by at least 10 reads (supplemental Table 3). Computations comparing single nucleotide variant calls by SNP 6.0 profiling with the calls made by our next-generation sequencing pipeline computed a sensitivity of 90.1 and a specificity of 99.4%.
Identification of somatic mutations in MLL2, CREBBP, TNFRSF14, EZH2, and BCL2
Recent reports have identified a high frequency of mutations in the genes MLL2 and CREBBP, and mutations in EZH2 and TNFRSF14 in NHL, and mutation data from various NHLs have been reported on BCL2.15,25,31,34,36,37 In our dataset, we confirm frequent mutations in all 5 genes in FL (supplemental Results).
Identification of novel recurrently mutated genes in FL
The bioinformatics pipeline nominated 33 genes to be mutated in ≥2 of 12 discovery cases; 30 of 33 of these genes were subjected to Sanger sequence validation. In addition, we selected genes nominated only once for Sanger sequence validation based on biological function. Overall, we verified mutations in 65 of the nominated genes (Tables 1-3 and supplemental Table 4). As expected, the genes that did not verify using Sanger sequencing were often represented as minor subclones in the discovery cases (low next-generation mutant tag frequencies), leaving open the possibility that some of these also constituted somatically acquired, albeit subclonal, mutations (supplemental Table 5).
Details of confirmed novel gene mutations occurring in ≥2 cases in the 11 FL and 1 t-FL discovery-set NHL cases
| FL case no. . | Gene . | Nucleotide (genomic) NCBI 36 Hg 18 . | Amino acid change (protein) . | Sanger sequencing result (T/N) . | SNP array-based aCNA/LOH/aUPD . | PolyPhen-2 prediction results . | SIFT prediction results . | Next-gen sequencing % mutant tags . |
|---|---|---|---|---|---|---|---|---|
| ML2 | ARID1A | chr1_26970196-26970196_G_T | Splice site acceptor | Mut-Heter/WT | None | 44 | ||
| ML30 | ARID1A | chr1_26972534-26972534_C_T | 1276R/X | Mut-Heter/WT | None | 27 | ||
| ML47 | CHRM3 | chr1_238138618-238138618_A_G | 415D/G | Mut-Heter/WT | Gain | Benign (0) | Tolerated (0.38) | 39 |
| ML66 | CHRM3 | chr1_238138543-238138543_A_T | 390Q/L | Mut-Heter/WT | Gain | Benign (0.087) | Tolerated (0.31) | 26 |
| ML51 | DMRT3 | chr9_980911-980911_G_A | 442C/Y | Mut-Heter/WT | None | Probably damaging (0.999) | Damaging (0.02) | 31 |
| ML90 | DMRT3 | chr9_980691-980691_C_A | 369P/T | Mut-Heter/WT | None | Probably damaging (1.0) | Damaging (0) | 43 |
| ML30 | GRM7 | chr3_6878496-6878496_C_T | 141P/S | Mut-Heter/WT | None | Benign (0.245) | Tolerated (1.0) | 23 |
| ML42 | GRM7 | chr3_6878401-6878401_G_T | 109C/F | Mut-Heter/WT | None | Probable damaging (0.977) | Damaging (0.00) | 40 |
| ML47 | GRM7 | chr3_6878556-6878556_G_A | 161V/I | Mut-Heter/WT | None | Probable damaging (0.992) | Damaging (0.02) | 42 |
| ML47 | HIST1H1E | chr6_26264736-26264736_G_A | 47A/T | Mut-Heter/WT | Gain | Possibly damaging (0.597) | Tolerated (0.08) | 21 |
| ML90 | HIST1H1E | chr6_26264905-26264905_G_A | 103A/T | Mut-Heter/WT | None | Probably damaging (1.0) | Damaging (0) | 55 |
| ML90 | HIST1H1E | chr6_26264991-26264991_G_A | 132G/D | Mut-Heter/WT | None | Benign (0) | Tolerated (0.56) | 41 |
| ML55 | IRF8 | chr16_85954737_85954737_G_ | Frameshift deletion | Mut-Heter/WT | None | 56 | ||
| ML51 | IRF8 | chr16_85954790_85954790_G_T | 395E>X | Mut-Homo/WT | aUPD | 82 | ||
| ML51 | KLHDC7B | chr22_49334644-49334644_C_T | 296R/X | Mut-Heter/WT | None | 32 | ||
| ML55 | KLHDC7B | chr22_49333831-49333831_T_A | 25S/T | Mut-Heter/WT | None | Benign (0.004) | Tolerated (0.78) | 33 |
| ML55 | LYST | chr1_234017235-234017235_C_A | 1584E/X | Mut-Heter/WT | None | 57 | ||
| ML66 | LYST | chr1_234005012-234005012_A_ | Splice site acceptor | Mut-Heter/WT | Loss | 42 | ||
| ML47 | MCL1 | chr1_150551543_150551543_C_G | 155S/T | Mut-Heter/WT | None | Benign (0.001) | Tolerated (0.19) | 46 |
| ML90 | MCL1 | chr1_150551543_150551543_C_T | 155S/N | Mut-Heter/WT | Gain | Benign (0.0) | Tolerated (0.21) | 19 |
| ML55 | PCLO | chr7_82422909-82422909_G_T | 1766L/I | Mut-Heter/WT | None | Possibly damaging (0.617) | NA | 50 |
| ML66 | PCLO | chr7_82421122-82421122_A_T | 2361F/L | Mut-Heter/WT | None | Benign (0.0) | NA | 16 |
| ML47 | OCT2 | chr19_47291387-47291387_A_ | Frameshift deletion | Mut-Heter/WT | None | 45 | ||
| ML90 | OCT2 | chr19_47291598-47291598_C_T | 282R/H | Mut-Heter/WT | None | Probably damaging (1.0) | Damaging (0) | 51 |
| ML47 | TNFRSF14 | chr1_2484552-2484552_T_A | 88N>I | Mut-Homo/WT | aUPD | Probably damaging (1.000) | Damaging (0) | 100 |
| ML55 | TNFRSF14 | chr1_2483126-2483126_C_T | 112S>F | Mut-Homo/WT | aUPD | Probably damaging (1.000) | Damaging (0) | 92 |
| ML93 | TP53 | chr17_7518947-7518948_TC_ | Frameshift deletion | Mut-Homo/WT | Loss | 80 | ||
| ML79 | TP53 | chr17_7517845-7517845_C_T | 273R>H | Mut-Homo/WT | Loss | Possibly damaging (0.831) | Damaging (0.01) | 100 |
| ML66 | VPS39 | chr15_40287584-40287584_A_C | 20 C/W | Mut-Heter/WT | None | Probable damaging (0.977) | Damaging (0.01) | 52 |
| ML87 | VPS39 | chr15_40270618-40270618_A_T | 82 L>I | Mut-Homo/WT | aUPD | Possibly damaging (0.929) | Tolerated (0.38) | 87 |
| ML66 | WDR64 | chr1_240031064-240031066_GAA_ | In-frame deletion | Mut-Heter/WT | Gain | 19 | ||
| ML93 | WDR64 | chr1_239913455-239913455_G_A | 213C/Y | Mut-Heter/WT | None | Probably damaging (1.0) | Tolerated (0.07) | 42 |
| ML55 | ZNF541 | chr19_52736020-52736020_C_T | 802G/S | Mut-Heter/WT | None | Probable damaging (1.0) | Damaging (0.00) | 13 |
| ML66 | ZNF541 | chr19_52739383-52739383_G_ | Frameshift deletion | Mut-Heter/WT | None | 47 |
| FL case no. . | Gene . | Nucleotide (genomic) NCBI 36 Hg 18 . | Amino acid change (protein) . | Sanger sequencing result (T/N) . | SNP array-based aCNA/LOH/aUPD . | PolyPhen-2 prediction results . | SIFT prediction results . | Next-gen sequencing % mutant tags . |
|---|---|---|---|---|---|---|---|---|
| ML2 | ARID1A | chr1_26970196-26970196_G_T | Splice site acceptor | Mut-Heter/WT | None | 44 | ||
| ML30 | ARID1A | chr1_26972534-26972534_C_T | 1276R/X | Mut-Heter/WT | None | 27 | ||
| ML47 | CHRM3 | chr1_238138618-238138618_A_G | 415D/G | Mut-Heter/WT | Gain | Benign (0) | Tolerated (0.38) | 39 |
| ML66 | CHRM3 | chr1_238138543-238138543_A_T | 390Q/L | Mut-Heter/WT | Gain | Benign (0.087) | Tolerated (0.31) | 26 |
| ML51 | DMRT3 | chr9_980911-980911_G_A | 442C/Y | Mut-Heter/WT | None | Probably damaging (0.999) | Damaging (0.02) | 31 |
| ML90 | DMRT3 | chr9_980691-980691_C_A | 369P/T | Mut-Heter/WT | None | Probably damaging (1.0) | Damaging (0) | 43 |
| ML30 | GRM7 | chr3_6878496-6878496_C_T | 141P/S | Mut-Heter/WT | None | Benign (0.245) | Tolerated (1.0) | 23 |
| ML42 | GRM7 | chr3_6878401-6878401_G_T | 109C/F | Mut-Heter/WT | None | Probable damaging (0.977) | Damaging (0.00) | 40 |
| ML47 | GRM7 | chr3_6878556-6878556_G_A | 161V/I | Mut-Heter/WT | None | Probable damaging (0.992) | Damaging (0.02) | 42 |
| ML47 | HIST1H1E | chr6_26264736-26264736_G_A | 47A/T | Mut-Heter/WT | Gain | Possibly damaging (0.597) | Tolerated (0.08) | 21 |
| ML90 | HIST1H1E | chr6_26264905-26264905_G_A | 103A/T | Mut-Heter/WT | None | Probably damaging (1.0) | Damaging (0) | 55 |
| ML90 | HIST1H1E | chr6_26264991-26264991_G_A | 132G/D | Mut-Heter/WT | None | Benign (0) | Tolerated (0.56) | 41 |
| ML55 | IRF8 | chr16_85954737_85954737_G_ | Frameshift deletion | Mut-Heter/WT | None | 56 | ||
| ML51 | IRF8 | chr16_85954790_85954790_G_T | 395E>X | Mut-Homo/WT | aUPD | 82 | ||
| ML51 | KLHDC7B | chr22_49334644-49334644_C_T | 296R/X | Mut-Heter/WT | None | 32 | ||
| ML55 | KLHDC7B | chr22_49333831-49333831_T_A | 25S/T | Mut-Heter/WT | None | Benign (0.004) | Tolerated (0.78) | 33 |
| ML55 | LYST | chr1_234017235-234017235_C_A | 1584E/X | Mut-Heter/WT | None | 57 | ||
| ML66 | LYST | chr1_234005012-234005012_A_ | Splice site acceptor | Mut-Heter/WT | Loss | 42 | ||
| ML47 | MCL1 | chr1_150551543_150551543_C_G | 155S/T | Mut-Heter/WT | None | Benign (0.001) | Tolerated (0.19) | 46 |
| ML90 | MCL1 | chr1_150551543_150551543_C_T | 155S/N | Mut-Heter/WT | Gain | Benign (0.0) | Tolerated (0.21) | 19 |
| ML55 | PCLO | chr7_82422909-82422909_G_T | 1766L/I | Mut-Heter/WT | None | Possibly damaging (0.617) | NA | 50 |
| ML66 | PCLO | chr7_82421122-82421122_A_T | 2361F/L | Mut-Heter/WT | None | Benign (0.0) | NA | 16 |
| ML47 | OCT2 | chr19_47291387-47291387_A_ | Frameshift deletion | Mut-Heter/WT | None | 45 | ||
| ML90 | OCT2 | chr19_47291598-47291598_C_T | 282R/H | Mut-Heter/WT | None | Probably damaging (1.0) | Damaging (0) | 51 |
| ML47 | TNFRSF14 | chr1_2484552-2484552_T_A | 88N>I | Mut-Homo/WT | aUPD | Probably damaging (1.000) | Damaging (0) | 100 |
| ML55 | TNFRSF14 | chr1_2483126-2483126_C_T | 112S>F | Mut-Homo/WT | aUPD | Probably damaging (1.000) | Damaging (0) | 92 |
| ML93 | TP53 | chr17_7518947-7518948_TC_ | Frameshift deletion | Mut-Homo/WT | Loss | 80 | ||
| ML79 | TP53 | chr17_7517845-7517845_C_T | 273R>H | Mut-Homo/WT | Loss | Possibly damaging (0.831) | Damaging (0.01) | 100 |
| ML66 | VPS39 | chr15_40287584-40287584_A_C | 20 C/W | Mut-Heter/WT | None | Probable damaging (0.977) | Damaging (0.01) | 52 |
| ML87 | VPS39 | chr15_40270618-40270618_A_T | 82 L>I | Mut-Homo/WT | aUPD | Possibly damaging (0.929) | Tolerated (0.38) | 87 |
| ML66 | WDR64 | chr1_240031064-240031066_GAA_ | In-frame deletion | Mut-Heter/WT | Gain | 19 | ||
| ML93 | WDR64 | chr1_239913455-239913455_G_A | 213C/Y | Mut-Heter/WT | None | Probably damaging (1.0) | Tolerated (0.07) | 42 |
| ML55 | ZNF541 | chr19_52736020-52736020_C_T | 802G/S | Mut-Heter/WT | None | Probable damaging (1.0) | Damaging (0.00) | 13 |
| ML66 | ZNF541 | chr19_52739383-52739383_G_ | Frameshift deletion | Mut-Heter/WT | None | 47 |
T/N, tumor/normal; WT, wild-type.
Details of confirmed recurrent gene mutations in the linker HIST1H1 genes B-E in 114 FL cases
| FL case no. . | Gene . | Sequence context of mutations . | cDNA changes . | Amino acid change (protein) . | Sanger sequencing result (T/N) . | PolyPhen-2 prediction results . | SIFT prediction results . |
|---|---|---|---|---|---|---|---|
| L55 | HIST1H1B | AAGAG(C/G)TTGGTG | c.267C>C/G | 89S/R | Mut-Heter/WT | Benign (0.226) | Damaging (0) |
| L55 | HIST1H1B | CCAAGG(G/A)CACTGG | c.302G>G/A | 101G/D | Mut-Heter/WT | Probably damaging (1.0) | Damaging (0) |
| L77 | HIST1H1B | CAAGA(G/A)CTTGG | c.266G>G/A | 89S/N | Mut-Heter/WT | Possibly damaging (0.616) | Damaging (0) |
| L90 | HIST1H1B | ACCTA(A>C)AGCTG | c.641A>C | 214K->T | Mut-Homo/WT | Probably damaging (0.998) | Damaging (0.01) |
| ML91 | HIST1H1B | CTAAG(AAG)GCCAA | c.397_399Het_Del AAG | In-frame deletion | Mut-Heter/WT | ||
| FL11 | HIST1H1B | CAAAG(C/A)CAAGA | c.368C>C/A | 123A/D | Mut-Heter)/WT | Possibly damaging (0.622) | Damaging (0.01) |
| FL24 | HIST1H1B | ATTAA(G/C)CTGGG | c.252G>G/C | 84K/N | Mut-Heter/WT | Possibly damaging (0.597) | Damaging (0) |
| FL55 | HIST1H1B | CGGTG(G/C)CTACG | c.217G>G/C | 73G/A | Mut-Heter/WT | Probably damaging (1.000) | Damaging (0.02) |
| L27 | HIST1H1C | CCAAG(C/T)CCAAG | c.352C>C/T | 118P/S | Mut-Heter/WT | Possibly damaging (0.518) | Tolerated (0.55) |
| L28 | HIST1H1C | AAGTG(G/C)CTAAG | c.511G>G/C | 171A/P | Mut-Heter/WT | Probably damaging (0.987) | Tolerated (0.42) |
| L77 | HIST1H1C | CTGCC(A/T)AAAGT | c.559A>A/T | 187K/X | Mut-Heter/WT | ||
| FL54 | HIST1H1C | ATCAA(A/T)CTTG(G/C)TCTCA | c.243A>A/T; c.248G>G/C | 81K/N; 83G/A | Mut-Heter/WT | Possibly damaging (0.927) | Damaging (0) |
| FL59 | HIST1H1C | CAGAG(C>G)TCATC | c.127C>G | 43L->V | Mut-Homo/WT | Possibly damaging (0.927) | Damaging (0) |
| FL75 | HIST1H1C | TGAAAAAA(A)GCGTTG | c.192Homo_InsA | Frameshift insertion | Mut-Homo/WT | ||
| L7 | HIST1H1D | CAACA(AACA)GCCGT | c.236Het_InsAACA | Frameshift insertion | Mut-Heter/WT | ||
| L27 | HIST1H1D | AAG(G/A)TA(C/T)CGG | c.275G>G/A c.278C>C/T | 99G/D 100T/I | Mut-Heter/WT | Probably damaging (0.999) | Damaging (0) |
| L46 | HIST1H1D | ATCAA(G/C)CTTGG | c.246G>G/C | 82K/N | Mut-Heter/WT | Possibly damaging (0.774) | Damaging (0) |
| L53 | HIST1H1D | CGGTG(C/T)TTCTG | c.305C>C/T | 102A/V | Mut-Heter/WT | Probably damaging (0.995) | Damaging (0) |
| L55 | HIST1H1D | GGCGG(C/G)CAAGC | c.602C>C/G | 201A/G | Mut-Heter/WT | Benign (0.191) | Tolerated (0.65) |
| L56 | HIST1H1D | AGAAAG(C/T)GCTTG | c.197C>C/T | 66A/V | Mut-Heter/WT | Possibly damaging (0.954) | Tolerated (0.34) |
| ML64 | HIST1H1D | GCTTG(G>A)CCTCA | c.251G>A | 84G->D | Mut-Homo/WT | Probably damaging (0.98) | Damaging (0.05) |
| ML90 | HIST1H1D | CAAAG(G/A)TACTC | c.275G>G/A | 92G/D | Mut-Heter/WT | Probably damaging (0.999) | Damaging (0) |
| FL6 | HIST1H1D | GCGCC(G/C)CTACC | c.436G>G/C | 146A/P | Mut-Heter/WT | Possibly damaging (0.769) | Tolerated (0.25) |
| FL39* | HIST1H1D | CAAAGG(*)TACCGG | c.300Het_Ins* | Frameshift insertion | Mut-Heter/WT | ||
| FL52 | HIST1H1D | AACAC(C/G)TCAGC | c.542C>C/G | 181P/R | Mut-Heter/WT | Benign (0.001) | Damaging (0) |
| L23 | HIST1H1E | AGCCC(A/G)AGAAG | c.415A>A/G | 139K/E | Mut-Heter/WT | Benign (0.024) | Tolerated (0.06) |
| L24 | HIST1H1E | CCAAG(C/G)CAAAA | c.541C>C/G | 181P/A | Mut-Heter/WT | Benign (0.096) | Tolerated (1) |
| L27 | HIST1H1E | CTGCA(G/A)CTGCT | c.490G>G/A | 164A/T | Mut-Heter/WT | Possibly damaging (0.917) | Tolerated (0.07) |
| L64 | HIST1H1E | GCCTA(A/G)GGCTA | c.356A>A/G | 120A/T | Mut-Heter/WT | Benign (0) | Tolerated (1) |
| L114 | HIST1H1E | GGTTC(C)TTCAA | c.312Het_DelC | Frameshift insertion | Mut-Heter/WT | ||
| ML47 | HIST1H1E | CTAAA(G/A)CTGTT | c.139G>G/A | 47A/T | Mut-Heter/WT | Possibly damaging (0.597) | Tolerated (0.08) |
| ML90 | HIST1H1E | GTCGG(G/A)TTCCT; | c.308G>G/A | 103G/D | Mut-Heter/WT | Probably damaging (1.0) | Damaging (0) |
| ML90 | HIST1H1E | AGCCA(G/A)CAGGA | c.394G>G/A | 132A/T | Mut-Heter/WT | Benign (0) | Tolerated (0.56) |
| FL7 | HIST1H1E | CATTA(C/G)TAAAG | c.134C>C/G | 45T/S | Mut-Heter/WT | Possibly damaging (0.795) | Damaging (0.04) |
| FL29 | HIST1H1E | GCGGC(G/A)TATCT | c.169G>G/A | 57V/I | Mut-Heter/WT | Possibly damaging (0.951) | Tolerated (0.15) |
| FL34 | HIST1H1E | AGACC(C/T)CAAAG | c.463C>C/T | 155P/S | Mut-Heter//WT | Possibly damaging (0.799) | Tolerated (0.46) |
| FL39 | HIST1H1E | GGGTT(G/T)CTTCA | c.311G>G/T | 104S/F | Mut-Heter/WT | Probably damaging (0.975) | Damaging (0) |
| FL39 | HIST1H1E | TGGAG(C/T)CAAAA | c.500C>C/T | 167A/V | Mut-Heter/WT | Benign (0.209) | Tolerated (0.24) |
| FL42 | HIST1H1E | CCAAG(C/T)CAAAA | c.541C>C/T | 181P/S | Mut-Heter/WT | Possibly damaging (0.798) | Tolerated (0.66) |
| FL61 | HIST1H1E | CTAAA(A/T)AGGCA | c.364A>A/T | 122K/X | Mut-Heter/WT |
| FL case no. . | Gene . | Sequence context of mutations . | cDNA changes . | Amino acid change (protein) . | Sanger sequencing result (T/N) . | PolyPhen-2 prediction results . | SIFT prediction results . |
|---|---|---|---|---|---|---|---|
| L55 | HIST1H1B | AAGAG(C/G)TTGGTG | c.267C>C/G | 89S/R | Mut-Heter/WT | Benign (0.226) | Damaging (0) |
| L55 | HIST1H1B | CCAAGG(G/A)CACTGG | c.302G>G/A | 101G/D | Mut-Heter/WT | Probably damaging (1.0) | Damaging (0) |
| L77 | HIST1H1B | CAAGA(G/A)CTTGG | c.266G>G/A | 89S/N | Mut-Heter/WT | Possibly damaging (0.616) | Damaging (0) |
| L90 | HIST1H1B | ACCTA(A>C)AGCTG | c.641A>C | 214K->T | Mut-Homo/WT | Probably damaging (0.998) | Damaging (0.01) |
| ML91 | HIST1H1B | CTAAG(AAG)GCCAA | c.397_399Het_Del AAG | In-frame deletion | Mut-Heter/WT | ||
| FL11 | HIST1H1B | CAAAG(C/A)CAAGA | c.368C>C/A | 123A/D | Mut-Heter)/WT | Possibly damaging (0.622) | Damaging (0.01) |
| FL24 | HIST1H1B | ATTAA(G/C)CTGGG | c.252G>G/C | 84K/N | Mut-Heter/WT | Possibly damaging (0.597) | Damaging (0) |
| FL55 | HIST1H1B | CGGTG(G/C)CTACG | c.217G>G/C | 73G/A | Mut-Heter/WT | Probably damaging (1.000) | Damaging (0.02) |
| L27 | HIST1H1C | CCAAG(C/T)CCAAG | c.352C>C/T | 118P/S | Mut-Heter/WT | Possibly damaging (0.518) | Tolerated (0.55) |
| L28 | HIST1H1C | AAGTG(G/C)CTAAG | c.511G>G/C | 171A/P | Mut-Heter/WT | Probably damaging (0.987) | Tolerated (0.42) |
| L77 | HIST1H1C | CTGCC(A/T)AAAGT | c.559A>A/T | 187K/X | Mut-Heter/WT | ||
| FL54 | HIST1H1C | ATCAA(A/T)CTTG(G/C)TCTCA | c.243A>A/T; c.248G>G/C | 81K/N; 83G/A | Mut-Heter/WT | Possibly damaging (0.927) | Damaging (0) |
| FL59 | HIST1H1C | CAGAG(C>G)TCATC | c.127C>G | 43L->V | Mut-Homo/WT | Possibly damaging (0.927) | Damaging (0) |
| FL75 | HIST1H1C | TGAAAAAA(A)GCGTTG | c.192Homo_InsA | Frameshift insertion | Mut-Homo/WT | ||
| L7 | HIST1H1D | CAACA(AACA)GCCGT | c.236Het_InsAACA | Frameshift insertion | Mut-Heter/WT | ||
| L27 | HIST1H1D | AAG(G/A)TA(C/T)CGG | c.275G>G/A c.278C>C/T | 99G/D 100T/I | Mut-Heter/WT | Probably damaging (0.999) | Damaging (0) |
| L46 | HIST1H1D | ATCAA(G/C)CTTGG | c.246G>G/C | 82K/N | Mut-Heter/WT | Possibly damaging (0.774) | Damaging (0) |
| L53 | HIST1H1D | CGGTG(C/T)TTCTG | c.305C>C/T | 102A/V | Mut-Heter/WT | Probably damaging (0.995) | Damaging (0) |
| L55 | HIST1H1D | GGCGG(C/G)CAAGC | c.602C>C/G | 201A/G | Mut-Heter/WT | Benign (0.191) | Tolerated (0.65) |
| L56 | HIST1H1D | AGAAAG(C/T)GCTTG | c.197C>C/T | 66A/V | Mut-Heter/WT | Possibly damaging (0.954) | Tolerated (0.34) |
| ML64 | HIST1H1D | GCTTG(G>A)CCTCA | c.251G>A | 84G->D | Mut-Homo/WT | Probably damaging (0.98) | Damaging (0.05) |
| ML90 | HIST1H1D | CAAAG(G/A)TACTC | c.275G>G/A | 92G/D | Mut-Heter/WT | Probably damaging (0.999) | Damaging (0) |
| FL6 | HIST1H1D | GCGCC(G/C)CTACC | c.436G>G/C | 146A/P | Mut-Heter/WT | Possibly damaging (0.769) | Tolerated (0.25) |
| FL39* | HIST1H1D | CAAAGG(*)TACCGG | c.300Het_Ins* | Frameshift insertion | Mut-Heter/WT | ||
| FL52 | HIST1H1D | AACAC(C/G)TCAGC | c.542C>C/G | 181P/R | Mut-Heter/WT | Benign (0.001) | Damaging (0) |
| L23 | HIST1H1E | AGCCC(A/G)AGAAG | c.415A>A/G | 139K/E | Mut-Heter/WT | Benign (0.024) | Tolerated (0.06) |
| L24 | HIST1H1E | CCAAG(C/G)CAAAA | c.541C>C/G | 181P/A | Mut-Heter/WT | Benign (0.096) | Tolerated (1) |
| L27 | HIST1H1E | CTGCA(G/A)CTGCT | c.490G>G/A | 164A/T | Mut-Heter/WT | Possibly damaging (0.917) | Tolerated (0.07) |
| L64 | HIST1H1E | GCCTA(A/G)GGCTA | c.356A>A/G | 120A/T | Mut-Heter/WT | Benign (0) | Tolerated (1) |
| L114 | HIST1H1E | GGTTC(C)TTCAA | c.312Het_DelC | Frameshift insertion | Mut-Heter/WT | ||
| ML47 | HIST1H1E | CTAAA(G/A)CTGTT | c.139G>G/A | 47A/T | Mut-Heter/WT | Possibly damaging (0.597) | Tolerated (0.08) |
| ML90 | HIST1H1E | GTCGG(G/A)TTCCT; | c.308G>G/A | 103G/D | Mut-Heter/WT | Probably damaging (1.0) | Damaging (0) |
| ML90 | HIST1H1E | AGCCA(G/A)CAGGA | c.394G>G/A | 132A/T | Mut-Heter/WT | Benign (0) | Tolerated (0.56) |
| FL7 | HIST1H1E | CATTA(C/G)TAAAG | c.134C>C/G | 45T/S | Mut-Heter/WT | Possibly damaging (0.795) | Damaging (0.04) |
| FL29 | HIST1H1E | GCGGC(G/A)TATCT | c.169G>G/A | 57V/I | Mut-Heter/WT | Possibly damaging (0.951) | Tolerated (0.15) |
| FL34 | HIST1H1E | AGACC(C/T)CAAAG | c.463C>C/T | 155P/S | Mut-Heter//WT | Possibly damaging (0.799) | Tolerated (0.46) |
| FL39 | HIST1H1E | GGGTT(G/T)CTTCA | c.311G>G/T | 104S/F | Mut-Heter/WT | Probably damaging (0.975) | Damaging (0) |
| FL39 | HIST1H1E | TGGAG(C/T)CAAAA | c.500C>C/T | 167A/V | Mut-Heter/WT | Benign (0.209) | Tolerated (0.24) |
| FL42 | HIST1H1E | CCAAG(C/T)CAAAA | c.541C>C/T | 181P/S | Mut-Heter/WT | Possibly damaging (0.798) | Tolerated (0.66) |
| FL61 | HIST1H1E | CTAAA(A/T)AGGCA | c.364A>A/T | 122K/X | Mut-Heter/WT |
Boldface text indicates the nucleotide change from WT (first) to mutated (second).
cDNA, complementary DNA; T/N, tumor/normal; WT, wild-type.
Full insert sequence: (AACAGCCGGTATCAAGCTTGGCCTCAAGAGCTTGGTGAGCAAAGGTACTCTGGTGCAGACCAAAGG).
Details of confirmed recurrent gene mutations in OCT2, IRF8, ARID1A, and MCL1 in 114 FL cases
| FL case no. . | Gene . | Sequence context of mutations . | cDNA changes . | Amino acid change (protein) . | Sanger sequencing result (T/N) . | PolyPhen-2 prediction results . | SIFT prediction results . |
|---|---|---|---|---|---|---|---|
| L46 | OCT2 | GCCAG(A/T)CGACC | c.667A>A/T | 223T/S | Mut-Heter/WT | Possibly damaging (0.949) | Damaging (0) |
| L55 | OCT2 | GCCTA(C/T)CTCAG | c.920C>C/T | 307T/I | Mut-Heter/WT | Probably damaging (1.0) | Damaging (0) |
| L62 | OCT2 | GCCAG(A/G)CGACC | c.667A>A/G | 223T/A | Mut-Heter/WT | Benign (0.26) | Damaging (0) |
| ML47 | OCT2 | AGTGA(T)CCGCG | c.974Het_DelT | Frameshift deletion | Mut-Heter/WT | ||
| ML90 | OCT2 | GAGAC(G/A)CAAGAA | c.845G>G/A | 282R/H | Mut-Heter/WT | Probably damaging (1.0) | Damaging (0) |
| FL42 | OCT2 | GCCAG(A/G)CGACC | c.667A>A/G | 223T/A | Mut-Heter/WT | Benign (0.26) | Damaging (0) |
| FL59 | OCT2 | CTGTG(G/C)GGACG | c.1174 G>G/C | 392G/R | Mut-Heter/WT | Probably damaging (0.99) | Tolerated (0.19) |
| FL69 | OCT2 | CTGTG(G/C)GGACG | c.1174 G>G/C | 392G/R | Mut-Heter/WT | Probably damaging (0.99) | Tolerated (0.19) |
| FL71 | OCT2 | GCCAG(A/T)CGACC | c.667A>A/T | 223T/S | Mut-Heter/WT | Possibly damaging (0.949) | Damaging (0) |
| L62 | IRF8 | CCGTC(T/A)AAGTG | c.1279T>A/T | 427X/K | Mut-Heter/WT | ||
| L62 | IRF8 | TTCCT(G/A)AGGAA | c.343G>G/A | 115E/K | Mut-Heter/WT | Benign (0.433) | Tolerated (0.42) |
| ML55 | IRF8 | AACTG(G)CAGAA | c.1132Het_DelG | Frameshift deletion | Mut-Heter/WT | ||
| ML51 | IRF8 | CCCCC(G>T)AGGAG | c.1183G>T | 395E>X | Mut-Homo/WT | ||
| FL3 | IRF8 | ATGCC(T/G)CCATT | c.163T>G/T | 55S/A | Mut-Heter/WT | Benign (0.041) | Tolerated (1) |
| FL19 | IRF8 | TTCCT(G/A)AGGAA | c.343G>A/G | 115E/K | Mut-Heter/WT | Benign (0.433) | Tolerated (0.42) |
| FL27 | IRF8 | TGATG(C/T)AGGCC | c.1174C>C/T | 392Q/X | Mut-Heter/WT | ||
| FL51 | IRF8 | ATGTT(T)CCAGA | c.1218Het_DelT | Frameshift deletion | Mut-Heter/WT | ||
| ML55/FL8 | IRF8 | GGTGA(C/T)GCGGA | c.800C>C/T | 267T/M | Mut-Heter/Mut-Heter | Possibly damaging (0.954) | Tolerated (0.06) |
| ML2 | ARID1A | ATACA(G/T)GTCAA | Splice site | No | Mut-Heter/WT | ||
| ML30 | ARID1A | GACCA(C/T)GACAG | c.3826C>C/T | 1276R/X | Mut-Heter/WT | ||
| ML66 | ARID1A | GCGGG(G/T)GAACT | c.853G>G/T | 285G/X | Mut-Heter/WT | ||
| ML69 | ARID1A | GGGGG(G)AACTC | c.854Het_InsG | Frameshift insertion | Mut-Heter/WT | ||
| L38 | ARID1A | ATCCT(C/T)AGCCC | c.1741C>C/T | 581Q/X | Mut-Heter/WT | ||
| L62 | ARID1A | AGGGG(G)CATGAA | c.2666Het_DelG | Frameshift deletion | Mut-Heter/WT | ||
| FL6 | ARID1A | TAACA(T/C)GGCCA | c.2615T>T/C | 872M/T | Mut-Heter/WT | Possibly damaging (0.688) | Tolerated (0.58) |
| FL8 | ARID1A | GCCTA(C/G)GGCTT | c.444C>C/G | 148Y/X | Mut-Heter/unknown | ||
| FL15 | ARID1A | GCATC(C/T)GAGGC | c.2077C>C/T | 693R/X | Mut-Heter/WT | ||
| FL46 | ARID1A | CTCGG(C>T)CCGGG | c.914C>T | 305A>V | Mut-Homo/WT | Possibly damaging (0.675) | Tolerated (0.29) |
| FL60 | ARID1A | TCCAC(C/T)AACAA | c.511C>C/T | 171Q/X | Mut-Heter/WT | ||
| FL68 | ARID1A | TCCAC(C/T)AACAA | c.511C>C/T | 171Q/X | Mut-Heter/WT | ||
| FL72 | ARID1A | GGGGG(G)ACACC | c.5548Het_InsG | Frameshift insertion | Mut-Heter/WT | ||
| ML33 | MCL1 | GCCAG(C/T)AGAGG | c.450C>C/T | 167 A/V | Mut-Heter/WT | Possibly damaging (0.454) | Tolerated (0.14) |
| ML47 | MCL1 | CACCA(G/C)TACGG | c.464G>G/C | 155 S/T | Mut-Heter/WT | Benign (0.001) | Tolerated (0.19) |
| ML90 | MCL1 | CACCA(G/A)TACGG | c.464G>G/A | 155 S/N | Mut-Heter/WT | Benign (0.0) | Tolerated (0.21) |
| FL case no. . | Gene . | Sequence context of mutations . | cDNA changes . | Amino acid change (protein) . | Sanger sequencing result (T/N) . | PolyPhen-2 prediction results . | SIFT prediction results . |
|---|---|---|---|---|---|---|---|
| L46 | OCT2 | GCCAG(A/T)CGACC | c.667A>A/T | 223T/S | Mut-Heter/WT | Possibly damaging (0.949) | Damaging (0) |
| L55 | OCT2 | GCCTA(C/T)CTCAG | c.920C>C/T | 307T/I | Mut-Heter/WT | Probably damaging (1.0) | Damaging (0) |
| L62 | OCT2 | GCCAG(A/G)CGACC | c.667A>A/G | 223T/A | Mut-Heter/WT | Benign (0.26) | Damaging (0) |
| ML47 | OCT2 | AGTGA(T)CCGCG | c.974Het_DelT | Frameshift deletion | Mut-Heter/WT | ||
| ML90 | OCT2 | GAGAC(G/A)CAAGAA | c.845G>G/A | 282R/H | Mut-Heter/WT | Probably damaging (1.0) | Damaging (0) |
| FL42 | OCT2 | GCCAG(A/G)CGACC | c.667A>A/G | 223T/A | Mut-Heter/WT | Benign (0.26) | Damaging (0) |
| FL59 | OCT2 | CTGTG(G/C)GGACG | c.1174 G>G/C | 392G/R | Mut-Heter/WT | Probably damaging (0.99) | Tolerated (0.19) |
| FL69 | OCT2 | CTGTG(G/C)GGACG | c.1174 G>G/C | 392G/R | Mut-Heter/WT | Probably damaging (0.99) | Tolerated (0.19) |
| FL71 | OCT2 | GCCAG(A/T)CGACC | c.667A>A/T | 223T/S | Mut-Heter/WT | Possibly damaging (0.949) | Damaging (0) |
| L62 | IRF8 | CCGTC(T/A)AAGTG | c.1279T>A/T | 427X/K | Mut-Heter/WT | ||
| L62 | IRF8 | TTCCT(G/A)AGGAA | c.343G>G/A | 115E/K | Mut-Heter/WT | Benign (0.433) | Tolerated (0.42) |
| ML55 | IRF8 | AACTG(G)CAGAA | c.1132Het_DelG | Frameshift deletion | Mut-Heter/WT | ||
| ML51 | IRF8 | CCCCC(G>T)AGGAG | c.1183G>T | 395E>X | Mut-Homo/WT | ||
| FL3 | IRF8 | ATGCC(T/G)CCATT | c.163T>G/T | 55S/A | Mut-Heter/WT | Benign (0.041) | Tolerated (1) |
| FL19 | IRF8 | TTCCT(G/A)AGGAA | c.343G>A/G | 115E/K | Mut-Heter/WT | Benign (0.433) | Tolerated (0.42) |
| FL27 | IRF8 | TGATG(C/T)AGGCC | c.1174C>C/T | 392Q/X | Mut-Heter/WT | ||
| FL51 | IRF8 | ATGTT(T)CCAGA | c.1218Het_DelT | Frameshift deletion | Mut-Heter/WT | ||
| ML55/FL8 | IRF8 | GGTGA(C/T)GCGGA | c.800C>C/T | 267T/M | Mut-Heter/Mut-Heter | Possibly damaging (0.954) | Tolerated (0.06) |
| ML2 | ARID1A | ATACA(G/T)GTCAA | Splice site | No | Mut-Heter/WT | ||
| ML30 | ARID1A | GACCA(C/T)GACAG | c.3826C>C/T | 1276R/X | Mut-Heter/WT | ||
| ML66 | ARID1A | GCGGG(G/T)GAACT | c.853G>G/T | 285G/X | Mut-Heter/WT | ||
| ML69 | ARID1A | GGGGG(G)AACTC | c.854Het_InsG | Frameshift insertion | Mut-Heter/WT | ||
| L38 | ARID1A | ATCCT(C/T)AGCCC | c.1741C>C/T | 581Q/X | Mut-Heter/WT | ||
| L62 | ARID1A | AGGGG(G)CATGAA | c.2666Het_DelG | Frameshift deletion | Mut-Heter/WT | ||
| FL6 | ARID1A | TAACA(T/C)GGCCA | c.2615T>T/C | 872M/T | Mut-Heter/WT | Possibly damaging (0.688) | Tolerated (0.58) |
| FL8 | ARID1A | GCCTA(C/G)GGCTT | c.444C>C/G | 148Y/X | Mut-Heter/unknown | ||
| FL15 | ARID1A | GCATC(C/T)GAGGC | c.2077C>C/T | 693R/X | Mut-Heter/WT | ||
| FL46 | ARID1A | CTCGG(C>T)CCGGG | c.914C>T | 305A>V | Mut-Homo/WT | Possibly damaging (0.675) | Tolerated (0.29) |
| FL60 | ARID1A | TCCAC(C/T)AACAA | c.511C>C/T | 171Q/X | Mut-Heter/WT | ||
| FL68 | ARID1A | TCCAC(C/T)AACAA | c.511C>C/T | 171Q/X | Mut-Heter/WT | ||
| FL72 | ARID1A | GGGGG(G)ACACC | c.5548Het_InsG | Frameshift insertion | Mut-Heter/WT | ||
| ML33 | MCL1 | GCCAG(C/T)AGAGG | c.450C>C/T | 167 A/V | Mut-Heter/WT | Possibly damaging (0.454) | Tolerated (0.14) |
| ML47 | MCL1 | CACCA(G/C)TACGG | c.464G>G/C | 155 S/T | Mut-Heter/WT | Benign (0.001) | Tolerated (0.19) |
| ML90 | MCL1 | CACCA(G/A)TACGG | c.464G>G/A | 155 S/N | Mut-Heter/WT | Benign (0.0) | Tolerated (0.21) |
OCT2 data based on transcript accession ID #CCDS33035.1. L46 also contains a HIST1H1D 82K/N mutation absent in FL71. L62 also contains an IRF8 427X/K and ARID1A fs mutation. FL42 contains a HIST1H1E 181P/S mutation. FL59 contains a HIST1H1C 43L/V mutation absent in FL69. IRF8 data are based on transcript accession ID CCDS10956.1. ARID1A data are based on transcript accession ID CCDS285.1. MCL1 data are based on transcript accession ID CCDS957.1. Boldface text indicates the nucleotide change from WT (first) to mutated (second).
cDNA, complementary DNA; T/N, tumor/normal; WT, wild-type.
Overall, in addition to MLL2, CREBBP, BCL2, and TNFRSF14, we validated 15 recurrently mutated genes in the discovery set of 11 FL and 1 t-FL cases, most of which have not been previously described as recurrently somatically mutated in FL. These include the genes HIST1H1E, IRF8, OCT2 (POU2F2), ARID1A, MCL1, TP53, VPS39, GRM7, ZNF541, LYST, CHRM3, DMRT3, WDR64, PCLO, and KLHDC7B; all of these but LYST and PCLO were significantly mutated when compared with either gene-specific Sanger-confirmed silent mutation rates or mutation rates across the entire exome (supplemental Table 6). Details of these mutations have been summarized in Table 1.
From these recurrently mutated genes, we selected IRF8, OCT2, ARID1A, MCL1, DMRT3, CHRM3, VPS39, and GRM7, and based on their functional similarity to HIST1H1E the 5 linker histone H1 family genes A to E (HIST1H1 A-E) for an initial expansion mutation analysis for a combined total of 55 FL cases (cohort I). The results for GRM7, VPS39, CHRM3, and DMRT3 are summarized in supplemental Table 7.
Finally, to obtain more precise estimates of the frequency of gene mutations in the linker histone H1 family genes B-E (HIST1H1 B-E), IRF8, OCT2, ARID1A, and MCL1 in FL, we analyzed an additional 59 FL cases (cohort II) for a combined total of 114 FL cases. These results are discussed in the next section.
Identification of high-frequency mutations in the linker histone genes HIST1H1 B-E in FL
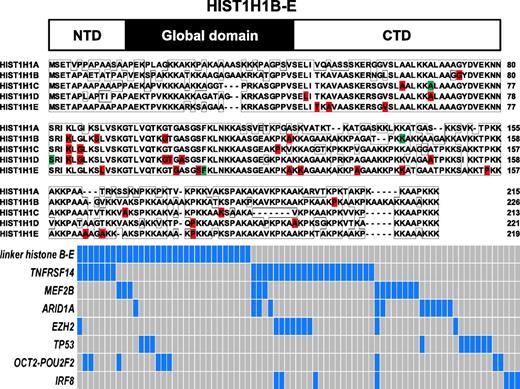
The linker histone genes serve functions in nucleosome spacing and gene regulation, in part involving alterations in gene methylation38 and in part involving direct protein-to-protein interactions.39,40 In this study, we identified 42 somatically acquired mutations and 8 somatically acquired synonymous nucleotide changes in 1 of 4 HIST1H1 genes (HIST1H1 B-E; HIST1H1A was not mutated) and 7 germline mutations. Overall, 27% (31/114) of FL cases carried mutations in HIST1H1 B-E. Of these, 6 FL cases carried 2 or 3 mutations, making this gene family one of the most frequently mutated gene targets in FL (Table 2). Although occasional indels and nonsense mutations were identified, the majority of HIST1H1 B-E mutations were of the missense type, and most were restricted to the C-terminal portion of these proteins (Figure 1).41 The latter finding provided evidence against a passenger status for these mutations (also note a ratio of nonsynonymous mutations and indels to synonymous mutations in HIST1B-E of 5.1:1 (higher than expected for random bystander mutagenesis) and that HIST1H1 A was not targeted for mutations).
Identification of novel mutations in the linker HIST1H1 genes B-E in FL and their association with other recurrent gene mutations. Top: schema of the HIST1H1 protein domain structure. Middle: details of somatic mutations identified in HIST1H1B-E in FL and comparative alignment of HIST1H1B-E amino acid residues. Red, location of missense mutations; green, location of indel mutation. Bottom: schema of the co-occurrence of linker histone HIST1H1B-E mutations and mutations in other genes. Cases with mutations are highlighted in blue.
Identification of novel mutations in the linker HIST1H1 genes B-E in FL and their association with other recurrent gene mutations. Top: schema of the HIST1H1 protein domain structure. Middle: details of somatic mutations identified in HIST1H1B-E in FL and comparative alignment of HIST1H1B-E amino acid residues. Red, location of missense mutations; green, location of indel mutation. Bottom: schema of the co-occurrence of linker histone HIST1H1B-E mutations and mutations in other genes. Cases with mutations are highlighted in blue.
A comparative review of the mutation status of other recurrently mutated genes in FL (TNFRSF14, MEF2B, ARID1A, EZH2, TP53, OCT2, and IRF8) uncovered that mutations in HIST1H1 B-E and in ARID1A/EZH2 combined or, alternatively, in IRF8, were largely nonoverlapping (Figure 1, bottom panel).
Given recent reports of physical binding of HIST1B-E proteins to DNMT1 and DNMT3B, we tested for these interactions in coimmunoprecipitations in transient transfection assays using all HIST1B-E WT proteins and many of the HIST1B-E mutants.40 Although we could not detect binding of DNMT1 to the HIST1B-E WT proteins, all of these linker histones co-immunoprecipitated DNMT3B. When testing the HIST1B-E mutant proteins for DNMT3B binding, 2 patterns of binding emerged: (1) absent binding for the indel mutants affecting the C-terminus (with frameshifts [fss] located N-terminal to amino acid residue ∼120) or (2) binding despite some quantitative differences (supplemental Figure 1A-D).
Mutations in the transcription factor OCT2 in FL
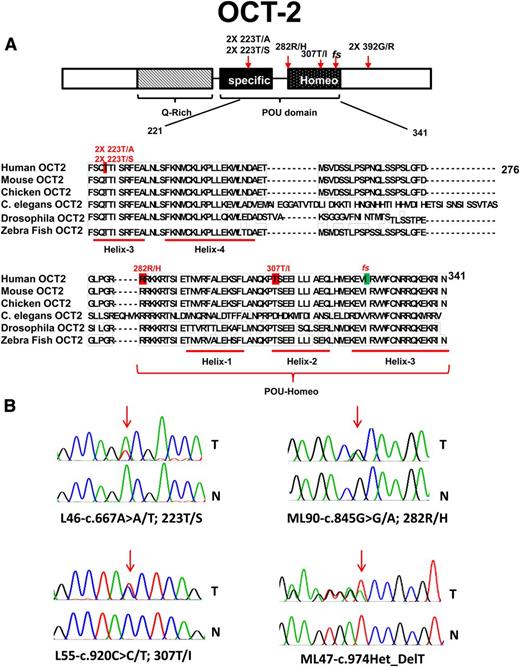
OCT2 is a member of the POU-homeodomain-containing family of transcription factors. The OCT2-POU domain is bipartite and consists of a POU-specific and POU-homeo subdomain, both of which interact with DNA via helix-turn-helix motifs (Figure 2).31,42,43 We have identified novel, somatically acquired heterozygous mutations in OCT2 in 8% (9/114) of FL cases; these were predominantly missense mutations located in evolutionarily conserved amino acid residues in the POU DNA–binding domain. In 1 case we observed a mono-allelic fs mutation (Table 3; Figure 2). Multiple OCT2 mutations recurrently targeted the amino acid residues Thr223 and Gly392.
Identification of novel mutations in OCT2 in FL. (A) Schema of the OCT2 protein domain structure, details of somatic mutations identified in OCT2 in FL, and a comparative alignment of OCT-2 amino acid residues across multiple species. Red, location of missense mutations; green, location of indel mutation. NTD, N-terminal domain; CTD, C-terminal domain. (B) OCT2 sequence traces of paired tumor (T) and normal (N) samples.
Identification of novel mutations in OCT2 in FL. (A) Schema of the OCT2 protein domain structure, details of somatic mutations identified in OCT2 in FL, and a comparative alignment of OCT-2 amino acid residues across multiple species. Red, location of missense mutations; green, location of indel mutation. NTD, N-terminal domain; CTD, C-terminal domain. (B) OCT2 sequence traces of paired tumor (T) and normal (N) samples.
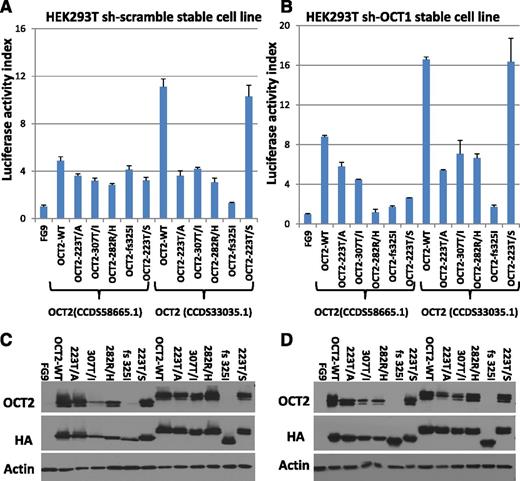
We tested the transactivation potential of epitope-tagged WT and mutant OCT2 in transient transfection assays using 2 isoforms of OCT2. Luciferase activity data normalized to protein input are summarized in Figure 3A and corresponding immunoblot data in Figure 3C; these indicate partial loss of transactivating function for most of the tested OCT2 mutations (note that BOB1, a binding partner for OCT1 and OCT2, is expressed in HEK293 cells; supplemental Figure 2F). To minimize interference in the assay by OCT1, the ubiquitously expressed OCT family member protein that can bind to the same DNA binding sites as OCT2, we generated a stable HEK293T cell line in which the expression of the OCT1 protein was knocked down (supplemental Figure 2E) and repeated the OCT2 transient transfection assays; these results are displayed in Figure 3B,D. Given that the FL-associated OCT2 mutations are mono-allelic, we co-transfected WT and mutant OCT2 plasmids in equal amounts. We measured a transactivation output that was below the expected value (defined as the sum of half of the output measured in the same experiments for WT and individual OCT2 mutants when transfected separately). These results and supporting data are displayed in supplemental Figure 2A-D.
Results of luciferase assays of OCT2 and OCT2 mutants. Two different isoforms of OCT2 were used as indicated. (A) Luciferase assays results in HEK293-T cells indexed to the vector control (FG9) and normalized to protein expression by densitometry. (B) Luciferase assays results in HEK293-T cells carrying stable knockdown of OCT1 indexed to the vector control (FG9) and normalized to protein expression by densitometry. (C) Immunoblot results for the data presented in (A). (D) Immunoblot results for the data presented in (B). (Please note that the anti-OCT2 antibody used does not recognize some of the OCT2 mutants well.)
Results of luciferase assays of OCT2 and OCT2 mutants. Two different isoforms of OCT2 were used as indicated. (A) Luciferase assays results in HEK293-T cells indexed to the vector control (FG9) and normalized to protein expression by densitometry. (B) Luciferase assays results in HEK293-T cells carrying stable knockdown of OCT1 indexed to the vector control (FG9) and normalized to protein expression by densitometry. (C) Immunoblot results for the data presented in (A). (D) Immunoblot results for the data presented in (B). (Please note that the anti-OCT2 antibody used does not recognize some of the OCT2 mutants well.)
A subsequent review of the crystal structure of OCT1 bound to an octamer site44 disclosed that the OCT2 fs 325 mutation disrupted helix 3, which is critical for binding the AAAT octamer subsite, explaining the lack of transactivation. OCT2 307T/I locates to helix 2, which forms a HTH motif with helix 3. Similarly, OCT2 residue 223 (the OCT1 Thr-45 equivalent) makes critical contacts with the octamer ATGC subsite, and OCT2 223T/A displayed reduced transactivation activity. Interestingly, OCT2 223 T/S did not show significant loss of transactivation activity, and Ser is found naturally at this site in selected POU domains.45 Finally, OCT2 residue 282 is involved in minor groove DNA contacts, and OCT2 R/H showed reduced transactivation activity as well.
The physiological targets of OCT2 in human B cells are essentially unknown. OCT2 is not required for surface immunoglobulin expression in the mouse,46 and no differences in surface immunoglobulin expression between FL cases with OCT2 WT or OCT2 mutants or lymphoma cell lines with transiently transfected OCT2 expression constructs (discussed below) were identified (supplemental Figure 3A).
Given the experimental and structural evidence for lowered functional activity of OCT2 mutants, we initially mimicked such lowered FL-associated OCT2 output through generation of a combined 8 LY1, LY7, and LY10 lymphoma cell lines that stably expressed validated OCT2 targeting short hairpin RNA (shRNA), as well as nontargeted shRNA (OCT2 knockdown mean of 87% and range 80%-90%; supplemental Figure 4A). RNA isolated from 8 lines and shRNA controls were analyzed using Affymetrix 2.1 ST arrays (supplemental Methods). Transcripts demonstrating >1.2-fold change and a false discovery rate (FDR) of <0.2 were further filtered for known OCT2 binding sites using ENCODE Chipseq data for OCT2 (supplemental Methods and supplemental Table 8). Overall, OCT2-dependent transcriptome changes were mild (less than twofold) and bidirectional, but a few novel OCT2-dependent, biologically interesting genes (FKBPL, GNG8, DOK3, BCL6B, and RACGAP1; CD36 or CRISP3 were not expressed in these lines) were identified.
Next, in separate experiments we used recombinant lentiviruses to transiently transfect OCI-LY1, LY7, LY10, and LY18, each expressing either HA-tagged OCT2 WT or OCT2 R282H or OCT2 fs 325. The expression levels of OCT1 and BOB1 and of BCL2 and BCL6 were not altered post-infection day 4 (supplemental Figure 4B-C).
We subjected RNA to Affymetrix 2.1 ST expression array analysis. Gene set enrichment analysis identified 3 significant (FDR <0.25) enriched sets for OCT2 WT transfected cells, 1 of which was also present in the OCT2 R282H analysis, and no enriched sets for the OCT2 fs 325 mutant (supplemental Figure 5). The gene set enrichment analysis did not identify networks specific to OCT2 R282H or OCT2 fs 325.
In a separate statistical analysis, we sought identification of individual genes that were significantly up- or downregulated in transiently transfected OCI-LY1, OCI-LY10, and OCI-LY18 lines. Expression changes for genes containing OCT2 binding sites at a FDR <0.2 and >1.5-fold change, by either WT OCT2, OCT2 282 R/H, or OCT2 325 fs were limited to a small number of genes with limited overlap (supplemental Table 9).
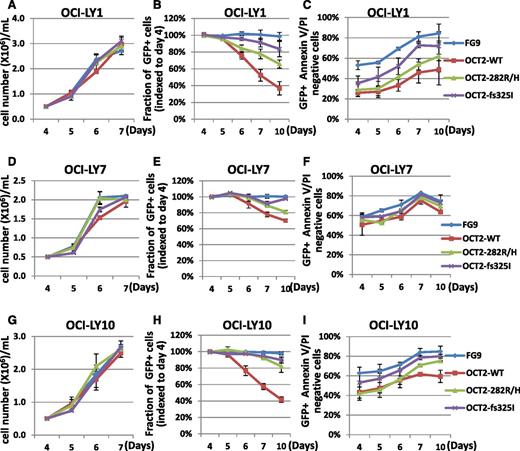
Next, in 2 separate experiments, we transiently transduced LY1, LY7, and LY10 lymphoma cells with OCT2 WT, OCT2 R282H, OCT2 325 fs mutants, or empty lentiviruses, and cultured cells for as many as 10 days post-infection. In these cultures, we measured preferential loss over time of green fluorescent protein (GFP)-positive cells in OCT2 WT–expressing lines and, to a lesser extent, in the OCT2 R282H lines compared with the OCT2 fs 325 mutant or FG9 vector–only lines, and higher levels of apoptotic cell death. These findings offer complementary in vivo evidence for beneficial effects of attenuating OCT2 activity through mutations in FL cells (Figure 4).
Effect of OCT2 expression on cell growth and survival in LY cell lines. (A-I) Analysis of transiently transfected OCI-LY1, OCI-LY7, and OCI-LY10 cells. (A,D,G) Serial cell density. (B,E,H) Serial analysis of the fraction of GFP-positive cells indexed to the fraction of GFP-positive cells per transfectant on day 4 post-viral innoculation. (C,F,I) Serial analysis of the percentage of Annexin V/propidium iodide–negative (live) cells.
Effect of OCT2 expression on cell growth and survival in LY cell lines. (A-I) Analysis of transiently transfected OCI-LY1, OCI-LY7, and OCI-LY10 cells. (A,D,G) Serial cell density. (B,E,H) Serial analysis of the fraction of GFP-positive cells indexed to the fraction of GFP-positive cells per transfectant on day 4 post-viral innoculation. (C,F,I) Serial analysis of the percentage of Annexin V/propidium iodide–negative (live) cells.
Inactivating mutations in ARID1A in FL
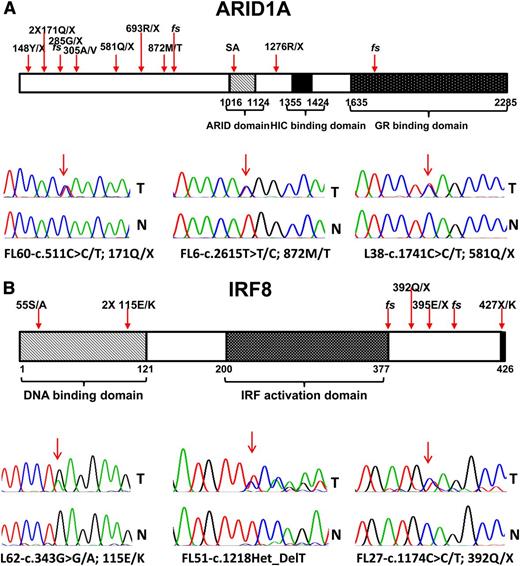
Mutations in ARID1A were identified in 11% (13/114) of FL cases.47 These mutations were predominantly mono-allelic and inactivating, and were distributed over the entire length of the gene (Table 3 and Figure 5A). By genetic criteria, these mutations support a tumor suppressor gene function of ARID1A in FL, similar to what has been proposed in many other cancers.48 Recurrent mutations in ARID1A in FL further strengthen the evidence for epigenetic deregulation as a major pathobiological principle in FL pathogenesis.
Identification of novel mutations in ARID1A and IRF8 in FL. Schema of the ARID1A and IRF8 protein domain structures and indication of FL-associated mutations, including sequence traces of paired tumor (T) and normal (N) samples.
Identification of novel mutations in ARID1A and IRF8 in FL. Schema of the ARID1A and IRF8 protein domain structures and indication of FL-associated mutations, including sequence traces of paired tumor (T) and normal (N) samples.
Inactivating mutations in the transcription factor IRF8 in FL
We identified 6% (7/114) of FL cases harboring a total of 7 IRF8 mutations and 2 cases with an IRF8 germline mutation (267T/M) that may be functionally relevant (Table 3 and Figure 5B).36 The IRF8 mutations comprised predominantly indels and nonsense mutations and a missense mutation converting the IRF8 stop codon into a codon for the amino acid lysine. One of the mutations was biallelic because of aUPD at chromosome 16. All truncating or fs mutations were located in the C-terminus of IRF8, potentially allowing for the expression of mutant IRF8 proteins with altered function.
Given the prior observations of epigenetic silencing of IRF8 in myeloid tumors, we proceeded with quantitative polymerase chain reaction–based measurements of IRF8 expression in cDNA made from RNA isolated from sorted lymphomatous B cells compared with sorted, lymph node–derived normal B cells. As is seen in supplemental Figure 6, although BCL2 expression was substantially higher in lymphoma vs nonlymphoma B cells, the mean expression levels of IRF8 (and of OCT2 and MCL1) were similar in comparison, although the lymphoma samples were characterized by a higher variability in the expression for all 3 genes than the normal B cells.
Novel PEST-domain mutations in MCL1 in FL
We detected 3% (3/114) of FL cases with novel somatically acquired missense mutations affecting amino acid residues 155 (155S/T and 155S/N) and 167 (167A/V) in the MCL1 PEST domain (supplemental Figure 7). One of these mutants occurred in the setting of a chromosomal gain on chromosome 1. Although the location of these mutations in the PEST domain suggested an effect on MCL1 protein stability, we could not detect substantial changes in the MCL1 half-life in 293 cell transfection studies after cycloheximide treatment (supplemental Figure 7C). We did detect a slight increase in the amount of USP9X that co-immunoprecipitated with the MCL1 mutants 155S/T and 155S/N (supplemental Figure 7D), but the absolute amount of USP9X that co-immunoprecipitated was <1% of the total.
Other gene mutations identified in the discovery panel of 11 FL cases and 1 t-FL case
We identified and confirmed many gene mutations occurring once or twice each in the discovery panel of 11 FL and 1 t-FL cases (supplemental Results, Tables 1-3, and supplemental Table 4), and some of these mutations are briefly mentioned here: multiple mutations in the histone genes HIST1H2AC and HIST1H2AM and a mutation in HIST1H4; missense mutations in TLR2 and TLR8; coexisting fs and missense mutations in EP300; a heterozygous nonsense mutation in TNFAIP3/A20 (a known tumor suppressor gene in a variety of NHLs and Hodgkin lymphoma that is located at 6q2312,49 ); a missense mutation in SYK at a known negative regulatory tyrosine residue (323Y>H) predicted to be activating; a heterozygous nonsense mutation in VAV1 located within a genomic deletion; a missense mutation in the proto-oncogene PRDM16; a heterozygous missense mutation predicted to be damaging in the tumor suppressor gene PTEN (deletions of 1 PTEN locus as part of interstitial deletions at 10q were detected in 3 FL cases, discussed below); a heterozygous missense mutation in CBL-B, predicted to be damaging as well; and, finally, various mutations in MAP kinase family members, phosphatases, signaling molecules, and DNA maintenance proteins.
aCNA and LOH in FL, and relation to gene mutations
Please see supplemental Results.
Discussion
In this study we present the combined results of next-generation sequencing and high-density SNP 6.0 array–based genomic profiling for 11 FL cases and 1 t-FL case and expansion of interesting findings into a combined total of 114 FL cases. Genomic profiling was done on DNA isolated from sorted cell populations, allowing for the highest signal-to-noise ratio for this type of analysis. We report on the identification of 15 novel significantly recurrently mutated genes in FL—the largest collection of novel gene targets in FL reported to date. Recurrently mutated genes were detected in FL samples that were derived from untreated or treated patients without any significant frequency differences, and therefore we identify multiple genes that appear to be directly involved in the pathogenesis of FL. While this work was under review, a study on the genetics of FL transformation identified mutations in some of the highlighted genes reported here (HIST1H1-E, IRF8, and ARID1A) in t-FL at frequencies not significantly different from unselected FL, suggesting (1) no causal role for these genes in the transformation process and (2) predominant occurrence of these mutations in a transformed progenitor common to FL and t-FL.50 Further, we extend prior knowledge of genomic copy number aberrations and LOH in FL and provide a listing of genes that are located within regions of aUPD—a lesion type implicated as being frequently associated with mutated genes in other cancers. Overall, these data have implications for basic and clinical research in FL.
The linker histone H1 family of genes (B-E) was mutated in 27% of FL, making this gene family one of the most frequently mutated genes identified in FL. Similar mutations have recently been described in DLBCL.25,33,51 The linker HIST1H1 B-E mutations were largely restricted to the C-terminus, and FL-associated HIST1H1 B-E C-terminal deletion mutants demonstrated a lack of interaction with DNMT3B. The functional consequences of the missense mutations in linker HIST1H1 B-E remain unresolved and may comprise loss-of-function, gain-of-function, or neomorphic functions. The reported phenotypes of cells with single HIST1H1 gene deletions in various experimental systems have been subtle,52 but published evidence in triple HIST1H1 gene knock-out cells suggests altered expression of a small set of genes and enrichment for genes subject to epigenetic regulation. Also observed were changes in chromatin and decreased nucleosome spacing, but only a subset of FL cases demonstrated more than 1 affected HIST1H1 gene.38 Others have made connections between certain linker HIST1H1 proteins and the p53 protein, but these findings do not readily explain the mutation pattern observed in FL.39 Interestingly, very recent observations of direct binding of linker HIST1H1 proteins to DNMT1 and DNMT3B and effects on epigenetic silencing suggest that such a connection may tie NHL-associated HIST1H1 B-E mutations in with frequent mutations in MLL2 and mutations in EZH2 and ARID1A (11% frequency in this study) as reported here and may suggest an even wider role for aberrant epigenetic regulation and nucleosome positioning in FL than was previously recognized.40,48,53
In this study we describe mono-allelic mutations in the transcription factor OCT2 in 8% of FL. These mutations were predominantly localized to highly conserved residues in the OCT2-DNA–binding domain, suggesting a structural, and likely damaging, effect on the OCT2 protein. In selected lymphoma cell lines, OCT2 has previously been identified as a positive regulator of the expression of BCL2, but functional data in primary human B-cell lymphoma samples are sparse54,55 and we did not detect effects on BCL2 expression in our experimental systems. Our results based on luciferase assays and structural comparisons of OCT2 with OCT1 suggest that most mutant OCT2 proteins are hypomorphs. The latter conclusion is consistent with the observations that (1) multiple lymphoma cell lines with engineered overexpression of WT OCT2 undergo apoptosis or grow less well than cells transfected with FL-associated OCT2 mutants, suggesting that attenuation of OCT2 function is beneficial to lymphoma cells; and (2) that profound knockdown of OCT2 in multiple lymphoma cell lines was well tolerated.
The expression changes of genes containing OCT2 binding sites after OCT2 knockdown or after forced OCT2 expression in lymphoma cell lines were relatively mild. Given the lack of a predefined OCT2-dependent transcriptional signature or well-defined OCT2 target genes in human B cells, our data provide novel OCT2 target genes for study but do not provide firm conclusions about genes critical for OCT2 function in FL. Finally, given that some amino acids in OCT2 were recurrently mutated and given the complex interaction involving OCT1 and BOB1 on specific genes, as-yet unidentified context-dependent functions of these mutant OCT2 proteins in FL B cells remain a formal possibility.56
The identification of recurrent predominantly inactivating or potentially transdominant negative IRF8 mutations in 6% of FL suggests that loss of IRF8 function in FL is pathobiologically relevant.36 IRF8 expression is frequently lost in myeloid leukemia, and IRF8-deficient mice develop a chronic myeloid leukemia-like illness followed by sporadic transformation to acute myeloid leukemia,57 but IRF8 mRNA expression was not substantially altered in FL. Recently, conditional targeting of IRF8 in B cells using a CD19-Cre mouse deleter strain resulted in an expansion phenotype in germinal center B cells. It is thus possible that by analogy, IRF8 loss in FL B cells contributes to lymphoma expansion.30
In summary, this study substantially adds to the knowledge about the FL-coding genome and suggests its basic and clinical research implications. It also identifies multiple novel genes with potential importance to FL biology and is likely to stimulate novel research endeavors in this important NHL subtype.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful for services provided by the genomics core of the University of Michigan Comprehensive Cancer Center and for the contribution of the hematological malignancy group of the University of Michigan. They also thank Dr Lynn Corcoran for helpful discussions and information and data sharing regarding OCT2 biology, and Mark Sausen at Personal Genome Diagnostics for performing the statistical analysis of passenger gene probabilities.
Funded in part by grants from the Weather Wax Foundation (M.K.). and Garden Fresh Company (S.N.M.). S.N.M. is a Scholar in Clinical Research of the Leukemia and Lymphoma Society.
Authorship
Contribution: M.S.K., H.F., K.J., D.B., M.S., D.L., L.M., V.C., A.E.C., and S.N.M. enrolled patients and analyzed clinical data; H.L., Y.L., P.O., M.Y., K.S.-C., and S.N.M. performed the laboratory research; D.R. assisted with fluorescence in-situ hybridization analysis; S.J. assisted with bioinformatics analysis of next generation sequencing data; K.S. assisted with statistical methods; and S.N.M. conceived the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sami N. Malek, Department of Internal Medicine, Division of Hematology and Oncology, University of Michigan, 1500 E Medical Center Dr, Ann Arbor, MI 48109-0936; e-mail: smalek@med.umich.edu.
References
Author notes
H.L. and M.S.K. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal