Key Points
Aberrantly diminished expression of miR-150 allows advanced CTCL to invade multiple organs with upregulation of CCR6.
MiR-150 inhibits IL-22-CCL20-CCR6 autocrine signaling in advanced CTCL.
Abstract
In this study, we show that microRNA-150 (miR-150) is significantly downregulated in advanced cutaneous T-cell lymphoma (CTCL), and that this downregulation is strongly associated with tumor invasion/metastasis. Inoculation of CTCL cell lines into nonobese diabetic/Shi-scid interleukin 2γ (IL-2γ) null mice led to CTCL cell migration to multiple organs; however, prior transfection of the cells with miR-150 substantially reduced the invasion/metastasis by directly downregulating CCR6, a specific receptor for the chemokine CCL20. We also found that IL-22 and its specific receptor subunit, IL22RA1, were aberrantly overexpressed in advanced CTCL, and that production of IL-22 and CCL20 was increased in cultured CTCL cells. IL22RA1 knockdown specifically reduced CCL20 production in CTCL cells, suggesting that IL-22 upregulation may activate the production of CCL20 and its binding to CCR6, thereby enhancing the multidirectional migration potential of CTCL cells. CTCL cells also exhibited nutrition- and CCL20-dependent chemotaxis, which were inhibited by miR-150 transfection or CCR6 knockdown. From these findings, we conclude that, in the presence of continuous CCR6 upregulation accompanied by miR-150 downregulation, IL-22 activation leads to continuous CCL20-CCR6 interaction in CTCL cells and, in turn, autocrine metastasis to distal organs. This suggests miR-150, CCL20, and CCR6 could be key targets for the treatment of advanced CTCL.
Introduction
MicroRNAs (miRNAs) are a class of small regulatory RNA molecules that play important roles in tumor development by pairing with the 3′ untranslated region (UTR) of target messenger RNAs to repress their productive translation.1 Thus, although the genes responsible for T-cell lymphomas remain largely unknown,2 in some lymphoma subtypes, miRNA dysregulation may contribute to tumorigenesis. For example, altered expression of various oncogenic or tumor suppressive miRNAs has been identified in lymphomas/leukemias and solid tumors.3-6
MicroRNA-150 (miR-150) has proven to be a key miRNA involved in the development of hematologic malignancies. For instance, Chang et al showed that c-myc negatively regulates several miRNAs, including miR-150. They found that ectopically expressed miR-150 acts as a tumor suppressor, and that injection of miR-150–transduced mouse lymphoma cells into immunodeficient mice produces fewer tumors than control cells.7 In addition, Jiang et al very recently reported that miR-150 is downregulated in acute myeloid leukemia, including mixed lineage leukemia-associated acute myeloid leukemia, and that downregulation of miR-150 leads to activation of c-Myb and Flt3, resulting in cell proliferation and an antiapoptotic effect.8 We also recently showed that miR-150 expression is diminished in natural killer (NK)/T-cell lymphomas, and that its ectopic expression leads to inhibition of tumor cell proliferation and cell senescence.9 In that context, we hypothesized that miR-150 plays a central role in the pathogenesis of T-cell lymphomagenesis. To test that idea, in the present study, we screened for miR-150 expression in peripheral T-cell lymphomas (PTCL) and advanced cutaneous T-cell lymphomas (CTCL) showing nodal involvement. We show here that miR-150 expression is significantly diminished in all examined T-cell lymphoma subtypes tested, and that it has the potential to inhibit tumor invasion and metastasis by targeting the CCR6 chemokine receptor.
Materials and methods
For additional information, see the supplemental Methods on the Blood Web site.
Cell lines
Detailed information about lymphoma/leukemia cell lines is provided in supplemental Table 1.
Primary T-cell lymphomas
We collected RNA samples from nodal PTCL and advanced CTCL cells. The 12 CTCLs included 2 cases of Sezary syndrome (SzS) and 10 cases of stage IV mycosis fungoides (MF). Notably, all 10 MF samples were obtained from lymph node biopsies. Two SzS samples were obtained from peripheral blood (malignant CD4+ T cells). Skin biopsies were also performed in all 12 CTCL cases. Individual lymphoma patient’s characteristics are shown separately in supplemental Table 2 (PTCL) and supplemental Table 3 (CTCL). Samples were collected under a protocol approved by the Institutional Review Boards of Akita and Kurume Universities. Written informed consent was obtained from all patients prior to collection of specimens, in keeping with all institutional policies and according to the Declaration of Helsinki.
Gene expression analysis
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE49308).
In vivo inoculation of lymphoma cells into immunodeficient mice
Transduced cell lines (2 × 105 each) were subcutaneously injected into 6- to 8-week-old female nonobese diabetic/Shi-scid interleukin 2γ (IL-2γ) null (NOG) mice (Central Institute for Experimental Animals, Kawasaki, Japan).10
Detailed information about premicro-RNA or siCCR6 constructs, and lentivirus or retrovirus infection, transient siRNA transfection, and others are described in the supplemental Methods.
Results
Aberrantly diminished miR-150 expression in mature T-cell lymphomas
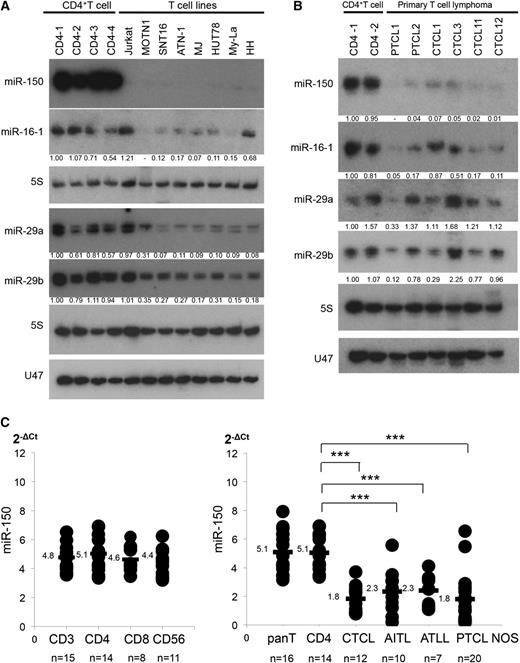
We recently reported that miR-150 expression is diminished in NK/T-cell lymphoma/leukemia.9 This led us to speculate that miR-150 may also be diminished in other lymphoma subtypes. Consistent with that idea, we detected diminished expression of miR-150 in several T-cell lymphoma and leukemia cell lines, as compared with normal CD4+ T cells (Figure 1A). We also examined 37 samples of primary CD4+ PTCL (20 PTCL not otherwise specified [PTCL NOS], 10 angio-immunoblastic T-cell lymphomas [AITL], 7 adult T-cell lymphomas [ATLL]), and 12 samples of advanced CTCL (10 MF and 2 SzS), which were obtained from lymph nodes (47 samples of PTCL NOS, AITL, ATLL, and MF) or peripheral blood (2 samples of SzS). Notably, the CTCL cases examined were from advanced MF with extensive nodal or visceral involvement (ISCL-EORTC stage IV or from SzS; supplemental Figure 1). Northern blot analysis of 6 cases of primary T-cell lymphoma (2 PTCL NOS, 2 MF, and 2 SzS) showed diminished expression of miR-150, as compared with normal CD4+ T cells (Figure 1B). Quantitative polymerase chain reaction (qPCR) also demonstrated that, while there were no significant differences in miR-150 expression among the different T-cell subsets, expression was significantly reduced in these primary samples (Figure 1C). These results indicate that miR-150 is aberrantly downregulated in PTCL and advanced CTCL.
Decreased expression of miR-150 in T-cell lymphomas. (A) Northern blot analysis of miR-150 in T-cell lymphoma/leukemia cell lines. Fold changes in miRNA levels were determined by densitometry and are shown below the gels after normalization to the level of 5S tRNA in normal CD4+ T cells (CD4-1), which were assigned a value of 1.00. Northern blots of miR-16-1, miR-29a, miR-29b, and U47 (another control for 5S tRNA) are also shown. Cell sorter was used to purify CD4+ T cells from lymph nodes or peripheral blood collected from primary samples (>95% purification). (B) Northern analysis of miR-150 in CD4+ T cells (n = 2) and primary samples of PTCL (2 cases of PTCL NOS), and 4 CTCL; 5S served as a control. Northern blots of miR-16-1, miR-29a, miR-29b, and U47 are also shown. (C) qPCR analysis of miR-150. (Left) qPCR analysis of miR-150 in normal CD3+ T cells (n = 15), CD4+ T cells (n = 14), CD8+ T cells (n = 8), and CD56+ T/NK cells (n = 11). (Right) qPCR analysis of miR-150 expression in 49 samples of CD4+ T-cell lymphoma, including CTCL (n = 12), AITL (n = 10), ATLL (n = 7), and PTCL NOS (n = 20). Y-axis: 2-ΔCt values for miRNA expression. Expression levels were normalized by data from PTCL2, which was assigned a value of 1.0; qPCR was conducted in duplicate, yielding similar results. P values were calculated using Mann-Whitney U test. Asterisks (*) indicate statistical significance: ***P < .001.
Decreased expression of miR-150 in T-cell lymphomas. (A) Northern blot analysis of miR-150 in T-cell lymphoma/leukemia cell lines. Fold changes in miRNA levels were determined by densitometry and are shown below the gels after normalization to the level of 5S tRNA in normal CD4+ T cells (CD4-1), which were assigned a value of 1.00. Northern blots of miR-16-1, miR-29a, miR-29b, and U47 (another control for 5S tRNA) are also shown. Cell sorter was used to purify CD4+ T cells from lymph nodes or peripheral blood collected from primary samples (>95% purification). (B) Northern analysis of miR-150 in CD4+ T cells (n = 2) and primary samples of PTCL (2 cases of PTCL NOS), and 4 CTCL; 5S served as a control. Northern blots of miR-16-1, miR-29a, miR-29b, and U47 are also shown. (C) qPCR analysis of miR-150. (Left) qPCR analysis of miR-150 in normal CD3+ T cells (n = 15), CD4+ T cells (n = 14), CD8+ T cells (n = 8), and CD56+ T/NK cells (n = 11). (Right) qPCR analysis of miR-150 expression in 49 samples of CD4+ T-cell lymphoma, including CTCL (n = 12), AITL (n = 10), ATLL (n = 7), and PTCL NOS (n = 20). Y-axis: 2-ΔCt values for miRNA expression. Expression levels were normalized by data from PTCL2, which was assigned a value of 1.0; qPCR was conducted in duplicate, yielding similar results. P values were calculated using Mann-Whitney U test. Asterisks (*) indicate statistical significance: ***P < .001.
In addition, we further validated candidate miRNAs that had been previously reported in PTCL and CTCL.11-15 Northern blot analysis was used to screen for 104 miRNAs in normal CD4+ T cells, CTCL cell lines, and primary CTCL samples (supplemental Table 4), and found a slight downregulation of miR-16-1, miR-29 family (miR-29a and miR-29b) in examined CTCL (Figure 1A-B),7,16,17 although qPCR analysis showed expression of the 3 miRNAs in primary lymphoma samples did not significantly differ from normal CD4+ T cells (data not shown).
MiR-150 acts as a tumor suppressor in T-cell lymphoma cells
To examine the function of miR-150 in T-cell lymphomagenesis, we first transfected green fluorescent protein (GFP)-empty-control or GFP–miR-150 into 5 T-cell lymphoma lines (ATN-1, MJ, HUT78, CD4+My-La, and HH), which led to upregulated expression of the miR-150 in the affected cells (Figure 2A-B). Recently, we demonstrated that human lymphoma cell lines could be grafted into immunodeficient nonobese diabetic/NOG mice.18 To examine if transduction of miR-150 might affect tumor formation, we subcutaneously inoculated miR-150–transfected or control cells (2 × 105; n = 5 each) into NOG mice, and found that cells expressing miR-150 produced significantly smaller tumors (My-La, MJ, and HUT78), or at least showed a trend toward smaller tumors (HH and ATN-1) (supplemental Figure 2). Furthermore, NOG mice inoculated with GFP-control My-La cells died within 31 to 37 days, while mice receiving GFP-control HH cells died within 55 to 70 days. By contrast, mice inoculated with GFP-miR-150 My-La cells survived 59 to 74 days, while those receiving GFP-miR-150 HH cells survived 105 days or longer. Thus, survival appeared to be substantially prolonged in mice receiving the miR-150 transfectants. To validate this, we carried out two additional independent inoculation experiments with two differently established transfectants, which yielded similar results. MiR-150 appeared to act as a tumor suppressor in T-cell lymphoma cells by increasing cell susceptibility to senescence and decreasing cell proliferation (supplemental Figure 3). Although miR-150 had only slight tumor suppressive effects on CTCL cells in culture, NOG mice inoculated with GFP-miR-150 CTCLs showed markedly greater survival than mice inoculated with GFP-CTCLs (Figure 2C). This strongly suggests that one or more other factors contribute to the prolonged survival.
Tumor-suppressive effect of miR-150 in T-cell lymphomas. (A) qPCR analysis of miR-150 expression in the indicated cell types transduced with GFP-empty (control) (red bars) vector or GFP-miR-150 (green bars). GFP-control or GFP-miRNA transfected cells were sorted and GFP-positive cells with >99% purification was obtained. X-axis: GFP-control or GFP-miR-150–transfected cell lines (ATN-1, MJ, HUT78, My-La, and HH). Y-axis: red and green bars depict −ΔΔCt values for miR-150 expression in the 5 transfected cell lines. Bars are means ± SEM of 3 independent experiments. (B) Northern blot analysis of the indicated miR-150 in indicated cells transfected with GFP-miR-150 (shown as “miR-150”) or empty control vector (shown as “control”). The transfected cell type is indicated at the top. Controls (5S tRNA) are shown in the bottom panel. (C) NOG mouse survival after engraftment of GFP-miR-150–transfected My-La or HH cells. Two independent inoculation assays are shown [My-La or HH-GFP-control(1), My-La or HH-GFP-control(2), My-La or HH-miR-150(1), My-La or HH-miR-150(2); n = 5 each]. X-axis: days from inoculation (day 0); Y-axis: survival rate. Significance (P) was determined using the Kaplan-Meier method.
Tumor-suppressive effect of miR-150 in T-cell lymphomas. (A) qPCR analysis of miR-150 expression in the indicated cell types transduced with GFP-empty (control) (red bars) vector or GFP-miR-150 (green bars). GFP-control or GFP-miRNA transfected cells were sorted and GFP-positive cells with >99% purification was obtained. X-axis: GFP-control or GFP-miR-150–transfected cell lines (ATN-1, MJ, HUT78, My-La, and HH). Y-axis: red and green bars depict −ΔΔCt values for miR-150 expression in the 5 transfected cell lines. Bars are means ± SEM of 3 independent experiments. (B) Northern blot analysis of the indicated miR-150 in indicated cells transfected with GFP-miR-150 (shown as “miR-150”) or empty control vector (shown as “control”). The transfected cell type is indicated at the top. Controls (5S tRNA) are shown in the bottom panel. (C) NOG mouse survival after engraftment of GFP-miR-150–transfected My-La or HH cells. Two independent inoculation assays are shown [My-La or HH-GFP-control(1), My-La or HH-GFP-control(2), My-La or HH-miR-150(1), My-La or HH-miR-150(2); n = 5 each]. X-axis: days from inoculation (day 0); Y-axis: survival rate. Significance (P) was determined using the Kaplan-Meier method.
MiR-150 inhibits tumor invasion and metastasis
We speculated that NOG mice inoculated with GFP-My-La or GFP-HH cells died due to tumor cell invasion of multiple visceral organs/tissues. To examine this possibility, we subcutaneously inoculated NOG mice with 2 × 105 GFP–miR-150 or GFP-empty (control) transduced CTCL (My-La and HH) (n = 10 each). Then, on the basis of our earlier findings (Figure 2C), the mice inoculated with My-La cells were sacrificed on day 31 and those receiving HH cells were sacrificed on day 55. As expected, all of the mice inoculated with GFP-control–My-La (GFP–My-La) cells showed invasion/metastasis of one or more visceral organs, with highly vascular organs such as the spleen, bone marrow (BM), lung, kidney, and liver (Figure 3A). In 7 of the 10 mice receiving GFP-control HH (GFP-HH) cells showed similar invasion (Figure 3A). By contrast, 7 of the 10 mice inoculated with GFP–miR-150–My-La cells showed no microscopically detectable tumor invasion/metastasis on day 31 (Figure 3B-C). And although the remaining 3 mice showed involvement of the liver and/or spleen, the affected regions were smaller than in the control mice. Similarly, 8 of the 10 mice inoculated with GFP–miR-150–HH cells showed no visually detectable involvement of the spleen, BM, kidney, liver, or lung on day 55 (Figure 3B-C). And while the remaining 2 mice showed liver involvement, the affected region was smaller than in the controls.
Transfection of miR-150 into T-cell lymphoma cell lines reduces invasion and metastasis. (A) Visceral invasion of CTCL cells in NOG mice. Photographs show invasion/metastasis into visceral organs by hematoxylin and eosin and GFP staining of liver (original magnification ×200) and kidney (original magnification ×100), GFP staining of lung (original magnification ×200) in GFP-empty (control) transduced My-La (shown as “GFP-My-La”), and HH (shown as “GFP-HH”) cells. Invading CTCL cells or the invaded region is surrounded by broken lines. (B) Invasion/metastasis by transfected My-La and HH (GFP-empty [control] or GFP-miR-150) in NOG mice. Photographs show spleen and BM from NOG mice inoculated with GFP-empty (control) (shown as “GFP-control”), or GFP-miR-150-CTCL cells. (C) Flow cytometric analysis of BM and spleen invasion by My-La and HH transfectants (GFP-empty control [n = 10 each] or GFP-miR-150 [n = 10 each]). Bars are means ± SEM of 3 independent experiments. Asterisks (*) indicate statistical significance: **.001 ≤ P < .01; ***P < .001. (D) Cell migration assay. Shown are RFUs (Ex 485/Em 538) from ATN-1, MJ, and HUT78 cells transfected with GFP-empty (control) (shown as “control”) or GFP-miR-150, and My-La and HH cells transfected with GFP-miR-16-1, GFP-miR-29a, or GFP-miR-150. Migration was stimulated by 2% HS in the lower chamber; no serum was added to the upper chamber. Incubation time was 16 hours and cell Lysis buffer were transferred to a 96-well plate, and fluorescence was measured by plate reader at 480 nm/520 nm. P values were calculated using Student t test. Asterisks (*) indicate statistical significance: ***P < .001. Bars are means ± SEM of 3 independent experiments. (E) A schematic illustration of the migration assay is shown besides the bar graphs. NS, not significant.
Transfection of miR-150 into T-cell lymphoma cell lines reduces invasion and metastasis. (A) Visceral invasion of CTCL cells in NOG mice. Photographs show invasion/metastasis into visceral organs by hematoxylin and eosin and GFP staining of liver (original magnification ×200) and kidney (original magnification ×100), GFP staining of lung (original magnification ×200) in GFP-empty (control) transduced My-La (shown as “GFP-My-La”), and HH (shown as “GFP-HH”) cells. Invading CTCL cells or the invaded region is surrounded by broken lines. (B) Invasion/metastasis by transfected My-La and HH (GFP-empty [control] or GFP-miR-150) in NOG mice. Photographs show spleen and BM from NOG mice inoculated with GFP-empty (control) (shown as “GFP-control”), or GFP-miR-150-CTCL cells. (C) Flow cytometric analysis of BM and spleen invasion by My-La and HH transfectants (GFP-empty control [n = 10 each] or GFP-miR-150 [n = 10 each]). Bars are means ± SEM of 3 independent experiments. Asterisks (*) indicate statistical significance: **.001 ≤ P < .01; ***P < .001. (D) Cell migration assay. Shown are RFUs (Ex 485/Em 538) from ATN-1, MJ, and HUT78 cells transfected with GFP-empty (control) (shown as “control”) or GFP-miR-150, and My-La and HH cells transfected with GFP-miR-16-1, GFP-miR-29a, or GFP-miR-150. Migration was stimulated by 2% HS in the lower chamber; no serum was added to the upper chamber. Incubation time was 16 hours and cell Lysis buffer were transferred to a 96-well plate, and fluorescence was measured by plate reader at 480 nm/520 nm. P values were calculated using Student t test. Asterisks (*) indicate statistical significance: ***P < .001. Bars are means ± SEM of 3 independent experiments. (E) A schematic illustration of the migration assay is shown besides the bar graphs. NS, not significant.
The prolonged survival of mice inoculated with GFP–miR-150–My-La cells could reflect, at least in part, miR-150–induced senescence or inhibition of proliferation. However, because the tumor suppressive effect of miR-150 seemed insufficient to substantially inhibit invasion/metastasis, we hypothesized that miR-150 acts by targeting one or more migration-related mediators. To test that idea, we carried out in vitro migration assays with My-La and HH transfectants plated on transwell plates. In this experiment, migration from the upper chamber to the lower chamber of the transwell plate was triggered by 2% human serum (HS) in lower chamber. Quantification of the migration based on relative fluorescence units (RFUs) showed that cells transfected with miR-150 migrated significantly less than control cells (Figure 3D-E). On the other hand, migration of My-La and HH cells was unaffected by transfection with two other tumor suppressive miRNAs (miR-16-1 and miR-29a), although these miRNAs showed cell proliferation inhibition (miR-16-1 and miR-29a) and senescence (miR-16-1) (supplemental Figure 4). Interestingly, transfection with GFP–miR-150 also significantly reduced migration of HUT78 cells. These results suggest that miR-150 has a specific ability to inhibit tumor cell migration via effects on undetermined target(s) whose dysregulation contributes to tumor invasion/metastasis.
The chemokine receptor CCR6 is the most likely target of miR-150 in metastatic CTCL
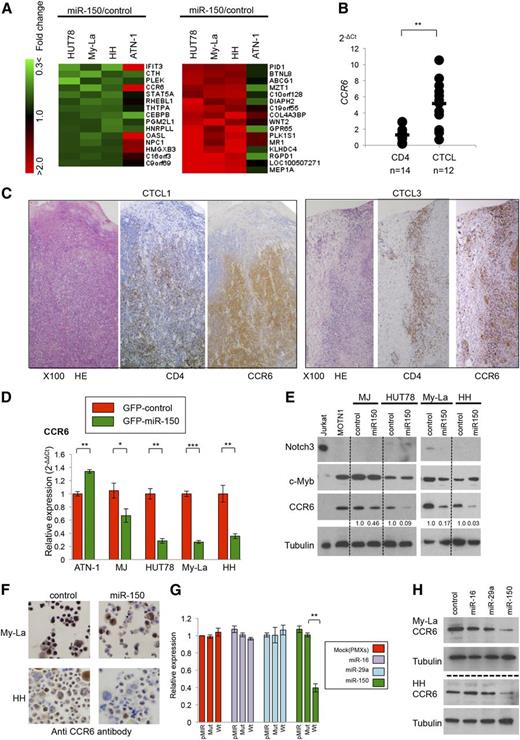
MiRNAs exert their inhibitory effects on expression by inhibiting translation of target proteins or by reducing the levels of target mRNAs.19 To detect miR-150 targets affecting invasion/metastasis, we analyzed gene expression in 3 CTCL cell lines whose migration in vitro was inhibited by miR-150 expression (Figure 3D). A total of 14 genes were commonly downregulated and 15 were commonly upregulated in the 3 CTCL cell lines (Figure 4A). Among these, we considered the CCR6 gene and the product (CCR6) to be the most likely direct target of miR-150 because CCR6 is a chemokine receptor whose aberrant overexpression is strongly associated with tumor invasion and metastasis in solid tumors,20-23 and upregulation of CCR6 was recently observed in advanced CTCL.24 We, therefore, used qPCR to examine CCR6 expression in 12 samples of primary metastatic CTCL and found it to be significantly higher than in normal CD4+ T cells (Figure 4B). Similarly, immunohistochemical analysis of skin samples from advanced primary MF patients showed CCR6 to be strongly expressed in malignant CD4+ T cells (Figure 4C).
CCR6 is a potential target of miR-150 in CTCL cells. (A) Detection of potential miR-150 target genes in GFP-miR-150 or GFP-empty (control) transduced CTCL cell lines. Left (downregulation): heat map of genes showing a GFP-miR-150 (shown as “miR-150”)/GFP-empty (shown as “control”) ratio <0.75. Right (upregulation): genes showing miR-150/control ratio >1.3. (B) CCR6 expression of normal CD4+ T cells (n = 14), CTCLs (10 cases of advanced MF and 2 cases of SzS). P values were calculated using the Mann-Whitney U test. Asterisks (*) indicate statistical significance: **.001 ≤ P < .01. (C) Immunohistochemistry of CCR6, CD4, and HE in primary CTCL (CTCL1 and CTCL3). (D) Taqman PCR analysis of CCR6 in the cells transfected with GFP-miR-150 or GFP-control. Y-axis: relative expression of CCR6 adjusted in GFP-transfected cells (2−ΔΔCt). P values were calculated using Student t test. Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 3 independent experiments. (E) Western blot analysis of Notch 3, c-Myb, CCR6, and tubulin in indicated transfected cell lines. Fold changes in CCR6 levels are shown below the gels. Control: GFP-empty (shown as “control”) transduced cells; miR-150: GFP-miR-150 transduced cells. Tubulin served as a control of western blot analysis. (F) Immunohistochemistry of CCR6 in My-La and HH cells transfected with or GFP-empty control (shown as “control”) or GFP-miR-150 (shown as “miR-150”). (G) Luciferase reporter assays showing CCR6 expression in Rat-1 fibroblasts transfected with miR-16-1, miR-29a, miR-150, or PMXs-puro control vector. Blots showing miR-150 expression are beside the bars: pMIR, empty vector (pMIR vector); Mut, CCR6 3′UTR mutation-inserted pMIR vector; and Wt, CCR6 3′UTR wild-type–inserted vector. P values were calculated using Student t test. Asterisks (*) indicate statistical significance: **.001 ≤ P < .01. Bars are means ± SEM of 3 independent experiments. (H) Western blot analysis of CCR6 and tubulin in My-La and HH cells transfected with GFP-control (control), GFP-miR-16-1 (miR-16), GFP-miR-29a (miR-29a), or GFP-miR-150 (miR-150).
CCR6 is a potential target of miR-150 in CTCL cells. (A) Detection of potential miR-150 target genes in GFP-miR-150 or GFP-empty (control) transduced CTCL cell lines. Left (downregulation): heat map of genes showing a GFP-miR-150 (shown as “miR-150”)/GFP-empty (shown as “control”) ratio <0.75. Right (upregulation): genes showing miR-150/control ratio >1.3. (B) CCR6 expression of normal CD4+ T cells (n = 14), CTCLs (10 cases of advanced MF and 2 cases of SzS). P values were calculated using the Mann-Whitney U test. Asterisks (*) indicate statistical significance: **.001 ≤ P < .01. (C) Immunohistochemistry of CCR6, CD4, and HE in primary CTCL (CTCL1 and CTCL3). (D) Taqman PCR analysis of CCR6 in the cells transfected with GFP-miR-150 or GFP-control. Y-axis: relative expression of CCR6 adjusted in GFP-transfected cells (2−ΔΔCt). P values were calculated using Student t test. Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 3 independent experiments. (E) Western blot analysis of Notch 3, c-Myb, CCR6, and tubulin in indicated transfected cell lines. Fold changes in CCR6 levels are shown below the gels. Control: GFP-empty (shown as “control”) transduced cells; miR-150: GFP-miR-150 transduced cells. Tubulin served as a control of western blot analysis. (F) Immunohistochemistry of CCR6 in My-La and HH cells transfected with or GFP-empty control (shown as “control”) or GFP-miR-150 (shown as “miR-150”). (G) Luciferase reporter assays showing CCR6 expression in Rat-1 fibroblasts transfected with miR-16-1, miR-29a, miR-150, or PMXs-puro control vector. Blots showing miR-150 expression are beside the bars: pMIR, empty vector (pMIR vector); Mut, CCR6 3′UTR mutation-inserted pMIR vector; and Wt, CCR6 3′UTR wild-type–inserted vector. P values were calculated using Student t test. Asterisks (*) indicate statistical significance: **.001 ≤ P < .01. Bars are means ± SEM of 3 independent experiments. (H) Western blot analysis of CCR6 and tubulin in My-La and HH cells transfected with GFP-control (control), GFP-miR-16-1 (miR-16), GFP-miR-29a (miR-29a), or GFP-miR-150 (miR-150).
We next used qPCR to assess CCR6 expression in transfected T-cell lymphoma cell lines, and found that the expression was significantly reduced in all miR-150–transfected CTCL lines tested (Figure 4D). In cells transfected with GFP-control or GFP–miR-150, western blot analysis showed CCR6 to be reduced in all CTCLs expressing miR-150 (Figure 4E), though expression of other potential mediators in immature T-cell development (c-Myb and Notch3) were unaffected.25,26 This might be because miR-150 could regulate different target(s) in immature T cells. Reduction of CCR6 was also confirmed by immunostaining against miR-150 transduced CTCL cells (Figure 4F).
Further, to verify the relevance of miR-150 to CCR6, we performed a miR-150 loss-of-function experiment using antisense oligonucleotides against normal CD4+ T cells. We found that antisense oligonucleotides targeting miR-150 induced upregulation of CCR6 in normal CD4+ T cells (n = 5). These results reveal a negative correlation between miR-150 and CCR6 (supplemental Figure 5).
To determine whether miR-150 interacts with the predicted seed sequence of CCR6 (TargetScan program),27 we carried out luciferase reporter assays using miR-150–(puromycin)-transfected Rat-1 fibroblasts (miR-150–Rat-1 cells). When the cells were transfected with the wild-type 3′ UTR of CCR6, we observed a significant reduction of luciferase activity that was not seen when the cells were transfected with a mutated 3′ UTR of CCR6. We also did not detect a significant reduction in luciferase activity in Rat-1 cells expressing miR-16-1 or miR-29a, which do not possess the CCR6 seed sequence (Figure 4G). Correspondingly, western blot analysis showed that expression of miR-16-1 or miR-29a in My-La and HH cells had no effect on CCR6 expression (Figure 4H). Taken together, these results strongly suggest that there is direct correlation between miR-150 and CCR6 in CD4+ T cells and CTCL.
CCR6 knockdown inhibits CTCL cell invasion and metastasis
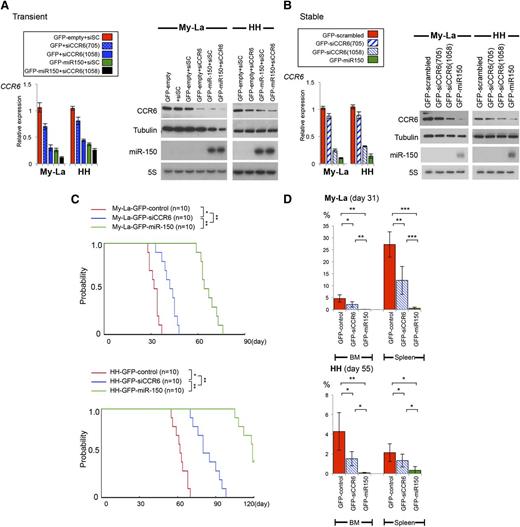
Although we demonstrated that negative regulation of CCR6 by miR-150 was likely to be deeply associated with tumor invasion/metastasis, association of CCR6 in tumor invasion/metastasis still remains unclear. Therefore, to examine whether CCR6 knockdown could inhibit metastasis and invasion, we established transient transfectant expressing siCCR6 (Figure 5A). As shown in qPCR, western and Northern blot analyses, enforced expression of miR-150 or transient CCR6 knockdown (siCCR6) each suppressed CCR6/CCR6 expression, but suppression was greatest in cells expressing miR-150 with siCCR6. Based on these results, we further established stable transfectants expressing siCCR6(1058) (Figure 5B). After establishing My-La and HH transfectants stably expressing GFP-siCCR6, we confirmed the reduced expression of CCR6, independent from miR-150. We inoculated these transfectants into NOG mice (n = 16 each): one group of survival observation (n = 10 each) and another for tumor invasion observation (n = 6 each). When we then inoculated NOG mice with My-La or HH cells expressing GFP-scrambled (control), GFP-siCCR6, or GFP-miR-150, mice receiving GFP-siCCR6–My-La or -HH cells showed significantly better survival than mice receiving control cells, although the best survival was achieved by mice receiving GFP–miR-150 transfectants (n = 10 each, Figure 5C).
Inoculation of GFP-siCCR6–transfected CTCL cells into NOG mice. (A) Expression of CCR6 in CTCL cell lines transiently transfected with siCCR6(705 or 1058). Left: relative expression of CCR6 in GFP-empty-CTCL (shown as “GFP-empty”) (My-La or HH), GFP-miR-150-CTCL cells transiently transfected with scrambled (shown as “+siSC”), or siCCR6 (shown as “+siCCR6”). Right: western blot analysis of CCR6 in GFP-empty (control) or GFP-miR-150 transduced CTCL cells transiently transduced with siCCR6(1058). Tubulin is the control for CCR6; 5S is the control for miR-150. (B) qPCR and western blot analyses of CCR6 in My-La and HH cells stably transfected with GFP-siCCR6. Relative expression of CCR6 in GFP-scrambled (control) CTCL (shown as “GFP-scrambled”), GFP-siCCR6 (705 or 1058)-CTCL, or GFP-miR-150-CTCL cells. GFP-siCCR6 expressing transfected cells were sorted and obtained with >99% purification. Northern blots against miR-150 are also shown below the western blot data. (C) Survival curves for NOG mice inoculated with My-La and HH transfected with GFP-scrambled (shown as “control”), GFP-siCCR6(1058), or GFP-miR-150 (n = 10 each). Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01. (D) Percentages of BM and spleen involvement in NOG mice inoculated CTCL cells transduced with GFP-control, GFP-siCCR6, or GFP-miR-150. NOG mice (n = 6 each) were sacrificed at day 31 (My-La) or day 55 (HH). P values were calculated using Student t test. Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 6 independent experiments.
Inoculation of GFP-siCCR6–transfected CTCL cells into NOG mice. (A) Expression of CCR6 in CTCL cell lines transiently transfected with siCCR6(705 or 1058). Left: relative expression of CCR6 in GFP-empty-CTCL (shown as “GFP-empty”) (My-La or HH), GFP-miR-150-CTCL cells transiently transfected with scrambled (shown as “+siSC”), or siCCR6 (shown as “+siCCR6”). Right: western blot analysis of CCR6 in GFP-empty (control) or GFP-miR-150 transduced CTCL cells transiently transduced with siCCR6(1058). Tubulin is the control for CCR6; 5S is the control for miR-150. (B) qPCR and western blot analyses of CCR6 in My-La and HH cells stably transfected with GFP-siCCR6. Relative expression of CCR6 in GFP-scrambled (control) CTCL (shown as “GFP-scrambled”), GFP-siCCR6 (705 or 1058)-CTCL, or GFP-miR-150-CTCL cells. GFP-siCCR6 expressing transfected cells were sorted and obtained with >99% purification. Northern blots against miR-150 are also shown below the western blot data. (C) Survival curves for NOG mice inoculated with My-La and HH transfected with GFP-scrambled (shown as “control”), GFP-siCCR6(1058), or GFP-miR-150 (n = 10 each). Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01. (D) Percentages of BM and spleen involvement in NOG mice inoculated CTCL cells transduced with GFP-control, GFP-siCCR6, or GFP-miR-150. NOG mice (n = 6 each) were sacrificed at day 31 (My-La) or day 55 (HH). P values were calculated using Student t test. Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 6 independent experiments.
GFP-siCCR6–inoculated NOG mice showed multiple visceral metastases on day 31 (My-La) or 55 (HH), but the percentages of tumor involvement were significantly smaller than control. On the other hand, the lowest levels of tumor involvement were seen in mice receiving miR-150 transfectants. For instance, percentage of involvement into BM and spleen of GFP-siCCR6–transduced CTCL cells were significantly smaller than GFP-control–transduced cells (Figure 5D). However, the percentage of involvement into BM and spleen of GFP–miR-150–transduced CTCL cells were significantly smaller than GFP-siCCR6–transduced cells. This significant difference between siCCR6 and miR-150 could be partially because of the tumor suppressive potential of miR-150.
Together, these results strongly suggest that CCR6 may be a key molecule of invasion/metastasis of CTCL, and therefore, miR-150–CCR6 axis is crucially involved in the pathogenesis of CTCL.
MiR-150 negatively regulates IL-22-CCL20-CCR6 autocrine pathway in advanced CTCL
CCR6 is the receptor for the chemokine CCL20, which is activated by the cytokines IL-22 and IL-17.28-30 CCL20 is expressed in the liver, lymph nodes, and skin,31 and it is well known that CCR6+ cells migrate to CCL20-producing organs/tissues, and that the interaction of CCL20 with CCR6 activates migration, proliferation, and cytokine production.22,32-35
We hypothesized that IL-22 and/or IL-17 produced by CTCL cells binds to receptors on the same cells or adjacent CCR6+ CTCL cells, activating intragenic CCL20 expression. To test this idea, we first used qPCR to assess IL-17A and IL-22 gene expressions in CTCL cells (n = 12) and normal CD4+ T cells (n = 14). Although we detected weaker expression of IL-17A in tumor cells than in normal CD4+ T cells, we detected stronger IL-22 expression than in normal CD4+ T cells (Figure 6A).
Negative regulation of miR-150 against autocrine IL-22-CCL20-CCR6 pathway in CTCL cells. (A) qPCR of IL-22 and IL-17A in CD4+ T cell (n = 14) and advanced CTCL samples (n = 12). P values were calculated using the Mann-Whitney U test. Asterisks (*) indicate statistical significance: **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 3 independent experiments. (B) Western blot of IL-22RA1 and CCR6 in 5 normal CD4+ T cell (CD4-1 to CD4-5), My-La and HH cell lines, and 5 primary CTCL cases. Jeko-1 (mantle cell lymphoma cell line) is positive control of IL-22RA1.38 (C) ELISA of IL-10, IL-17, IL-22, and CCL20 in CTCL cell lines. Cells (1 × 104) were cultured for 24 hours or 48 hours in 48-well dishes. Each well contained 500 μL of culture medium supplemented with 2% HS or HS free [HS(-)]. Y-axis: pg/mL. ELISA of CCL20 in CTCL cell lines. (D) Effect of IL22RA1 knockdown on IL-22 and CCL20. Left: expression of IL22RA1 in CTCL cell lines transiently transfected with siIL22RA1(744 and 1571). Right: western blot analysis of IL22RA1 in CTCL cells transiently transduced with siIL22RA1(1571) and control [+siSC(scrambled)]. Tubulin served as a control for IL22RA1. ELISA of IL-22 and CCL20 in CTCL cells transiently transduced with siIL22RA1(1571). These cells were cultured for 12 hours or 24 hours. P values were calculated using Student t test. Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 6 independent experiments. (E) ELISA of IL-22 and CCL20 in GFP-empty (control) or GFP-miR-150–transduced CTCL cells transiently transduced with siCCR6 (shown as “+siCCR6”). ELISA of IL-22 and CCL20 in GFP-siCCR6–transduced CTCL cells are also shown. These cells were cultured for 24 hours or 48 hours. Asterisks (*) indicate statistical significance: **.001 ≤ P < .01. Bars are means ± SEM of 3 independent experiments. NS: not significant.
Negative regulation of miR-150 against autocrine IL-22-CCL20-CCR6 pathway in CTCL cells. (A) qPCR of IL-22 and IL-17A in CD4+ T cell (n = 14) and advanced CTCL samples (n = 12). P values were calculated using the Mann-Whitney U test. Asterisks (*) indicate statistical significance: **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 3 independent experiments. (B) Western blot of IL-22RA1 and CCR6 in 5 normal CD4+ T cell (CD4-1 to CD4-5), My-La and HH cell lines, and 5 primary CTCL cases. Jeko-1 (mantle cell lymphoma cell line) is positive control of IL-22RA1.38 (C) ELISA of IL-10, IL-17, IL-22, and CCL20 in CTCL cell lines. Cells (1 × 104) were cultured for 24 hours or 48 hours in 48-well dishes. Each well contained 500 μL of culture medium supplemented with 2% HS or HS free [HS(-)]. Y-axis: pg/mL. ELISA of CCL20 in CTCL cell lines. (D) Effect of IL22RA1 knockdown on IL-22 and CCL20. Left: expression of IL22RA1 in CTCL cell lines transiently transfected with siIL22RA1(744 and 1571). Right: western blot analysis of IL22RA1 in CTCL cells transiently transduced with siIL22RA1(1571) and control [+siSC(scrambled)]. Tubulin served as a control for IL22RA1. ELISA of IL-22 and CCL20 in CTCL cells transiently transduced with siIL22RA1(1571). These cells were cultured for 12 hours or 24 hours. P values were calculated using Student t test. Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 6 independent experiments. (E) ELISA of IL-22 and CCL20 in GFP-empty (control) or GFP-miR-150–transduced CTCL cells transiently transduced with siCCR6 (shown as “+siCCR6”). ELISA of IL-22 and CCL20 in GFP-siCCR6–transduced CTCL cells are also shown. These cells were cultured for 24 hours or 48 hours. Asterisks (*) indicate statistical significance: **.001 ≤ P < .01. Bars are means ± SEM of 3 independent experiments. NS: not significant.
Upon IL-22 binding to its specific receptor complex, IL-22RA1/IL-10RB, and CCL20 are upregulated.36 It is well known that the IL-22 receptor subunit, IL-22RA1, is normally expressed on epithelial cells but not in immune cells, whereas IL-10RB is ubiquitously expressed in almost all organs and tissues.36 However, recent studies have shown that IL-22RA1 is aberrantly expressed in malignant lymphomas,37,38 suggesting it could also be aberrantly expressed in CTCL. We, therefore, assessed IL-22RA1 expression in normal CD4+ T cells, CTCL cell lines, and primary CTCL samples. We found that IL-22RA1 is expressed in both CTCL lines and primary CTCLs, but not in any of the normal CD4+ T-cell samples examined (n = 5) (Figure 6B). This suggests that IL-22 acts in CTCL cells themselves by binding to the IL-22RA1/IL-10RB complex. In addition, we also examined CCR6 for CTCL cells and primary samples, and confirmed the aberrant overexpression (Figure 6B).
With that in mind, we used enzyme-linked immunosorbent assays (ELISAs) to assess the levels of IL-10, IL-17, IL-22, and CCL20 in medium conditioned by CTCL cells and found high levels of both IL-22 and CCL20 in medium conditioned by My-La and HH cells, which showed strong migration. Moreover, the levels of IL-22 and CCL20 increased with culture duration (24 hours, 48 hours) (Figure 6C). By contrast, levels of IL-10 and IL-17 were much lower than the IL-22 levels, and they did not change with culture duration. In addition, IL-22 and CCL20 expressions were not significantly different in CTCL cells between cultured in HS-free medium and in cultured HS. These results strongly suggested that IL-22–CCL20 pathway might be crucially involved in advanced CTCL, and there might be a positive correlation between IL-22 and CCL20. To examine whether IL-22 could affect CCL20 production through IL22 to IL22RA1 binding, we conducted transient knockdown of IL22RA1 against CTCL cells (Figure 6D). We found that IL22RA1 knockdown reduced CCL20 levels but not IL-22 levels in examined CTCL cells. This suggested that CCL20 production could be induced by IL-22-IL22RA1 axis in CTCL cells.
Next, to clarify the relationship among IL-22, CCL20, CCR6, and miR-150 in CTCL cells, we tested whether miR-150 or CCR6 knockdown might affect the production of IL-22 or CCL20. When we measured IL-22 and CCL20 in medium conditioned for 24 hours or 48 hours by cells expressing GFP-control or GFP–miR-150, with or without siCCR6 (Figure 6E), CCR6 knockdown had no significant effect on IL-22 or CCL20 levels under any conditions. On the other hand, IL-22 and CCL20 levels were significantly reduced in medium conditioned by cells expressing miR-150. This may reflect the broad spectrum of potential miR-150 targets (ie, miR-150 might not only negatively regulate CCR6, but also be undetermined upstream regulator(s) of IL-22).
Ectopic expression of miR-150 and/or CCR6 knockdown inhibits CCL20- and/or nutrition-dependent chemotaxis of CTCL cells
We have shown nutrition-dependent chemotaxis of CTCL cells by use of transwell assay system (Figure 3D). In this experiment, we used HS as a trigger of the cell migration from upper to lower chamber. As previously mentioned, CCR6+ cells were likely to show chemotaxis onto CCL20+-producing organs/tissues,19,20,31 after which the interaction of CCL20 and CCR6 can initiate further migration.21,22,32-35 Thus, CCL20 can also be a cue for CTCL migration in the transwell assay. To determine whether the migration capacity of CTCL cells is affected by the presence or absence of CCL20 or HS, we again used the transwell system to assess the migration of My-La and HH cells (Figure 7A). After incubating the cells for 16 hours, we measured the migration index (RFUs) to the lower chamber. We found that when cells in the upper chamber were incubated with the medium containing CCL20, HS, or CCL20 + HS in the lower chamber, the migration was significantly greater than when the lower chamber contained HS(-)/CCL20(-) medium (Figure 7B).
MiR-150 and siCCR6 collaborate to inhibit CTCL cell migration. (A) Migration of My-La (top) and HH (bottom) cells in the presence of different extracellular stimuli; 5 × 105 cells in 100 μL of medium without HS and CCL20 in the upper chamber through a coated basement membrane toward different concentrations of CCL20 (0.1∼10 ng/mL), with or without 2% HS at lower chamber (500 μL). The lower chamber contained HS free or CCL20, HS, CCL20, or CCL20 + HS. P values were calculated using Student t test. Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 6 independent experiments. (B) A schematic illustration of the migration assay described above. (C) ELISA of CCL20 in upper and lower chambers (transwell assays) by use of GFP-empty (shown as “control”), GFP-siCCR6 (shown as “siCCR6”), or GFP-miR-150 (shown as “miR-150”) transduced CTCL cells in indicated condition in the lower chamber. (D) A schematic illustration of the ELISA assay (HS + CCL20 condition in the lower chamber). (E) Migration of CTCL cells [GFP-empty+siSC (control), GFP-empty+siCCR6(1058) (transient), GFP-siCCR6(1058)+siSC, GFP-miR-150+siSC, and GFP-miR-150 +siCCR6(1058)] in the presence or absence of CCL20 (0 or 0.5 ng/mL), or HS (2%) in the lower chamber. P values were calculated using Student t test. Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 3 independent experiments. (F) A schematic diagram of the relationship between IL-22, CCL20, CCR6, and miR-150 in CTCL. NS: not significant.
MiR-150 and siCCR6 collaborate to inhibit CTCL cell migration. (A) Migration of My-La (top) and HH (bottom) cells in the presence of different extracellular stimuli; 5 × 105 cells in 100 μL of medium without HS and CCL20 in the upper chamber through a coated basement membrane toward different concentrations of CCL20 (0.1∼10 ng/mL), with or without 2% HS at lower chamber (500 μL). The lower chamber contained HS free or CCL20, HS, CCL20, or CCL20 + HS. P values were calculated using Student t test. Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 6 independent experiments. (B) A schematic illustration of the migration assay described above. (C) ELISA of CCL20 in upper and lower chambers (transwell assays) by use of GFP-empty (shown as “control”), GFP-siCCR6 (shown as “siCCR6”), or GFP-miR-150 (shown as “miR-150”) transduced CTCL cells in indicated condition in the lower chamber. (D) A schematic illustration of the ELISA assay (HS + CCL20 condition in the lower chamber). (E) Migration of CTCL cells [GFP-empty+siSC (control), GFP-empty+siCCR6(1058) (transient), GFP-siCCR6(1058)+siSC, GFP-miR-150+siSC, and GFP-miR-150 +siCCR6(1058)] in the presence or absence of CCL20 (0 or 0.5 ng/mL), or HS (2%) in the lower chamber. P values were calculated using Student t test. Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 3 independent experiments. (F) A schematic diagram of the relationship between IL-22, CCL20, CCR6, and miR-150 in CTCL. NS: not significant.
Because CTCL cells spontaneously produce CCL20, its concentration also could be another “index” of cell migration. Thus, we used ELISAs for GFP-empty (control), GFP-siCCR6, and GFP-miR-150–transduced CTCL cells to measure CCL20 in the upper and lower chambers after 2, 4, 6, 8, and 16 hours (Figure 7C). The lower chamber initially contained a combination of CCL20 (0.5 ng/mL), HS (2%), or CCL20 + HS. Under these conditions, in GFP-control cells, the CCL20 concentration in the upper chamber increased over time, and the CCL20 concentration in the lower chamber also increased when CCL20 and/or HS was present. Because of intragenic CCL20 production from migrated cells in the lower chamber, CCL20 concentration in the lower chamber became greater than in the upper chamber during the migration assay. These results indicate that chemokine (CCL20)- or nutrition (HS)- gradient could be a cue of chemotaxis of CTCL cells. Notably, the difference of CCL20 concentration between the lower and upper chambers in GFP-siCCR6 or GFP-miR-150–transduced CTCL cells were remarkably decreased when compared with that of GFP-control (Figure 7C-D).
When migration assay was performed using transfecting cells with GFP-siCCR6 (stable), siCCR6 (transient), GFP-miR-150 (stable), or GFP–miR-150 (stable) + siCCR6 (transient), these transfectants showed significant reduction of cell migration toward CCL20 (0.5 ng/mL), HS, or CCL20 + HS in the lower chamber (Figure 7E). Notably, siCCR6-mediated (transient, stable) migration was inhibited to the greatest degree in cells expressing miR-150. This might be because CCR6 expression was suppressed to the greatest degree in the CTCL cells transduced with both siCCR6 and miR-150. In addition, we examined migration assay for GFP-miR-150 or GFP-siCCR6–transduced CTCL cells in the presence of other chemokines (CCL17, CCL22, and CCL27) in the lower chamber. However, we could not detect any migration inhibition in the presence of these chemokines (supplemental Figure 6).
Taken together, these findings enable us to conclude that, along with its own tumor suppressive potential, miR-150 can reduce CCL20- or nutrition-dependent chemotactic capabilities of advanced CTCL by down-regulating the IL-22-CCL20-CCR6 pathway (Figure 7F).
Discussion
Recent investigations of miRNA expression in CTCL (SzS and MF) revealed altered expression of a number of miRNAs.11-14 Among those, downregulation of miR-150 was reported in advanced CTCL11 but not in early or low grade CTCL, which might reflect a lack of advanced CTCL cases in the earlier reports. Nonetheless, aggressive CTCL (SzS and stage IV MF) showed aggressive clinical behavior and often involved multiple visceral organs.39-41 More often, miRNA dysregulation added to underlying genetic events, and the resultant aberrations enhanced tumorigenicity through activation of additional signaling pathways.3-6 Our earlier work also demonstrated miRNA expression in aggressive lymphomas, suggesting that such alterations may be deeply associated with the progression of malignant lymphomas.9,18,42-44 This makes it tempting to suggest that miR-150 downregulation is a crucial component of advanced CTCL. That remains to be tested, however.
Our identified miR-150 target was CCR6, a member of the β-chemokine receptor family, which is also known as CD196 surface antigen. CCR6 is preferentially expressed in immature dendritic cells and naive T cells, and plays important roles in B-lineage maturation and antigen-driven B-cell differentiation,19,20 regulating the migration and recruitment of dendritic and T cells during inflammatory and immunologic responses.19,20,31,45,46 Notably, CCR6 is strongly associated with tumorigenesis, especially in advanced and metastatic cancer.23,33-35 In colorectal cancer, for example, upregulation of CCR6 in the tumor cells strongly correlates with liver metastasis and has proven to be an independent prognostic marker of the tumor entity.22 These findings prompted us to hypothesize that CCR6 is a key regulator of invasion/metastasis in CTCL, and our results are consistent with that idea.
The origin of CTCL is skin-homing CD4+ T cells.39,40 At present, CD4+ T-helper cells are divided into the TH1, TH2, TH17, and TH22 subsets. Of those, the TH22 subset produces only IL-22, while the TH17 subset produces both IL-17 and IL-22.28,30,45-47 Both IL-17 and IL-22 are known to activate CCL20.45,46 Two groups previously demonstrated that skin-homing CD4+ T cells produce IL-22 but not IL-17, and the cell is expressing CCR6, CCR10, and CCR4 receptors.28,30 Likewise, we found that advanced CTCL cells also produce IL-22 but not IL-17. Together, these findings suggest that CTCL is likely derived from the TH22 CD4+ subset. Miyagaki et al also recently demonstrated that expression of IL-22 (but not IL-17), CCL20, and CCR6 is clearly elevated in both the serum and skin lesions of patients with advanced CTCL.24 They hypothesized that the production of IL-22 in dendritic cells (but not CTCL cells) might promote CCL20 production in keratinocytes, which could in turn, enhance the chemotactic capability of CTCL cells via CCR6+. In this scenario, CCR6+ CTCL cells likely migrated toward CCL20-rich regions such as the skin. However, these investigators did not mention the possibility that CTCL cells may be capable of autocrine migration, mediated by their own production of IL-22 and CCL20. From our in vivo experiments, it appears that the CCL20-dependent chemotaxis is not crucially involved in CTCL cell migration, as it is unlikely that mouse CCL20 affects human CTCL cells. Moreover, because CCL20 production in advanced CTCL cells may be much stronger than in adjacent organs or cells, extragenic CCL20 would not be expected to be the trigger for their migration. We therefore, conclude that advanced stage of CTCL cell migration mainly reflects an autocrine mechanism, as follows: IL-22 derived from CTCL cells binds to IL-22RA/IL-10RB receptors on the same CTCL cells or neighboring CCR6+ (CTCL, dendritic, and keratinocyte) cells, which induce the cells to produce CCL20. The subsequent interaction of CCL20-CCR6 along with the upregulation of undetermined downstream targets would then lead to the activation of multidirectional CTCL migration capability, after which a nutrition-dependent chemotaxis may become the cue for CTCL cells to invade blood vessels and blood-flow-rich organs.
In summary, our findings suggest that miR-150, CCL20, and CCR6 are potential therapeutic targets in metastatic CTCL. However, the following still remains unknown: which downstream transcript factor(s) of IL-22 could lead to produce CCL20; which downstream factor(s) of CCL20-CCR6 interaction might be associated with migration of CTCL cells; or which downstream direct target of miR-150 affects IL-22. Further analysis will be required to determine the precise downstream mechanism underlying these interactions in CTCL.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank E. Kobayashi, H. Kataho, and Y. Abe for their outstanding technical assistance; Dr T. Nanjo (Akita University, Akita, Japan) and Prof R. Ichinohasama (Tohoku University, Sendai, Japan) for their histologic diagnosis of lymphoma; and Prof M. Manabe and Dr N. Hasunuma (Akita University, Graduate School of Medicine, Department of Dermatology and Plastic Surgery, Akita, Japan) for clinical diagnosis of CTCL.
This work was supported by the Japan Society for the Promotion of Science (Grant-in-Aid for Scientific Research–KAKENHI, grant number 25461405) (H.T.).
Authorship
Contribution: M.I., K.T., S.I., and A.K. performed experiments and analyzed data; M.I. and S.I. generated figures; A.W., M.N., J.Y., K.O., and K.S. analyzed data; and H.T. designed experiments, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hiroyuki Tagawa, Department of Hematology, Nephrology, and Rheumatology, Akita University Graduate School of Medicine, 1-1-1 Hondo, Akita 0108543, Japan; e-mail: htagawa0279jp@yahoo.co.jp or htagawa@doc.med.akita-u.ac.jp.

![Figure 2. Tumor-suppressive effect of miR-150 in T-cell lymphomas. (A) qPCR analysis of miR-150 expression in the indicated cell types transduced with GFP-empty (control) (red bars) vector or GFP-miR-150 (green bars). GFP-control or GFP-miRNA transfected cells were sorted and GFP-positive cells with >99% purification was obtained. X-axis: GFP-control or GFP-miR-150–transfected cell lines (ATN-1, MJ, HUT78, My-La, and HH). Y-axis: red and green bars depict −ΔΔCt values for miR-150 expression in the 5 transfected cell lines. Bars are means ± SEM of 3 independent experiments. (B) Northern blot analysis of the indicated miR-150 in indicated cells transfected with GFP-miR-150 (shown as “miR-150”) or empty control vector (shown as “control”). The transfected cell type is indicated at the top. Controls (5S tRNA) are shown in the bottom panel. (C) NOG mouse survival after engraftment of GFP-miR-150–transfected My-La or HH cells. Two independent inoculation assays are shown [My-La or HH-GFP-control(1), My-La or HH-GFP-control(2), My-La or HH-miR-150(1), My-La or HH-miR-150(2); n = 5 each]. X-axis: days from inoculation (day 0); Y-axis: survival rate. Significance (P) was determined using the Kaplan-Meier method.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/10/10.1182_blood-2013-09-527739/4/m_1499f2.jpeg?Expires=1769082560&Signature=0gTWDEqs---Ec4Fs0VEX00Y9tT6gQy2P1quUefxnrNLvJwRMmKc0gCsjWGphfXdx6zfLv2KI5PQmW6Y2kY2tAAS~nd6xpJgj85Vb6IHI~GVDtePhNQmPJw~O-wwfmiyMBdTWcO~IjbKnZKVbjnfgwWb4yXs5gg2REVISbzWoj-rEnkn5e8tdrwUlgDfnwRcWl5Qb4uPrjicR53ux9LhfLGnRARm3168ZqY~e7ubJ2nLX5yTISK68-oviVIBES9Cxi92rF32-zMEZAfL4aZqLeZuNiSFRBSfsT07Db2ZrLcjcDdz0YAU720bgO7AaXCeJXOJbssmXaESgR8InKx0lIg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Transfection of miR-150 into T-cell lymphoma cell lines reduces invasion and metastasis. (A) Visceral invasion of CTCL cells in NOG mice. Photographs show invasion/metastasis into visceral organs by hematoxylin and eosin and GFP staining of liver (original magnification ×200) and kidney (original magnification ×100), GFP staining of lung (original magnification ×200) in GFP-empty (control) transduced My-La (shown as “GFP-My-La”), and HH (shown as “GFP-HH”) cells. Invading CTCL cells or the invaded region is surrounded by broken lines. (B) Invasion/metastasis by transfected My-La and HH (GFP-empty [control] or GFP-miR-150) in NOG mice. Photographs show spleen and BM from NOG mice inoculated with GFP-empty (control) (shown as “GFP-control”), or GFP-miR-150-CTCL cells. (C) Flow cytometric analysis of BM and spleen invasion by My-La and HH transfectants (GFP-empty control [n = 10 each] or GFP-miR-150 [n = 10 each]). Bars are means ± SEM of 3 independent experiments. Asterisks (*) indicate statistical significance: **.001 ≤ P < .01; ***P < .001. (D) Cell migration assay. Shown are RFUs (Ex 485/Em 538) from ATN-1, MJ, and HUT78 cells transfected with GFP-empty (control) (shown as “control”) or GFP-miR-150, and My-La and HH cells transfected with GFP-miR-16-1, GFP-miR-29a, or GFP-miR-150. Migration was stimulated by 2% HS in the lower chamber; no serum was added to the upper chamber. Incubation time was 16 hours and cell Lysis buffer were transferred to a 96-well plate, and fluorescence was measured by plate reader at 480 nm/520 nm. P values were calculated using Student t test. Asterisks (*) indicate statistical significance: ***P < .001. Bars are means ± SEM of 3 independent experiments. (E) A schematic illustration of the migration assay is shown besides the bar graphs. NS, not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/10/10.1182_blood-2013-09-527739/4/m_1499f3.jpeg?Expires=1769082560&Signature=Ea2zrJ0JBLEebYtz-a4sSaiZpnfj0NNF~RcfAMuTPrdm0-kVhwOuYd8JI7~cQP0CBld462c3WCStMnkTBqLV1yiUW21tyoNGYvUHvy6qlznoUETwFisd~qUj7Mib3cxsZ8eFd5MuT1h9h5ZwVWkfZkiHmx6d7UIb2KjhMJLW02fvYr3KBZ1XZ58LekPizdORSW7EPWyYKvLPLDmpwyKx3J6f0Ke64gNmw-M9x64Ec1rJYjUomA4VOSIHTcGBDNmp9GLViyvw1BbGUateFgxzvn8ptePvSCC1MLt2zHZ4PNJfnUnO8d7HESjufCb~XaM8dDlJ6VdsZxLFu7LqJ0gWbw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Negative regulation of miR-150 against autocrine IL-22-CCL20-CCR6 pathway in CTCL cells. (A) qPCR of IL-22 and IL-17A in CD4+ T cell (n = 14) and advanced CTCL samples (n = 12). P values were calculated using the Mann-Whitney U test. Asterisks (*) indicate statistical significance: **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 3 independent experiments. (B) Western blot of IL-22RA1 and CCR6 in 5 normal CD4+ T cell (CD4-1 to CD4-5), My-La and HH cell lines, and 5 primary CTCL cases. Jeko-1 (mantle cell lymphoma cell line) is positive control of IL-22RA1.38 (C) ELISA of IL-10, IL-17, IL-22, and CCL20 in CTCL cell lines. Cells (1 × 104) were cultured for 24 hours or 48 hours in 48-well dishes. Each well contained 500 μL of culture medium supplemented with 2% HS or HS free [HS(-)]. Y-axis: pg/mL. ELISA of CCL20 in CTCL cell lines. (D) Effect of IL22RA1 knockdown on IL-22 and CCL20. Left: expression of IL22RA1 in CTCL cell lines transiently transfected with siIL22RA1(744 and 1571). Right: western blot analysis of IL22RA1 in CTCL cells transiently transduced with siIL22RA1(1571) and control [+siSC(scrambled)]. Tubulin served as a control for IL22RA1. ELISA of IL-22 and CCL20 in CTCL cells transiently transduced with siIL22RA1(1571). These cells were cultured for 12 hours or 24 hours. P values were calculated using Student t test. Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 6 independent experiments. (E) ELISA of IL-22 and CCL20 in GFP-empty (control) or GFP-miR-150–transduced CTCL cells transiently transduced with siCCR6 (shown as “+siCCR6”). ELISA of IL-22 and CCL20 in GFP-siCCR6–transduced CTCL cells are also shown. These cells were cultured for 24 hours or 48 hours. Asterisks (*) indicate statistical significance: **.001 ≤ P < .01. Bars are means ± SEM of 3 independent experiments. NS: not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/10/10.1182_blood-2013-09-527739/4/m_1499f6.jpeg?Expires=1769082560&Signature=C44Il0z8DIl2PzkZuFzx8S3bYtR9yNTfVmng7SGazsisgxumNJxFqHsvVWAc67QjesXT74OgoOIyzqINtECraT1gnCf90XlKboiuWBwk13NQ14s9t5GbvG6sSTs6euphEppWuMIS14Nbly1BXjJFZiCg5cqoqgxt~TGGgPdov0YqFJ5nHDSoQMqp1ga44o9gXhKC400lnpn6BVCBfeEExU9vCSguFiEdJhVBhqNLZ7cdTnJyhhKLk3QbA3RVNbz7mtIYOMDyWgt5dXD8C-NUyLNjR3YW8lvzxeoZly705LzechTR0PNbC~5VLv6SU5rDRMjn1uzbAGPgJk64xeVcdQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. MiR-150 and siCCR6 collaborate to inhibit CTCL cell migration. (A) Migration of My-La (top) and HH (bottom) cells in the presence of different extracellular stimuli; 5 × 105 cells in 100 μL of medium without HS and CCL20 in the upper chamber through a coated basement membrane toward different concentrations of CCL20 (0.1∼10 ng/mL), with or without 2% HS at lower chamber (500 μL). The lower chamber contained HS free or CCL20, HS, CCL20, or CCL20 + HS. P values were calculated using Student t test. Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 6 independent experiments. (B) A schematic illustration of the migration assay described above. (C) ELISA of CCL20 in upper and lower chambers (transwell assays) by use of GFP-empty (shown as “control”), GFP-siCCR6 (shown as “siCCR6”), or GFP-miR-150 (shown as “miR-150”) transduced CTCL cells in indicated condition in the lower chamber. (D) A schematic illustration of the ELISA assay (HS + CCL20 condition in the lower chamber). (E) Migration of CTCL cells [GFP-empty+siSC (control), GFP-empty+siCCR6(1058) (transient), GFP-siCCR6(1058)+siSC, GFP-miR-150+siSC, and GFP-miR-150 +siCCR6(1058)] in the presence or absence of CCL20 (0 or 0.5 ng/mL), or HS (2%) in the lower chamber. P values were calculated using Student t test. Asterisks (*) indicate statistical significance: *.01 ≤ P < .05; **.001 ≤ P < .01; ***P < .001. Bars are means ± SEM of 3 independent experiments. (F) A schematic diagram of the relationship between IL-22, CCL20, CCR6, and miR-150 in CTCL. NS: not significant.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/10/10.1182_blood-2013-09-527739/4/m_1499f7.jpeg?Expires=1769082560&Signature=2XRvF2FSmwYb84wQU8ZCvl35v5f89mbad6THTM9pNzF1kt-P4tmglSoHZ982UvxnhjRz4caZ2EUnKoh1YdG7T4u--MhlcA~-BACSIdTjIjLVlvbT93D9YCcyIGMo0yA8GqYHILZoEYDqs6TBEgY7ELvIq-fBGgWF9Oz4ohhlJHR-DR~qWcBI-nO6HQqhl308xCv33RoTAqqsNL5B535TZRrNnmthNlisZhIJMztoV317EY~Q3eDj4gbgGsrBKpG8O-YwGUvXiCAF4SRxgeSjDGcCveX-HAKY0vNAtnbcwMhZyHPUPI1dEVxUBFCnO0ZSL2wopAakJ~TeNj6UlUPBdQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal