Key Points
Notch signaling controls hematopoiesis of human pluripotent stem cells.
Abstract
Notch signaling regulates several cellular processes including cell fate decisions and proliferation in both invertebrates and mice. However, comparatively less is known about the role of Notch during early human development. Here, we examined the function of Notch signaling during hematopoietic lineage specification from human pluripotent stem cells of both embryonic and adult fibroblast origin. Using immobilized Notch ligands and small interfering RNA to Notch receptors we have demonstrated that Notch1, but not Notch2, activation induced hairy and enhancer of split 1 (HES1) expression and generation of committed hematopoietic progenitors. Using gain- and loss-of-function approaches, this was shown to be attributed to Notch-signaling regulation through HES1, which dictated cell fate decisions from bipotent precursors either to the endothelial or hematopoietic lineages at the clonal level. Our study reveals a previously unappreciated role for the Notch pathway during early human hematopoiesis, whereby Notch signaling via HES1 represents a toggle switch of hematopoietic vs endothelial fate specification.
Introduction
The Notch pathway is evolutionarily conserved from invertebrates to mammals and was first recognized as a determinant of cell fate decisions between epithelial and neural lineages in Drosophila.1-3 Notch signaling has also been shown to play an important role in the regulation of endothelial and hematopoietic cell fates in both invertebrates and mice.4,5 Perturbation of Notch signaling has deleterious effects on embryonic hematopoiesis and vascular development.6-11 In mouse models, Notch1 was shown to be required for generation of hematopoietic cells from bipotent hemogenic endothelial precursors, known as hemangioblasts.7 In humans, a critical role for Notch signaling, through the Notch ligand Jagged1, has been demonstrated in the proliferation and differentiation of normal primitive human hematopoietic progenitors.12-14 However, these studies were mostly performed using adult hematopoietic cells or established hematopoietic cell lines. Although the effect of Notch signaling in human embryos or embryonic stem cells (hESCs) has been investigated previously,15,16 the role of Notch signaling in cellular specification and hematopoietic or endothelial commitment during early human development remains largely unknown.
The relationship and cellular organization of hESC-derived angiogenic and hematopoietic precursors have been established.17 These studies led to the identification of CD45–PECAM1+fetal liver kinase-1+VE-cadherin+ (CD45negPFV) cells, a primitive bipotent endothelial-like precursor population differentiated from hESCs that is exclusively responsible for hematopoietic fate. This system provides a less complex and more experimentally tractable platform to investigate endo-hematopoietic precursors. The generation of induced pluripotent stem cell (iPSC) lines18-20 has provided additional opportunities to understand the fundamental processes of human cell fate decisions in the context of tissue regeneration and human disease. Using hESCs and iPSCs generated from human dermal fibroblasts (hFib-iPSCs) as a model of hematopoietic and endothelial development, we investigated the potential role of Notch signaling on the induction and specification of hematopoiesis during hESC and hFib-iPSC differentiation. Our study reveals that Notch signaling acts as a molecular determinant to induce hematopoiesis from human hemogenic precursors at the expense of endothelial commitment.
Materials and methods
Maintenance and hematopoietic differentiation of hESCs and iPSCs
Modulation of Notch signaling
Notch signaling was activated upon immobilization of soluble Jagged1 (Jag1; 10 μg/mL) or Delta1 (Dll1; 10 μg/mL) with Matrigel and was blocked by γ-secretase inhibitor (GSI; DAPT 10 μM, Cat# D5942; Sigma-Aldrich) or hairy and enhancer of split 1 (HES1) small interfering RNA (siRNA) (100 nM; Dharmacon; sequences: ACGAGAGCAAGAAUAAAU, AGGCUGGAGAGGCGGCUAA, UCAACACGACACCGGAUAA, and ACUGCAUGACCCAGAUCAA) in developing embryoid bodies (EBs) and isolated hemogenic precursors.14,17,23 Additionally, Notch signaling was altered by retroviral HES1 overexpression as we previously published24 or short hairpin RNA (shRNA) knockdown in EBs. Four 29-mer shRNAs to target HES1 were generated using HuSH pGFP-V-RS plasmid vector (TG312478, Origene). The sequences are as follows: tatgttcatattggattgcgcctttgtat, gagagctgtattaagtgactgaccatgca, gcttcagcgagtgcatgaacgaggtgacc, and tctgagcacagaaagtcatcaaagcctat. HuSH pGFP-V-RS plasmid vector containing noneffective 29-mer shGFP sequence was used as an experimental control.
Hemato-endothelial commitment of precursors by Notch
To examine the effect of Notch signaling on hematopoietic or endothelial lineage specification of individual hemogenic precursors, isolated single hemogenic precursors from hPSCs were directly dispensed into 96-well plates. Differentiation toward hematopoietic and/or endothelial lineages was induced for 2 weeks with a 50:50 mixture of hematopoietic and endothelial culture medium in the presence or absence of GSI or Jag1. Specified cells toward hematopoietic and/or endothelial lineages upon Notch inhibition (GSI) or activation (Jag1) were distinguished by immunocytochemical staining of CD45 (BD) and endothelial nitric oxide synthase (Abcam). Expression was measured on a microplate reader (PHERAstar).
Molecular profiling of Notch signals
Quantitative polymerase chain reaction was performed using SYBR Green Master Mix Kit (Applied Biosystems) on a thermal cycler (Stratagene). Primers can be found in supplemental Table 1 (see the Blood Web site).
For microarray analysis (Gene Expression Omnibus accession number: GSE47466), total RNA was extracted from undifferentiated hESCs, hemogenic precursors, and the remaining cells in day10 hEBs. Amplified-labeled RNA was hybridized to HG-U133AB chips (Affymetrix). Data analysis was performed by using GeneSpring 6.0 (Silicon Genetics) including per array and per gene normalization, as previously described.17 The expression level of hemogenic precursors and remaining cells in day10 hEBs was calculated relative to undifferentiated hESCs.
Western blot analysis
Twenty micrograms of proteins extracted from hPSCs and hEBs was transferred to PVDF membrane (Amersham) and probed with full-length Notch1 (Santa Cruz), cleaved Notch1 (Notch intracellular domain [NICD]; Cell Signaling), HES1 (Chemicon), or β-actin (Santa Cruz) antibodies. Horseradish peroxidase–conjugated secondary antibodies were used to detect the signals.
Immunocytochemistry
Whole mount hEBs and frozen hEB sections were incubated with either mouse IgG isotype (5 μg/mL; BD Pharmingen) or primary antibodies against platelet endothelial cell adhesion molecule 1 (PECAM1), Jagged1, Notch1 (Santa Cruz), and HES1 (Chemicon) and then subsequently visualized with fluorescent secondary antibodies. Characterization of endothelial cells was performed by Dil-Ac-LDL (1,1′-dioctadecyl-3,3,3′,3′-tetramethyl-indocarbocyanine perchlorate) uptake and vascular endothelial (VE)–cadherin staining as previously reported.17 All the cells were counterstained with 4,6-diamidino-2-phenylindole. Morphometrical analysis was performed by Image Pro (Media Cybernetics).
Flow cytometry
Single cells dissociated from hEBs were stained with fluorescent-conjugated hematopoietic markers including PECAM1-phycoerythrin, CD34–fluorescein isothiocyanate, and CD45-allophycocyanin (BD) or endothelial markers VE-cadherin–phycoerythrin and fetal liver kinase-1–allophycocyanin (BD).17 During hematopoietic differentiation, PECAM1+CD45–, CD34+CD45+, and CD45+ fractions were identified as hemogenic precursor, primitive blood, and blood cells, respectively. Notch1 (Santa Cruz) expression in EBs upon Jag1 stimulation was examined. Live cells were identified by exclusion of 7-Aminoactinomycin. Cells were analyzed for surface marker expression on FACSCalibur (BDIS) and FlowJo (Treestar).
siRNA treatment
Target gene knockdown studies were performed by siRNA treatment following the manufacturer’s instruction (Dharmacon). Notch1 (ON-TARGETplus SMARTpool L-007771) and Notch2 (ON-TARGETplus SMARTpool L-012235) were knocked down by siRNAs (100 nM) for 5 days (short term) or 15 days (long term). We treated HES1 siRNA (ON-TARGETplus SMARTpool L-007770) at 3 time points (days 0, 4, and 9) based on the expression of HES1 during hematopoietic hEB differentiation. A scrambled siRNA (ON-TARGETplus Non-targeting Pool) was used as an experimental control.
Hematopoietic CFU and endothelial tube formation assay
Colony-forming unit (CFU) assays were performed with day 10 and day 15 hEBs cultured in the presence or absence of Jag1. Dissociated single cells (15 000 or 10 000) were plated into methylcellulose supplemented with hematopoietic cytokines. Additionally, isolated hemogenic precursors were also examined for clonogenic colony-formation capacity with cells (10 000 or 5000) upon Jag1, GSI, or siRNA treatment.
Endothelial cells that emerged from either hEBs or hemogenic precursors by GSI-, siRNA-, or shRNA-mediated HES1 suppression were harvested and expanded in endothelial-specific medium (Lonza). Tube formation assay was conducted by plating hPSC-derived endothelial cells onto a thin-layer Matrigel-coated plate and incubating for 48 hours in endothelial culture medium. Human umbilical vein endothelial cell (HUVEC) was used as a positive control.
Statistical analysis
Statistical analysis was performed by Student t test.
Results
Notch ligands via HES1 induce hematopoietic differentiation
In mice, Notch signaling is activated through the interaction of Notch receptors (Notch1-4) with Notch ligands (Delta1, Delta3, Delta4, Jagged1, and Jagged2) to control embryonic development toward hemangioblasts.6-11 In the hematopoietic system, Jagged125 and Delta114,26 are key regulators of Notch signaling. Accordingly, to understand the role of Notch signaling in human, we examined whether Jagged1 (Jag1) and Delta1 (Dll1) contribute to hematopoietic development from hPSCs.
As it has previously been shown that immobilized Notch ligand induces direct ligand-receptor interactions,14,16,27 we tested both soluble and immobilized Jag1 and Dll1 during hPSC hematopoiesis. Given the effectiveness of immobilization using the FLAG-tagging method,28,29 soluble Jag1 or Dll1 was immobilized directly to hESC cultures through the addition of Matrigel16 prior to EB formation and hematopoietic differentiation (supplemental Figure 1A) to mimic the FLAG-tagging method. Augmentation of CD34+CD45+ primitive blood cells (supplemental Figure 1B-C) and CD45+ blood cells (Figure 1A and supplemental Figure 1D) from EBs was only evident in the presence of Jag1 or Dll1 when the ligands were immobilized via addition to Matrigel, where HES1 expression was induced in both hESC-derived (Figure 1B) and iPSC-derived EBs (Figure 1C). These results demonstrate that Notch ligands immobilized with the addition to Matrigel (hereafter referred to as Jag1 and Dll1) could enhance blood differentiation of hPSCs along with increased HES1 expression. As such, all subsequent studies were performed using this strategy of immobilization of Notch ligands.
Notch signaling is involved in the hematopoietic development of hFib-hPSCs. (A) Notch ligand immobilization augmented hematopoietic differentiation of hFib-hESCs. EBs treated with Matrigel immobilized with soluble Jagged1 (sJag1) or Delta1 (sDll1) during phase 2 of EB development had increased hematopoietic CD45+ blood development. *P < .05 (n = 3). (B-C) HES1 expression was upregulated by Jag1 and Dll1 phase 2 treatment in developing EBs formed from hESCs (B) and hFib-iPSCs (C) compared with IgG-treated control EBs (CTRL). However, control EBs showed higher HES1 expression than undifferentiated hESCs (B) and hFib-iPSCs (C). **P < .01; *P < .05 (n = 3). (D) Time course expression of Notch1, Notch2, and HES1 during hematopoietic EB development. The expression level of Notch1 and HES1 reached steady state by day 6 and then increased gradually and peaked at day 15. However, only minor changes were observed in Notch2 expression. Two different periods, phase 1 (0-5 days) and phase 2 (6-15 days), were identified based on the expression pattern of Notch signals and previous studies. (E) Notch1-mediated signaling regulated blood development of hESCs. Either the transient (phase 1 [0-5 days] or phase 2 [6-15 days]) or continuous (phases 1 and 2 [0-15 days]) suppression of Notch1 (N1) but not Notch2 (N2) by siRNA treatment decreased blood development relative to control scrambled siRNA treatment. *P < .05 (n = 3). (F-I) Hematopoietic development of EBs is regulated by the status of Notch signaling. Notch-signaling activation by adding Jag1 (F,H) or Dll1 (G,I) to developing EBs results in augmented blood differentiation (F-G). Increased or decreased blood formation coincided with up- or downregulation of HES1 (H-I) by either ligand treatment or Notch1 knockdown (siNotch1). Hematopoiesis and Notch signaling could be recovered in only phase 1 Notch1 knockdown EBs by adding Jag1 (F,H) or Dll1 (G,I), whereas neither the hematopoiesis (F-G) nor the expression of HES1 (H-I) could be recovered after long-term (phase 2 only or phases 1 and 2) suppression of Notch1 (+). **P < .01; *P < .05 (n = 3).
Notch signaling is involved in the hematopoietic development of hFib-hPSCs. (A) Notch ligand immobilization augmented hematopoietic differentiation of hFib-hESCs. EBs treated with Matrigel immobilized with soluble Jagged1 (sJag1) or Delta1 (sDll1) during phase 2 of EB development had increased hematopoietic CD45+ blood development. *P < .05 (n = 3). (B-C) HES1 expression was upregulated by Jag1 and Dll1 phase 2 treatment in developing EBs formed from hESCs (B) and hFib-iPSCs (C) compared with IgG-treated control EBs (CTRL). However, control EBs showed higher HES1 expression than undifferentiated hESCs (B) and hFib-iPSCs (C). **P < .01; *P < .05 (n = 3). (D) Time course expression of Notch1, Notch2, and HES1 during hematopoietic EB development. The expression level of Notch1 and HES1 reached steady state by day 6 and then increased gradually and peaked at day 15. However, only minor changes were observed in Notch2 expression. Two different periods, phase 1 (0-5 days) and phase 2 (6-15 days), were identified based on the expression pattern of Notch signals and previous studies. (E) Notch1-mediated signaling regulated blood development of hESCs. Either the transient (phase 1 [0-5 days] or phase 2 [6-15 days]) or continuous (phases 1 and 2 [0-15 days]) suppression of Notch1 (N1) but not Notch2 (N2) by siRNA treatment decreased blood development relative to control scrambled siRNA treatment. *P < .05 (n = 3). (F-I) Hematopoietic development of EBs is regulated by the status of Notch signaling. Notch-signaling activation by adding Jag1 (F,H) or Dll1 (G,I) to developing EBs results in augmented blood differentiation (F-G). Increased or decreased blood formation coincided with up- or downregulation of HES1 (H-I) by either ligand treatment or Notch1 knockdown (siNotch1). Hematopoiesis and Notch signaling could be recovered in only phase 1 Notch1 knockdown EBs by adding Jag1 (F,H) or Dll1 (G,I), whereas neither the hematopoiesis (F-G) nor the expression of HES1 (H-I) could be recovered after long-term (phase 2 only or phases 1 and 2) suppression of Notch1 (+). **P < .01; *P < .05 (n = 3).
Notch1 specifically regulates hematopoietic differentiation from EBs
We have previously shown that hematopoietic development from hEBs can be divided into 2 phases: phase 1 (0-5 days), specification (into hemogenic precursors from hPSCs); and phase 2 (6-15 days), commitment (toward hematopoietic progenitors).30 To further dissect the role of Notch signaling during hPSC hematopoiesis, we examined the expression of Notch regulators during 15 days of EB development. Endogenous Notch1 and HES1 expression levels increased marginally during the first 5 days but were more substantially upregulated after day 6, whereas Notch2 expression remained unchanged throughout (Figure 1D). To determine the specific role that Notch signaling may play during the phases of hPSC hematopoietic induction, we knocked down Notch1 and Notch2 by siRNAs during phase 1, 2, or both periods (phases 1 and 2) of EB differentiation (Figure 1E). Notch1 downregulation only in either phase 1 or 2 or both phases together decreased hematopoietic differentiation (Figure 1E) compared with scrambled siRNA control treatments. These results support a critical role of Notch1, but not Notch2, throughout hEB-based hematopoietic differentiation. To determine the critical phase of hPSC hematopoiesis regulated by Notch signaling, we treated EBs with Jag1 or Dll1 coupled with Notch1 siRNA treatment (Figure 1F-I; supplemental Figure 1E-F). Consistently, reduced hematopoiesis from hPSCs was induced by knockdown of Notch1 (Figure 1F-G) in addition to a reduction in expression of HES1 (Figure 1H-I) and siRNA target Notch1 (supplemental Figure 1E-F) relative to scrambled siRNA controls. This reduction caused by knockdown of Notch1 could be recovered by addition of Jag1 and, to a lesser extent, Dll1 but was limited to phase 1 only. This effective rescue by Jag1 and Dll1 did not occur when Notch1 was knocked down at phase 1 or combined phase 1 and phase 2 (Figure 1F-I; supplemental Figure 1E-F). In contrast, Notch2 modulation did not affect blood differentiation and expression of Notch1 and HES1 (data not shown). These results suggest that Notch1-mediated blood differentiation and Notch-signaling activation in response to Jag1 and Dll1 is critical during the commitment phase of hPSC hematopoiesis (phase 2), whereas Jag1 and Dll1 can functionally compensate for Notch1 reduction during the specification phase of hPSC hematopoiesis (phase 1).
Jagged1 activates Notch signaling within developing EBs
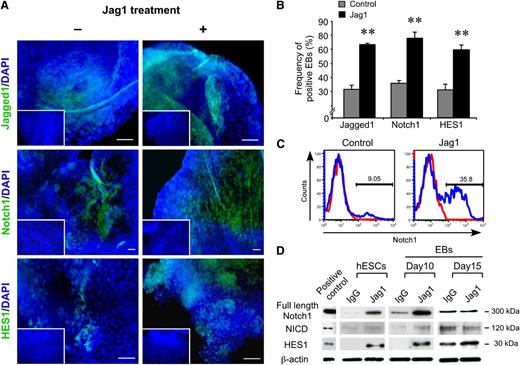
As Jagged1 seemingly represents a more potent inducer of Notch signaling during hPSC hematopoiesis in our system, we focused our additional studies on Jag1. To further investigate Notch signaling during hematopoiesis, EB protein levels of Jagged1, Notch1, and HES1 were examined in the presence of Jag1. Jagged1, Notch1, and HES1 were expressed at low levels in day 10 EBs (Figure 2A-B) and in unstimulated hESC cultures (supplemental Figure 2A) but were upregulated upon Jag1 induction (Figure 2A,D). It is believed that Jagged1 is unable to activate Notch1, as exemplified in mouse aorta-gonad-medonephros25 , where exceptions include cells devoid of fringe or potential activation via Jagged2. It is thus important to consider that effects of Jag1 could be indirect and act via other cell types in heterogeneous systems. Even though little is known about amplification of Notch expression in target cells by inducer or inhibitor treatments,31-33 quantitative analysis of single EBs demonstrated that Jag1 stimulation elicited a significant increase in the number of EBs expressing Jagged1, Notch1, and HES1 during hPSC hematopoietic differentiation (Figure 2B). Quantification of Notch1 expression by fluorescence-activated cell sorter (FACS) analysis following Jag1 stimulation demonstrated a nearly fourfold increase in Notch1 expression compared with control (Figure 2C). Confirming that Notch signaling has been activated in EBs in response to Jag1 by western blot analysis revealed the presence of both full-length and cleaved NICD, as well as HES1 (Figure 2D). As predicted based on phase 1 to phase 2 steps of hematopoietic induction from hPSCs,30 HES1 and NICD were only detectable in day10 EBs in the presence of Jag1, and levels did not increase with persistent Jag1 stimulation to day 15 (Figure 2D), indicating that maximum effects were achieved. The timing of NICD and HES1 regulation coincided with the emergence of hemogenic precursors (CD31+CD45–), which are known to be responsible for hematopoietic fate. This correlation of hemogenic precursor emergence, together with simultaneous upregulation of HES1 and NICD, provided further support for the importance of Notch signaling specifically during the later stages of hematopoietic commitment from hPSCs that warranted further investigation.
Activation of Notch signaling within developing EBs by Jagged1. (A) The activation of Notch signaling during hematopoietic hEB development was assessed using whole mount immunofluorescent staining of hEBs stimulated with (+) or without (–) Jag1, to detect Jagged1 (upper panels), Notch1 (middle panels), and HES1 (bottom panels). Higher numbers of positive cells for each factor were observed in Jag1-treated hEBs (+, right panels). Scale bars represent 100 μm. (B) Quantification of Jagged1-, Notch1-, or HES1-positive hEBs in the presence or absence of Jag1. **P < .01 (n = 3). (C) Flow cytometric analysis represented increase of Notch1-expressing cells in day 10 EBs upon Jag1 stimulation compared with control. (D) Western blot analysis of Notch-signaling pathway components, including full-length Notch1, NICD, and HES1 in undifferentiated hESCs and developing hematopoietic EB development, in the presence or absence of Jag1. Notch1-positive brain extract was used as a positive control.
Activation of Notch signaling within developing EBs by Jagged1. (A) The activation of Notch signaling during hematopoietic hEB development was assessed using whole mount immunofluorescent staining of hEBs stimulated with (+) or without (–) Jag1, to detect Jagged1 (upper panels), Notch1 (middle panels), and HES1 (bottom panels). Higher numbers of positive cells for each factor were observed in Jag1-treated hEBs (+, right panels). Scale bars represent 100 μm. (B) Quantification of Jagged1-, Notch1-, or HES1-positive hEBs in the presence or absence of Jag1. **P < .01 (n = 3). (C) Flow cytometric analysis represented increase of Notch1-expressing cells in day 10 EBs upon Jag1 stimulation compared with control. (D) Western blot analysis of Notch-signaling pathway components, including full-length Notch1, NICD, and HES1 in undifferentiated hESCs and developing hematopoietic EB development, in the presence or absence of Jag1. Notch1-positive brain extract was used as a positive control.
Notch signaling regulates subsequent blood commitment during EB development
To dissect the Notch-signaling mechanisms associated with commitment (phase 2) of hematopoiesis from hPSCs, we formed hEBs from both hESCs and hFib-iPSCs (Figure 3A-D) in the presence of Jag1, HES1 siRNA, or GSI33-35 that prevents Notch cleavage and downstream signaling. Total numbers and frequencies of primitive blood cells (CD34+CD45+), committed hematopoietic cells (CD45+), and primitive hematopoietic CFUs were analyzed (Figure 3E-L) for each treatment.
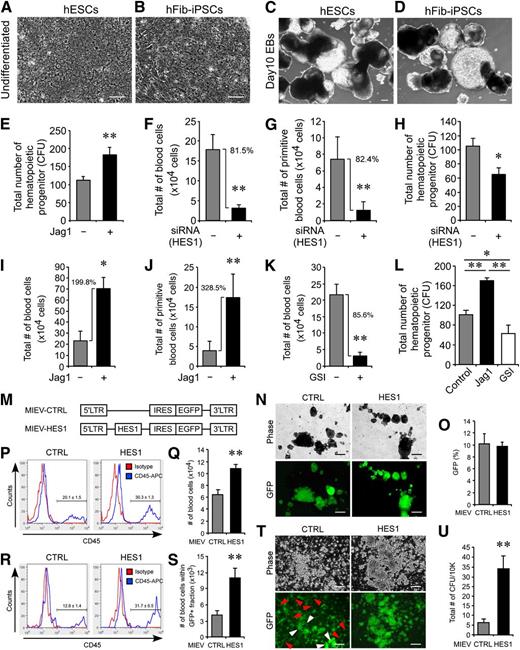
Modulation of Notch signaling alters blood commitment of developing hEBs. (A-D) Morphologic similarity was observed between hESC (A,C) and hFib-iPSC (B,D) in undifferentiated (A-B) and day 10 developing EBs (C-D). Scale bars represent 100 μm (A-B) and 250 μm (C-D). (E) Total number of hematopoietic colonies was higher in Jag1-treated hEBs vs control (181.8 ± 20.8 vs 111.8 ± 9.9). **P < .01 (n = 4). (F-G) Knockdown HES1 using siRNA treatment reduced 81.5% of committed (F) and 82.4% of primitive blood (G) cells in hematopoietic hESC-EB development relative to siRNA controls. **P < .01 (n = 3). (H) Hematopoietic colony formation (CFU) was performed to examine the effect of HES1 knockdown on hematopoietic progenitors. siRNA-mediated knockdown of HES1 led to a reduction in the number of hematopoietic colonies (64.8 ± 9.1 vs 104.8 ± 11.5) relative to scrambled siRNA controls. *P < .05 (n = 4). (I-J) During hFib-iPSC hematopoiesis, Jag1-activated Notch1 signaling increased the number of committed (I, 199.8% increase) and primitive blood cells (J, 328.5% increase) compared with controls. *P < .05; **P < .01 (n = 3). (K) Inhibition of hFib-iPSC blood commitment by GSI. EBs treated with GSI showed 85.6% reduced blood commitment relative to controls. **P < .01 (n = 3). (L) Alterations in Notch signaling control the generation of CFU-forming hematopoietic progenitors. Jag1-induced Notch signaling increased the number of CFUs (170.25 ± 5.47), whereas GSI-treated EBs formed fewer CFUs (63.25 ± 16.58) relative to controls (101.25 ± 8.66). *P < .05; **P < .01 (n = 4). (M-N) MIEV retroviral vectors for overexpression of HES1 (M) and GFP signals in EBs transduced with control (CTRL, left panels) or HES1 (right panels) retroviral overexpression vectors (N). Scale bars represent 250 μm. (O) Transduction efficiency of both control (10.12 ± 1.7%) and HES1 (9.81 ± 0.6%) retroviruses at day 15 of EB development by FACS. (P-S) Augmentation of blood in total EBs (P-Q) or within the GFP+ cell fraction (R-S) at day 15 of EB development. Both frequency based on FACS analysis (P, 30.3 ± 1.3% vs 20.1 ± 1.5%) and total number of blood cells (Q, 10.9 ± 0.6 × 104 cells vs 6.6 ± 0.8 × 104 cells) are higher in developing hEBs transduced by HES1 overexpression vs control. Within the GFP+ cell fraction, higher frequency (R, 31.7 ± 6.5% vs 12.8 ± 1.4%) and number of blood cells (S, 11.1 ± 1.73 × 103 cells vs 4.2 ± 0.71 × 103 cells) are observed in HES1 overexpression. **P < .01 (n = 4). (T) Contribution of GFP+ cells in hematopoietic CFUs from both control and HES1 overexpression. GFP+ cells from control CFUs contributed in both hematopoietic colonies (white arrowhead) and attached endothelial colonies similarly (red arrowhead), whereas the majority of GFP+ cells are restricted to hematopoietic colonies upon HES1 overexpression. Scale bars represent 100 μm. (U) HES1 overexpression increased total number of hematopoietic colonies over the control (34.3 ± 6.4 vs 6.7 ± 1.5). **P < .01 (n = 3).
Modulation of Notch signaling alters blood commitment of developing hEBs. (A-D) Morphologic similarity was observed between hESC (A,C) and hFib-iPSC (B,D) in undifferentiated (A-B) and day 10 developing EBs (C-D). Scale bars represent 100 μm (A-B) and 250 μm (C-D). (E) Total number of hematopoietic colonies was higher in Jag1-treated hEBs vs control (181.8 ± 20.8 vs 111.8 ± 9.9). **P < .01 (n = 4). (F-G) Knockdown HES1 using siRNA treatment reduced 81.5% of committed (F) and 82.4% of primitive blood (G) cells in hematopoietic hESC-EB development relative to siRNA controls. **P < .01 (n = 3). (H) Hematopoietic colony formation (CFU) was performed to examine the effect of HES1 knockdown on hematopoietic progenitors. siRNA-mediated knockdown of HES1 led to a reduction in the number of hematopoietic colonies (64.8 ± 9.1 vs 104.8 ± 11.5) relative to scrambled siRNA controls. *P < .05 (n = 4). (I-J) During hFib-iPSC hematopoiesis, Jag1-activated Notch1 signaling increased the number of committed (I, 199.8% increase) and primitive blood cells (J, 328.5% increase) compared with controls. *P < .05; **P < .01 (n = 3). (K) Inhibition of hFib-iPSC blood commitment by GSI. EBs treated with GSI showed 85.6% reduced blood commitment relative to controls. **P < .01 (n = 3). (L) Alterations in Notch signaling control the generation of CFU-forming hematopoietic progenitors. Jag1-induced Notch signaling increased the number of CFUs (170.25 ± 5.47), whereas GSI-treated EBs formed fewer CFUs (63.25 ± 16.58) relative to controls (101.25 ± 8.66). *P < .05; **P < .01 (n = 4). (M-N) MIEV retroviral vectors for overexpression of HES1 (M) and GFP signals in EBs transduced with control (CTRL, left panels) or HES1 (right panels) retroviral overexpression vectors (N). Scale bars represent 250 μm. (O) Transduction efficiency of both control (10.12 ± 1.7%) and HES1 (9.81 ± 0.6%) retroviruses at day 15 of EB development by FACS. (P-S) Augmentation of blood in total EBs (P-Q) or within the GFP+ cell fraction (R-S) at day 15 of EB development. Both frequency based on FACS analysis (P, 30.3 ± 1.3% vs 20.1 ± 1.5%) and total number of blood cells (Q, 10.9 ± 0.6 × 104 cells vs 6.6 ± 0.8 × 104 cells) are higher in developing hEBs transduced by HES1 overexpression vs control. Within the GFP+ cell fraction, higher frequency (R, 31.7 ± 6.5% vs 12.8 ± 1.4%) and number of blood cells (S, 11.1 ± 1.73 × 103 cells vs 4.2 ± 0.71 × 103 cells) are observed in HES1 overexpression. **P < .01 (n = 4). (T) Contribution of GFP+ cells in hematopoietic CFUs from both control and HES1 overexpression. GFP+ cells from control CFUs contributed in both hematopoietic colonies (white arrowhead) and attached endothelial colonies similarly (red arrowhead), whereas the majority of GFP+ cells are restricted to hematopoietic colonies upon HES1 overexpression. Scale bars represent 100 μm. (U) HES1 overexpression increased total number of hematopoietic colonies over the control (34.3 ± 6.4 vs 6.7 ± 1.5). **P < .01 (n = 3).
In addition to the increase in frequency and total number of primitive blood cells (CD34+CD45+) and mature blood cells (CD45+) following Jag1 treatment (Figure 1A; supplemental Figure 1B-D), functional assessment of primitive clonogenic hematopoietic progenitors (CFUs) revealed that Notch activation by Jag1 induced an increase in hematopoietic progenitors (Figure 3E). Conversely, inhibition of Notch signaling during EB development by HES1 downregulation (supplemental Figure 3A) reduced the number of committed (Figure 3F) and primitive blood cells (Figure 3G) in hEBs, as well as the total number of hematopoietic progenitors (CFUs; Figure 3H), compared with scrambled siRNA controls. These results demonstrate that independent of ligand stimulation, Notch signaling via HES1 is critical for the development of hematopoietic progenitors and mature blood cells.
Hematopoietic development of hFib-iPSCs could also be regulated by Notch signaling. Upon Jag1 stimulation, hFib-iPSCs showed an increase in committed (Figure 3I) and primitive (Figure 3J) blood cell production. These Jag1-induced effects could be reduced by the inhibition of Notch processing through the use of GSI (Figure 3K). Similar to functional hematopoietic colony formation from hESCs, hematopoietic progenitor development from hFib-iPSCs was regulated by Notch signaling (Figure 3L). Activated Notch upon Jag1 stimulation enhanced not only primitive blood augmentation but also more definitive blood commitment in the erythrocyte lineage progenitors specifically (supplemental Figure 3B-C). FACS plots shown in supplemental Figure 3D represent the effect of Notch-signaling effectors in hPSC hematopoiesis. These results demonstrate that activation of HES1 by Jag1 treatment augments hematopoietic development following definitive blood commitment by inducing blood fate specification in both embryonic and adult skin-derived sources of hPSCs.
To understand the independent role of HES1 in this process, HES1 overexpression (using retroviral transduction) was also used to test the effect of activated Notch signaling on hESC hematopoiesis (Figure 3M-U). Control and HES1 retroviral vectors (Figure 3M) showed similar transduction efficiency during EB development (Figure 3N-O). Emergence of CD45+ blood cells was increased in EBs transduced with HES1 compared with control (Figure 3P-Q), with more than threefold increase in HES1 (supplemental Figure 3E). The total number of transduced (GFP+) CD45+ blood cells was increased upon HES1 overexpression (Figure 3R-S), as well as the total number of CFUs (Figure 3T-U). These data indicate that Notch activation targeting through HES1 alone enhances hematopoietic commitment of hPSCs.
HES1-mediated Notch activation targets hemogenic precursor development of hPSCs
The augmentation of both hematopoietic progenitors and mature blood cells during the commitment phase suggested that HES1-mediated Notch activation may act on hemogenic precursors arising after hPSC specification to the hematopoietic fate. Accordingly, we evaluated the exact frequency and number of hemogenic precursors emerging at day 10 and day 15 (phase 2, commitment step) in the presence or absence of Jag1 (Figure 4A-B). Jag1 increased the hemogenic precursor frequency at day 10 of EB development showing active Notch1 and HES1 (supplemental Figure 4A) relative to controls. However, by day 15, the frequency of hemogenic precursors in Jag1-treated EBs had declined, in contrast to the continued increase in hemogenic precursors from untreated EBs (Figure 4A). The total numbers of hemogenic cells observed at both day 10 and day 15 following Jag1 treatment followed the same trend (Figure 4B). An increase in hematopoietic progenitors and commitment to mature blood cell output (supplemental Figure 4B-C) corresponded to the subsequent loss by day 15 (Figure 4A-B). These results indicate that Notch signaling through Jag1 facilitates hemogenic precursor commitment that in turn increases the output of mature hematopoietic cells as evidenced by subsequent exhaustion of hemogenic precursors at day 15.
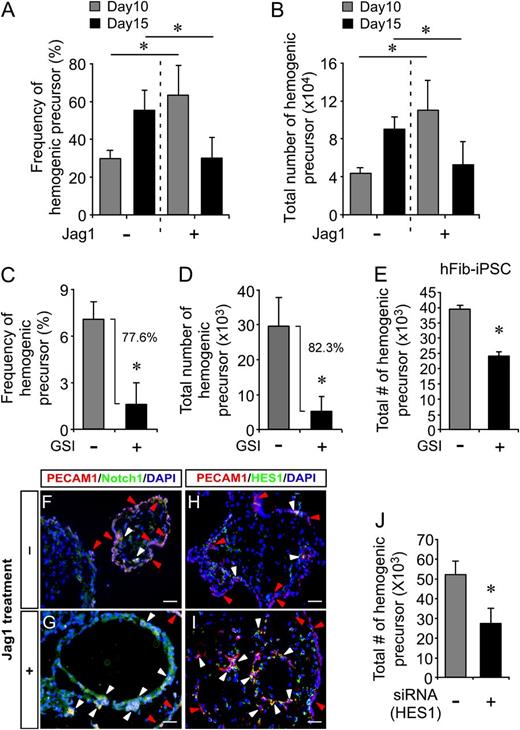
Regulation of hPSC hemogenic precursor specification by Notch signaling. (A-B) Effect of Jag1-mediated Notch activation on hemogenic precursor specification of hESCs. Increased frequency (A, 63.1 ± 15.6% vs 29.6 ± 4.5%) and total number (B, 11 ± 3.2 × 104 cells vs 4.4 ± 0.4 × 104 cells) of hemogenic precursors were observed in hEBs treated with Jag1 relative to controls on day 10. In contrast, on day 15, the frequency (A, 29.7 ± 10.8% vs 55.1 ± 10.6%) and total number (B, 5.3 ± 2.5 × 104 cells vs 9.1 ± 1.2 × 104 cells) of hemogenic precursors in Jag1-treated hEBs had significantly decreased. *P < .05 (n = 3). (C-D) Effect of GSI on the emergence of hemogenic precursors during hEB development. hEBs treated with a GSI showed decreased frequency (C, 1.58 ± 1.36% vs 7.08 ± 1.08%) and total number (D, 5.2 ± 4.1 × 103 cells vs 29.6 ± 8.0 × 103 cells) of hemogenic precursor relative to controls on day 10. *P < .05 (n = 3). (E) GSI inhibited hemogenic precursor augmentation from hEBs formed with hFib-iPSCs (24.3 ± 1.1 × 103 cells vs 39.6 ± 1.2 × 103 cells). *P < .05 (n = 3). (F-I) Effect of Notch signaling on the emergence of hemogenic precursors from hESCs was confirmed by colocalization of PECAM1 with Notch1 (F-G) and HES1 (H-I) by immunofluorescence. Day 10 hEBs stimulated by Jag1 (G,I) were compared with control hEBs (F,H). In EBs stimulated with Jag1, the majority of Notch1+ cells (G) and HES1+ cells (I) coexpressed hemogenic precursor marker PECAM1 (white arrowhead), whereas lower numbers of PECAM1+ cells (red arrowheads) are predominant in control hEBs. Scale bars represent 100 μm. (J) HES1 knockdown during hematopoietic hEB development impaired hemogenic precursor specification. HES1 siRNA–treated hEBs exhibited reduced numbers of hemogenic precursors compared with the scrambled siRNA control (27.6 ± 7.4 × 103 cells vs 52.1 ± 7.1 × 103 cells). *P < .05 (n = 3).
Regulation of hPSC hemogenic precursor specification by Notch signaling. (A-B) Effect of Jag1-mediated Notch activation on hemogenic precursor specification of hESCs. Increased frequency (A, 63.1 ± 15.6% vs 29.6 ± 4.5%) and total number (B, 11 ± 3.2 × 104 cells vs 4.4 ± 0.4 × 104 cells) of hemogenic precursors were observed in hEBs treated with Jag1 relative to controls on day 10. In contrast, on day 15, the frequency (A, 29.7 ± 10.8% vs 55.1 ± 10.6%) and total number (B, 5.3 ± 2.5 × 104 cells vs 9.1 ± 1.2 × 104 cells) of hemogenic precursors in Jag1-treated hEBs had significantly decreased. *P < .05 (n = 3). (C-D) Effect of GSI on the emergence of hemogenic precursors during hEB development. hEBs treated with a GSI showed decreased frequency (C, 1.58 ± 1.36% vs 7.08 ± 1.08%) and total number (D, 5.2 ± 4.1 × 103 cells vs 29.6 ± 8.0 × 103 cells) of hemogenic precursor relative to controls on day 10. *P < .05 (n = 3). (E) GSI inhibited hemogenic precursor augmentation from hEBs formed with hFib-iPSCs (24.3 ± 1.1 × 103 cells vs 39.6 ± 1.2 × 103 cells). *P < .05 (n = 3). (F-I) Effect of Notch signaling on the emergence of hemogenic precursors from hESCs was confirmed by colocalization of PECAM1 with Notch1 (F-G) and HES1 (H-I) by immunofluorescence. Day 10 hEBs stimulated by Jag1 (G,I) were compared with control hEBs (F,H). In EBs stimulated with Jag1, the majority of Notch1+ cells (G) and HES1+ cells (I) coexpressed hemogenic precursor marker PECAM1 (white arrowhead), whereas lower numbers of PECAM1+ cells (red arrowheads) are predominant in control hEBs. Scale bars represent 100 μm. (J) HES1 knockdown during hematopoietic hEB development impaired hemogenic precursor specification. HES1 siRNA–treated hEBs exhibited reduced numbers of hemogenic precursors compared with the scrambled siRNA control (27.6 ± 7.4 × 103 cells vs 52.1 ± 7.1 × 103 cells). *P < .05 (n = 3).
Inhibition of Notch cleavage using a GSI reduced both the frequency and total number of hemogenic precursors from hESCs (Figure 4C-D) and hFib-iPSCs (Figure 4E). To identify the cellular target of Jag1-induced Notch signaling during hematopoietic development of hESCs, we used immunocytochemical colocalization for components of the Notch pathway combined with markers of hemogenic precursors (Figure 4F-I). The vast majority of Notch1+ and HES1+ cells were PECAM1+ hemogenic precursors in Jag1-stimulated EBs (Figure 4G,I), compared with untreated controls (Figure 4F,H), suggesting that Jag1-induced Notch-signaling activation regulates the emergence of hemogenic precursors through HES1 expression. To determine the functional significance of Jag1-induced HES1-expressing hemogenic precursors within hEBs, we targeted HES1 by siRNA-mediated knockdown relative to scrambled siRNA control. Using real-time polymerase chain reaction analysis, we demonstrated successful knockdown of HES1 expression in day 15 EBs (supplemental Figure 3A). Knockdown of HES1 resulted in reduced hemogenic precursor development (Figure 4J). These data indicate that Notch signaling controls emergence of hemogenic precursors directly through HES1.
Notch signaling mediates hematopoietic vs endothelial cell fate
To determine whether Notch signaling directly acts on hPSC-derived hemogenic precursors, we cultured dissociated single cells from untreated day10 hEBs in previously optimized serum-free hematopoietic culture-inducing conditions (Hem-culture).17 GSI-mediated inhibition of Notch signaling during extended Hem-culture of untreated hEBs resulted in diminished numbers of blood cells (Figure 5A-B). Based on these observations, we hypothesized that modulation of Notch signaling could influence subsequent lineage specification directly from hemogenic precursors and thus prospectively isolated bipotent hemogenic precursors17 for in vitro culture and assessment of Notch-signaling regulation (Figure 5C). As hemogenic precursors are bipotent,17 we investigated whether HES1-mediated Notch signaling induced hematopoietic cell fate determination and affected endothelial differentiation. To test this hypothesis, CFU assays for hematopoietic or endothelial progenitors, together with immunostaining, were used to quantitatively assess the potential for hematopoietic vs endothelial development arising from isolated hemogenic precursors derived from day10 hEBs (Figure 5C).
Effect of Notch signaling on the hematopoietic commitment of purified hemogenic precursors. (A-B) GSI-mediated inhibition of Notch signaling in dissociated single cells from unsorted total EBs resulted in an 89.3% reduction of CD45+ blood cell frequency (A) and immunocytochemical staining for CD45 on single cells after extended hematopoietic culture (Hem-culture) revealed lower numbers of CD45+ cells in the GSI-treated group (B). *P < .05 (n = 3). (C) Schematic summary to examine the effect of up- or downregulated Notch signaling on blood commitment of isolated hemogenic precursors. (D) Emergence of CD45+ blood cells could be modulated by controlling Notch signaling in isolated hemogenic precursors. GSI-mediated Notch downregulation inhibited blood commitment (5.38 ± 1.45%) over controls (28.2 ± 12%). Conversely, Jag1-stimulated hemogenic precursors enhanced blood augmentation (78.2 ± 18%). *P < .05; **P < .01 (n = 3). (E) GSI-mediated inhibition of Notch signaling on isolated hemogenic precursors resulted in a significant reduction of CD45+ blood cells by immunocytochemical staining. (F-G) Total number of hematopoietic colonies (F) and endothelial colonies (G) formed from isolated hemogenic precursors by modulating Notch signaling during extended Hem-culture. GSI-mediated Notch downregulation decreased the number of hematopoietic colonies (F, 4.66 ± 2.51), whereas it increased the number of endothelial colonies (G, 7.33 ± 1.52). On the contrary, upregulated Notch upon Jag1 treatment increased the number of hematopoietic colonies (F, 26.67 ± 4.73) but decreased endothelial colonies (G, 1.33 ± 0.58) compared with the number of hematopoietic (F, 14.67 ± 2.52) and endothelial colonies (G, 3 ± 1) in the control. *P < .05; **P < .01 (n = 3). (H) Effect of up- or downregulation of Notch on endothelial-specific marker expression was analyzed by flow cytometry. Expression level of PECAM1 and VE-cadherin was evaluated in isolated hemogenic precursors with activated or suppressed Notch signaling by Jag1 or GSI treatment, respectively. GSI-treated hemogenic precursors showed higher frequency of PECAM1+VE-cadherin+ (54.73 ± 6.17%) over controls (27.94 ± 7.59%) and Jag1 treatment (9.94 ± 3.82%). *P < .05; **P < .01 (n = 3). (I) Effect of Jag1 on blood commitment of isolated hemogenic precursors. Total CD45+ blood cells increased by 3.97-fold relative to controls following extended Hem-culture upon Jag1 stimulation. *P < .05 (n = 3). (J-K) Reduction of CD45+ blood commitment from isolated hemogenic precursors after HES1 knockdown relative to scrambled siRNA control; frequency (J, 64.9% reduction) and total blood cells (K, 82.6% reduction). *P < .05 (n = 3).
Effect of Notch signaling on the hematopoietic commitment of purified hemogenic precursors. (A-B) GSI-mediated inhibition of Notch signaling in dissociated single cells from unsorted total EBs resulted in an 89.3% reduction of CD45+ blood cell frequency (A) and immunocytochemical staining for CD45 on single cells after extended hematopoietic culture (Hem-culture) revealed lower numbers of CD45+ cells in the GSI-treated group (B). *P < .05 (n = 3). (C) Schematic summary to examine the effect of up- or downregulated Notch signaling on blood commitment of isolated hemogenic precursors. (D) Emergence of CD45+ blood cells could be modulated by controlling Notch signaling in isolated hemogenic precursors. GSI-mediated Notch downregulation inhibited blood commitment (5.38 ± 1.45%) over controls (28.2 ± 12%). Conversely, Jag1-stimulated hemogenic precursors enhanced blood augmentation (78.2 ± 18%). *P < .05; **P < .01 (n = 3). (E) GSI-mediated inhibition of Notch signaling on isolated hemogenic precursors resulted in a significant reduction of CD45+ blood cells by immunocytochemical staining. (F-G) Total number of hematopoietic colonies (F) and endothelial colonies (G) formed from isolated hemogenic precursors by modulating Notch signaling during extended Hem-culture. GSI-mediated Notch downregulation decreased the number of hematopoietic colonies (F, 4.66 ± 2.51), whereas it increased the number of endothelial colonies (G, 7.33 ± 1.52). On the contrary, upregulated Notch upon Jag1 treatment increased the number of hematopoietic colonies (F, 26.67 ± 4.73) but decreased endothelial colonies (G, 1.33 ± 0.58) compared with the number of hematopoietic (F, 14.67 ± 2.52) and endothelial colonies (G, 3 ± 1) in the control. *P < .05; **P < .01 (n = 3). (H) Effect of up- or downregulation of Notch on endothelial-specific marker expression was analyzed by flow cytometry. Expression level of PECAM1 and VE-cadherin was evaluated in isolated hemogenic precursors with activated or suppressed Notch signaling by Jag1 or GSI treatment, respectively. GSI-treated hemogenic precursors showed higher frequency of PECAM1+VE-cadherin+ (54.73 ± 6.17%) over controls (27.94 ± 7.59%) and Jag1 treatment (9.94 ± 3.82%). *P < .05; **P < .01 (n = 3). (I) Effect of Jag1 on blood commitment of isolated hemogenic precursors. Total CD45+ blood cells increased by 3.97-fold relative to controls following extended Hem-culture upon Jag1 stimulation. *P < .05 (n = 3). (J-K) Reduction of CD45+ blood commitment from isolated hemogenic precursors after HES1 knockdown relative to scrambled siRNA control; frequency (J, 64.9% reduction) and total blood cells (K, 82.6% reduction). *P < .05 (n = 3).
GSI-mediated Notch-signaling suppression impaired blood commitment, whereas Jag1 stimulation enhanced hematopoiesis of isolated hemogenic precursors during extended Hem-culture (Figure 5D-E). Activated Notch upon Jag1 stimulation enhanced hematopoietic colonies but decreased endothelial colonies (Figure 5F-G). Furthermore, expression of endothelial-specific markers, PECAM1 and VE-cadherin, was highly increased in GSI-mediated Notch suppression, whereas Jag1 decreased expression of endothelial markers (Figure 5H). A similar reduction of endothelial differentiation was found using retroviral HES1 overexpression (supplemental Figure 5A-B). Jag1 increased the total number of blood cells from isolated hemogenic precursors in response to Jag1-induced Notch activation (Figure 5I). Conversely, inhibition of Notch signaling through HES1 siRNA dramatically impaired the hematopoietic output from hemogenic precursors isolated from untreated hEBs relative to scrambled siRNA control (Figure 5J-K). These data demonstrate that active Notch signaling specifically regulates hemogenic precursors from hPSCs at the expense of endothelial precursors and provides definitive evidence that the Notch pathway is critical for the commitment phase of early human hematopoiesis.
Upon siRNA (Figure 6A) or retroviral shRNA-mediated (Figure 6B-C) HES1 knockdown, hematopoietic progenitor formation was reduced for all erythroid and myeloid progenitors (Figure 6D). The fate of hematopoietic progenitors was confirmed by cell surface expression of myeloid or erythroid lineage markers (data not shown). In contrast to the attenuation of hematopoietic progenitors, downregulation of HES1 increased endothelial cell capacity (Figure 6E-G). These endothelial progenitors were characterized by examining morphology, Dil-Ac-LDL uptake, staining of VE-cadherin, and functional endothelial tube formation assay (Figure 6H). The molecular signature of hematopoietic- and endothelial-specific genes was also investigated in the colonies upon Jag1 or GSI treatment (Figure 6I-L). Upon Jag1 stimulation, expression levels of Runx1 (Figure 6J) and TAL1 (Figure 6K) were highly increased in hematopoietic colonies compared with endothelial colonies that emerged following GSI treatment. Conversely, GSI-mediated Notch suppression induced higher expression of endothelial-specific genes KDR, VE-cadherin, and Tie2 vs Jag1-stimulated Notch activation (Figure 6L). Consistently, HES1 knockdown achieved by retroviral transduction (supplemental Figure 6A-C) induced endothelial commitment (supplemental Figure 6D). These data reveal that HES1-mediated Notch signaling specifies hematopoietic cell fate at the expense of endothelial progenitor commitment from bipotent hematopoietic and endothelial precursors.
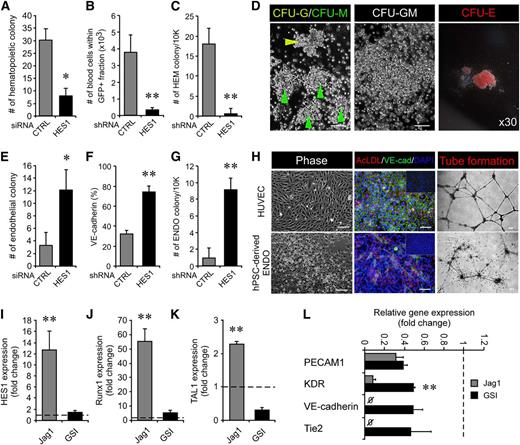
HES1 is a key modulator of commitment to the hematopoietic or endothelial fate during development. (A) HES1 knockdown achieved by siRNA in hemogenic precursors formed fewer hematopoietic colonies (8 ± 2.9 vs 30.3 ± 4.4). *P < .05 (n = 4). (B-C) Retroviral knockdown of HES1 impaired blood production (B, 0.36 ± 0.12 × 103 cells vs 3.85 ± 0.98 × 103 cells) and hematopoietic colony formation (C, 15.5 ± 1.29 vs 25.25 ± 3.59). **P < .01 (n = 4). (D) Colonies formed after HES1 siRNA knockdown represented typical hematopoietic colony subtypes, including granulocytes (CFU-G, yellow arrowhead in left panel), macrophages (CFU-M, green arrowhead in left panel), granulocyte macrophages (CFU-GM, middle panel), and erythrocytes (CFU-E, right panel). Scale bars represent 100 μm. (E) siRNA-mediated HES1 knockdown in hemogenic precursors increased the number of endothelial colonies (12.0 ± 3.2 vs 3.3 ± 2.1) relative to controls. *P < .05 (n = 4). (F-G) Retroviral HES1 suppression induced a higher level of endothelial-specific VE-cadherin expression (F, 74.3 ± 5.89% vs 31.73 ± 3.07%) and total number of endothelial colonies (G, 9.25 ± 1.26 vs 1 ± 1.15) than the control. **P < .01 (n = 4). (H) Characterization of hPSC-derived endothelial cells in HES1 knockdown cultures shown by morphology (bottom left panel), VE-cadherin staining with Dil-Ac-LDL uptake (bottom middle panel), and functional in vitro tube formation (bottom right panel) compared with HUVEC positive controls (upper row). Scale bars represent 100 μm (left and middle panels) and 250 μm (right panels). (I-K) Relative expression of HES1 (I) and hematopoietic markers, Runx1 (J) and TAL1 (K), in hematopoietic and endothelial colonies harvested from CFU assays upon Jag1 and GSI stimulation, respectively. Taken together with HES1 (I, 12.74 ± 3.39-fold), higher expression levels of Runx1 (J, 55.12 ± 8.88-fold) and TAL1 (K, 2.3 ± 0.05-fold) were observed in hematopoietic colonies vs endothelial colonies (1.44 ± 0.24-fold; F, 5.44 ± 1.01-fold; G, 0.3 ± 0.06-fold). HUVEC was used as a control. **P < .01 (n = 3). (L) Quantitative analysis of endothelial-specific gene expression relative to HUVEC demonstrating GSI-mediated Notch knockdown increased endothelial-specific KDR (0.49 ± 0.02-fold vs 0.08 ± 0.02-fold), VE-cadherin (0.46 ± 0.2-fold vs nondetected), and Tie2 (0.48 ± 0.1-fold vs nondetected) gene expression over Jag1 stimulation. Expression level of PECAM1 was similar in both GSI-treated (0.38 ± 0.03-fold) and Jag1-treated (0.31 ± 0.08-fold) cells.
HES1 is a key modulator of commitment to the hematopoietic or endothelial fate during development. (A) HES1 knockdown achieved by siRNA in hemogenic precursors formed fewer hematopoietic colonies (8 ± 2.9 vs 30.3 ± 4.4). *P < .05 (n = 4). (B-C) Retroviral knockdown of HES1 impaired blood production (B, 0.36 ± 0.12 × 103 cells vs 3.85 ± 0.98 × 103 cells) and hematopoietic colony formation (C, 15.5 ± 1.29 vs 25.25 ± 3.59). **P < .01 (n = 4). (D) Colonies formed after HES1 siRNA knockdown represented typical hematopoietic colony subtypes, including granulocytes (CFU-G, yellow arrowhead in left panel), macrophages (CFU-M, green arrowhead in left panel), granulocyte macrophages (CFU-GM, middle panel), and erythrocytes (CFU-E, right panel). Scale bars represent 100 μm. (E) siRNA-mediated HES1 knockdown in hemogenic precursors increased the number of endothelial colonies (12.0 ± 3.2 vs 3.3 ± 2.1) relative to controls. *P < .05 (n = 4). (F-G) Retroviral HES1 suppression induced a higher level of endothelial-specific VE-cadherin expression (F, 74.3 ± 5.89% vs 31.73 ± 3.07%) and total number of endothelial colonies (G, 9.25 ± 1.26 vs 1 ± 1.15) than the control. **P < .01 (n = 4). (H) Characterization of hPSC-derived endothelial cells in HES1 knockdown cultures shown by morphology (bottom left panel), VE-cadherin staining with Dil-Ac-LDL uptake (bottom middle panel), and functional in vitro tube formation (bottom right panel) compared with HUVEC positive controls (upper row). Scale bars represent 100 μm (left and middle panels) and 250 μm (right panels). (I-K) Relative expression of HES1 (I) and hematopoietic markers, Runx1 (J) and TAL1 (K), in hematopoietic and endothelial colonies harvested from CFU assays upon Jag1 and GSI stimulation, respectively. Taken together with HES1 (I, 12.74 ± 3.39-fold), higher expression levels of Runx1 (J, 55.12 ± 8.88-fold) and TAL1 (K, 2.3 ± 0.05-fold) were observed in hematopoietic colonies vs endothelial colonies (1.44 ± 0.24-fold; F, 5.44 ± 1.01-fold; G, 0.3 ± 0.06-fold). HUVEC was used as a control. **P < .01 (n = 3). (L) Quantitative analysis of endothelial-specific gene expression relative to HUVEC demonstrating GSI-mediated Notch knockdown increased endothelial-specific KDR (0.49 ± 0.02-fold vs 0.08 ± 0.02-fold), VE-cadherin (0.46 ± 0.2-fold vs nondetected), and Tie2 (0.48 ± 0.1-fold vs nondetected) gene expression over Jag1 stimulation. Expression level of PECAM1 was similar in both GSI-treated (0.38 ± 0.03-fold) and Jag1-treated (0.31 ± 0.08-fold) cells.
Notch regulation clonally governs hemato-endothelial lineages from hPSCs
Our results indicated that subsequent hematopoiesis from isolated bipotent hemogenic precursors is controlled by Notch regulation (Figure 5), whereas Notch inhibition blocked hematopoiesis but activated endothelial commitment of hPSCs by either ligand treatment or genetic modifications (Figure 6). To examine these effects on hemato-endothelial lineage specification at the clonal level, isolated single bipotent hemogenic precursors were differentiated toward either hematopoietic or endothelial lineages with activation or inhibition of Notch signaling upon Jag1 or GSI treatment. Immunocytochemical staining for CD45 and eNOS was used to distinguish hematopoietic, endothelial, and/or bipotent cell lineages. Individual wells were classified into hematopoietic (HEM), endothelial (ENDO), or bipotent cells that exhibit hemangioblastic properties (Bipotent) with CD45 and/or eNOS expression (Figure 7A). The majority of hematopoietic wells (HEM), mostly observed in Jag1 treatment, consisted of small and round cells expressing CD45 (left panels), whereas endothelial wells (ENDO) consisted of larger cells expressing eNOS (middle panels) largely upon GSI treatment. Additionally, wells having a similar ratio of CD45 and eNOS were defined as bipotent (right panels) and were reduced by GSI treatment (Figure 7B). The frequency of individual precursors exhibiting an endothelial fate was increased upon GSI-induced Notch inhibition (Figure 7D), whereas individual hPSC-derived cells displayed an increase in hematopoietic potential when Notch is activated via addition to Jag1 (Figure 7E) compared with basal conditions (Figure 7C). Taken together, these data indicate that Notch is a key regulator of hemato-endothelial balance operating at the single-cell level.
Notch signaling clonally controls the hematopoietic vs endothelial lineage specification of hPSC-derived hemogenic precursors and proposed model for hemato-endothelial lineage specification of hPSCs via HES1. (A) Schematic for the experiment performed to determine the role of Notch signaling in hematopoietic vs endothelial lineage specification of isolated hemogenic precursors at the clonal level. Isolated hemogenic precursors were directly dispensed into 96-well plates and then underwent hemato-endothelial specification in the presence or absence of GSI or Jag1. The specified cells were distinguished by immunocytochemical staining with CD45 and eNOS. The number of wells having hematopoietic, endothelial, or bipotent cells was quantified on a plate reader. (B) Morphologic and phenotypic distinctions among hematopoietic, endothelial, and bipotent cells specified from single hemogenic precursors upon GSI or Jag1 treatment. Each well was counted as hematopoietic (HEM), endothelial (ENDO), or bipotent (Bipotent) cells depending on whether the majority of the cells in the wells were positive for CD45+ (HEM), eNOS+ (ENDO), or CD45+/eNOS+ (Bipotent). Scale bars represent 100 μm. White arrowhead, CD45+ cell; yellow arrowhead, eNOS+ cell. (C-E) Lineage specification of isolated single precursors upon Notch inhibition (GSI, D) or Notch activation (Jag1, E) compared with endogenous developmental potential (Basal, C). Frequencies of wells positive for CD45+ (HEM), eNOS+ (ENDO), and CD45+/eNOS+ (Bipotent) were significantly different in GSI (D) or Jag1 (E) treatments compared with controls. *P < .05; **P < .01 (n = 3). (F) The proposed model suggests that HES1, the downstream effector of Notch signaling, is a key regulator of hemogenic specification and subsequent commitment of hemogenic precursors to hematopoietic or endothelial lineages in hPSCs.
Notch signaling clonally controls the hematopoietic vs endothelial lineage specification of hPSC-derived hemogenic precursors and proposed model for hemato-endothelial lineage specification of hPSCs via HES1. (A) Schematic for the experiment performed to determine the role of Notch signaling in hematopoietic vs endothelial lineage specification of isolated hemogenic precursors at the clonal level. Isolated hemogenic precursors were directly dispensed into 96-well plates and then underwent hemato-endothelial specification in the presence or absence of GSI or Jag1. The specified cells were distinguished by immunocytochemical staining with CD45 and eNOS. The number of wells having hematopoietic, endothelial, or bipotent cells was quantified on a plate reader. (B) Morphologic and phenotypic distinctions among hematopoietic, endothelial, and bipotent cells specified from single hemogenic precursors upon GSI or Jag1 treatment. Each well was counted as hematopoietic (HEM), endothelial (ENDO), or bipotent (Bipotent) cells depending on whether the majority of the cells in the wells were positive for CD45+ (HEM), eNOS+ (ENDO), or CD45+/eNOS+ (Bipotent). Scale bars represent 100 μm. White arrowhead, CD45+ cell; yellow arrowhead, eNOS+ cell. (C-E) Lineage specification of isolated single precursors upon Notch inhibition (GSI, D) or Notch activation (Jag1, E) compared with endogenous developmental potential (Basal, C). Frequencies of wells positive for CD45+ (HEM), eNOS+ (ENDO), and CD45+/eNOS+ (Bipotent) were significantly different in GSI (D) or Jag1 (E) treatments compared with controls. *P < .05; **P < .01 (n = 3). (F) The proposed model suggests that HES1, the downstream effector of Notch signaling, is a key regulator of hemogenic specification and subsequent commitment of hemogenic precursors to hematopoietic or endothelial lineages in hPSCs.
Discussion
In vitro differentiation of hPSCs provides a useful surrogate for understanding the cellular and molecular mechanisms that govern lineage specification in the developing human embryo. Although Notch signaling has been implicated in a variety of human hematopoietic diseases and disorders,35-38 its role in early human development is largely unknown. Our current study identifies a critical role for Notch signaling during early human hematopoiesis and specifies a temporal window by which Notch signaling dictates blood differentiation at the expense of endothelial commitment (Figure 7F), as demonstrated by Notch ligand effects, gain- and loss-of-function approaches, and integration at the clonal level.
Although this report is the first to reveal a role for Notch signaling in early human embryonic hematopoiesis, similar mechanisms have been shown to regulate hematopoietic differentiation in other species. For example, gene deletion studies in both zebra fish and mice have demonstrated a potential role for Notch signaling in hematopoietic and endothelial development.4-6,8,9,25,39 In addition, both Notch1 and Jagged1 are implicated in the induction of definitive over primitive hematopoiesis.4,16,25 Notch1-deficient mouse embryonic stem cells contribute to primitive yolk-sac hematopoiesis in chimeric mice but are incapable of adult blood development,4 whereas Jagged1 is required for the establishment of definitive mouse hematopoietic precursors.25
Controlling lineage specification from hPSCs remains a primary goal of regenerative medicine research; however, few reports have used exogenous factors to control cell fate commitment to one lineage over another. Similar to the previously proposed positive feedback loop involving Notch signaling in hematopoietic stem cells,40 our study reveals that Notch1 activation by Jag1 augments hematopoietic development from both embryonic- and adult-induced forms of hPSCs and that signaling via HES1 plays a critical role in early human hematopoietic vs endothelial cell commitment (Figure 7F). In particular, direct manipulation of isolated hemogenic precursors showed that Notch signaling represents a molecular toggle switch between endothelial and hematopoietic cell fate choices from hPSC-derived precursors (Figure 7). Notably, similar results were obtained for both hESC-derived and hFib-iPSC–derived hematopoiesis suggesting conservation among hPSCs states. This is important as adult cells likely represent the more attractive and easily accessible cell source for generation of sufficient hematopoietic cell numbers to be used clinically.
Consistent with previous reports,16,33 the Notch pathway appeared dormant in hPSC cultures. However, stimulation of the pathway with Jagged1 induced Notch signaling during hematopoietic EB differentiation. This suggests that, despite its dormancy at steady state in undifferentiated hPSCs, the Notch-signaling pathway remains accessible to hPSCs for the purpose of controlling lineage development. We propose that dormancy of the Notch pathway in hPSCs may constitute a mechanism to sustain the pluripotent state by preventing activation of pathways related to hemato-endothelial lineage progression. Accordingly, our report provides a rare example of how stages of cell fate specification during hPSC differentiation can be controlled using exogenously soluble ligands. Our study validates the use of hPSCs to investigate early events in human blood specification. This will aid in our understanding of disorders affecting endothelial-hematopoietic tissues in adults where patient-specific hFib-iPSCs can be derived and used as disease models.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Kathy Xu, Marilyne Levadoux-Martin, Monica Graham, and Stacey Dozono for technical assistance with staining, flow cytometry, culture of cells, and experimental assistance, respectively.
This work was supported by a grant from the Canadian Cancer Society Research Institute (M.B.). M.B. holds a Canada Research Chair in human stem cell biology.
Authorship
Contribution: J.B.L. designed the research, performed experiments, and wrote the manuscript; T.E.W.-O., J.-H.L., A.S., and S.-H.H. performed experiments; B.A.S.M., I.-H.P., I.D.B., and G.Q.D. discussed the paper; and M.B. conceived and designed the research, supervised the whole project, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mickie Bhatia, Stem Cell and Cancer Research Institute (SCC-RI), MDCL 5029, Michael G. DeGroote School of Medicine, McMaster University, 1200 Main St West, Hamilton, ON, Canada, L8N 3Z5; e-mail: mbhatia@mcmaster.ca.

![Figure 1. Notch signaling is involved in the hematopoietic development of hFib-hPSCs. (A) Notch ligand immobilization augmented hematopoietic differentiation of hFib-hESCs. EBs treated with Matrigel immobilized with soluble Jagged1 (sJag1) or Delta1 (sDll1) during phase 2 of EB development had increased hematopoietic CD45+ blood development. *P < .05 (n = 3). (B-C) HES1 expression was upregulated by Jag1 and Dll1 phase 2 treatment in developing EBs formed from hESCs (B) and hFib-iPSCs (C) compared with IgG-treated control EBs (CTRL). However, control EBs showed higher HES1 expression than undifferentiated hESCs (B) and hFib-iPSCs (C). **P < .01; *P < .05 (n = 3). (D) Time course expression of Notch1, Notch2, and HES1 during hematopoietic EB development. The expression level of Notch1 and HES1 reached steady state by day 6 and then increased gradually and peaked at day 15. However, only minor changes were observed in Notch2 expression. Two different periods, phase 1 (0-5 days) and phase 2 (6-15 days), were identified based on the expression pattern of Notch signals and previous studies. (E) Notch1-mediated signaling regulated blood development of hESCs. Either the transient (phase 1 [0-5 days] or phase 2 [6-15 days]) or continuous (phases 1 and 2 [0-15 days]) suppression of Notch1 (N1) but not Notch2 (N2) by siRNA treatment decreased blood development relative to control scrambled siRNA treatment. *P < .05 (n = 3). (F-I) Hematopoietic development of EBs is regulated by the status of Notch signaling. Notch-signaling activation by adding Jag1 (F,H) or Dll1 (G,I) to developing EBs results in augmented blood differentiation (F-G). Increased or decreased blood formation coincided with up- or downregulation of HES1 (H-I) by either ligand treatment or Notch1 knockdown (siNotch1). Hematopoiesis and Notch signaling could be recovered in only phase 1 Notch1 knockdown EBs by adding Jag1 (F,H) or Dll1 (G,I), whereas neither the hematopoiesis (F-G) nor the expression of HES1 (H-I) could be recovered after long-term (phase 2 only or phases 1 and 2) suppression of Notch1 (+). **P < .01; *P < .05 (n = 3).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/7/10.1182_blood-2012-12-471649/4/m_1162f1.jpeg?Expires=1769168849&Signature=gnaEznClCTp0Ao1QwWv6V2MmgH-QzY~A98-7Ruy7-W6p3hNx~an5z-k1cUSzKrHbY8bbOtMAkHuOIufOlc1gtgbvvm0VpL8nRNmIICQo5dYixBsfnujicUITFlUBHpNvn4B~9HXHmUhqxKMCKSl9T6MM2ANiIgdJWzQz4D5AeD4rbVzmP~cbuGUXz6Mv2A9oLCiaH0-wQnTkMLga2P4kkU9lt-M96IS2ztk0AfOyvoXOxoyvNYBjDVuaIEe6a6aT4iNPhcHeuCTZ4TAXdDVwSjcZ9qu3JzWIAd585KA71JXGJhnRcE-7TmSt1kdo-zhVWzJGk4ygzsoE6Za52NmCNg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal