Key Points
EPZ-5676 is a potent DOT1L inhibitor that causes tumor regressions in a rat xenograft model of MLL-rearranged leukemia.
Abstract
Rearrangements of the MLL gene define a genetically distinct subset of acute leukemias with poor prognosis. Current treatment options are of limited effectiveness; thus, there is a pressing need for new therapies for this disease. Genetic and small molecule inhibitor studies have demonstrated that the histone methyltransferase DOT1L is required for the development and maintenance of MLL-rearranged leukemia in model systems. Here we describe the characterization of EPZ-5676, a potent and selective aminonucleoside inhibitor of DOT1L histone methyltransferase activity. The compound has an inhibition constant value of 80 pM, and demonstrates 37 000-fold selectivity over all other methyltransferases tested. In cellular studies, EPZ-5676 inhibited H3K79 methylation and MLL-fusion target gene expression and demonstrated potent cell killing that was selective for acute leukemia lines bearing MLL translocations. Continuous IV infusion of EPZ-5676 in a rat xenograft model of MLL-rearranged leukemia caused complete tumor regressions that were sustained well beyond the compound infusion period with no significant weight loss or signs of toxicity. EPZ-5676 is therefore a potential treatment of MLL-rearranged leukemia and is under clinical investigation.
Introduction
Rearrangements in the MLL gene at position 11q23 occur in 5% to 10% of acute leukemias of lymphoid, myeloid, or mixed/indeterminant lineage and are especially common in infant acute leukemias and in secondary acute myeloid leukemias arising in patients following treatment of other malignancies with topoisomerase II inhibitors.1-4 Acute leukemias bearing MLL rearrangements are aggressive diseases. Current treatment options are limited to chemotherapy and allogeneic hematopoietic stem cell transplantation; however, these have significant side effects and outcomes remain poor. As a result, there is intense interest in developing novel therapeutic strategies for this disease. The MLL gene encodes a large multidomain protein (MLL) that regulates transcription of developmental genes including the HOX genes.1 The amino terminal portion of the protein contains regions that target MLL to DNA directly, whereas the carboxyl terminal portion of the protein contains a Su(Var)3-9, Enhancer of zeste and Trithorax domain with methyltransferase activity specific for lysine 4 of histone H3 (H3K4).5-9 MLL rearrangements result in the loss of the carboxy-terminal methyltransferase domain and an in-frame fusion of the amino-terminal region of MLL to 1 of more than 60 potential fusion partners.1-3 The vast majority of translocations result in oncogenic fusion proteins in which the native methyltransferase domain is replaced by sequences derived from AF4, AF9, AF10, and ENL, which interact with DOT1L directly or indirectly in complexes that promote transcriptional elongation.10-18 DOT1L is a histone methyltransferase enzyme that targets lysine 79 in the globular domain of histone H3 (H3K79) for mono-, di-, or trimethylation (H3K79me1, me2, or me3).19,20 As a result, MLL-fusion proteins gain the ability to recruit DOT1L to MLL target genes where the resulting hypermethylation at H3K79 leads to aberrant expression of a characteristic set of genes including HOXA9 and MEIS1 that drive leukemogenesis.14,15,21-27 Several recent studies have used genetic ablation or small molecule inhibitors to demonstrate that DOT1L methyltransferase activity is required for MLL-fusion–mediated leukemogenesis in preclinical models of MLL-rearranged leukemia.15,21,26,28-33 Overall, these studies have established pharmacological inhibition of DOT1L enzymatic activity as a promising therapeutic strategy for the treatment of MLL-rearranged leukemias. We recently developed EPZ004777, a small molecule inhibitor of DOT1L H3K79 methyltransferase activity that demonstrates selective killing of MLL-rearranged leukemia cells in culture and prolonged survival in a mouse model of MLL-rearranged leukemia.29-31 Although this molecule established the feasibility of developing potent selective DOT1L inhibitors as therapies for MLL-rearranged leukemia, the pharmacokinetic properties of EPZ004777 limit its effectiveness in vivo and render it unsuitable for clinical development. Here we report the identification of EPZ-5676, a DOT1L inhibitor with improved potency and drug-like properties that has recently entered clinical evaluation as a therapy for MLL-rearranged leukemia. We describe the characterization of the EPZ-5676 with respect to its inhibitory activity in enzymatic assays, its interaction with DOT1L protein and its pharmacologic, pharmacokinetic, and pharmacodynamic activity in preclinical models of MLL-rearranged leukemia.
Materials and methods
Reagents and cell lines
EPZ-5676 was synthesized by Epizyme. Stock solutions (50 or 10 mM) were prepared in dimethylsulfoxide (DMSO) and stored at −20°C. Human leukemia cell lines MV4-11 (CRL-9591), RS4;11 (CRL-1873), Kasumi-1 (CRL-2724), HL-60 (CCL-240), and Jurkat (TIB-152) were obtained from the ATCC. SEM (ACC 546), Molm-13 (ACC 554), NOMO-1 (ACC 542), KOPN-8 (ACC 552), REH (ACC 22), and 697 (ACC 42) were obtained from the DSMZ. All cell lines were grown in the recommended cell culture media at 37°C in 5% CO2.
Biochemical enzyme inhibition assays and X-ray crystal structure determination.
Biochemical enzyme inhibition assays were performed as previously described.30 The enzyme inhibition constant (Ki) value for EPZ-5676 was determined by fitting inhibition data to the Morrison quadratic equation.34 Residence times for EPZ-5676 and EPZ004777 were calculated as the reciprocal of the enzymatic-ligand dissociation rate, determined by surface plasmon resonance using methods described previously.35 The X-ray crystal structure of EPZ-5676 in complex with the human DOT1L methyltransferase domain was determined using methods previously described.35 Atomic coordinates and structure factors for the EPZ-5676:DOT1L crystal structure have been deposited in the Protein Data Bank (accession number 4HRA).
Immunoblot analysis of inhibition of histone methylation by EPZ-5676
To analyze inhibition of cellular H3K79 methylation by EPZ-5676, exponentially growing MV4-11 cells were incubated in the presence of 0.2% DMSO or increasing concentrations of EPZ-5676 for 96 hours. Cells were processed and histones extracted and analyzed by immunoblotting with antibodies to H3K79me2 or total H3 as previously described.30 To analyze the selectivity of inhibition of H3K79 methylation in MV4-11 cells following treatment with 1 µM EPZ-5676, histones were immunoblotted with a panel of methyl-specific antibodies as described in supplemental Methods (available on the Blood website). To analyze inhibition of H3K79 methylation in peripheral blood mononuclear cells (PBMCs) from rats dosed with EPZ-5676, 20 μL of PBMC whole cell lysate was immunoblotted with antibodies to H3K79me2 or total H3 as described in supplemental Methods.
ELISA analysis of inhibition of H3K79me2 by EPZ-5676
H3K79me2 and total histone H3 levels in histone samples from cell culture and in vivo experiments were quantified in H3K79me2 and total histone H3 enzyme-linked immunosorbent assays (ELISA), respectively. A detailed description of each assay can be found in supplemental Methods. H3K79me2 values were normalized to total histone H3 in each sample by dividing the H3K79me2 ELISA optical density reading by the total H3 optical density reading for the same sample. Cell treatments before histone extraction and ELISA analysis were as follows. For the determination of half-maximal inhibitory concentration (IC50) values for EPZ-5676 inhibition of cellular H3K79 methylation, exponentially growing cells were plated in 6-well plates at 2 × 105 cells/mL and incubated in the presence of 0.2% DMSO or EPZ-5676 for 96 hours. Histones were extracted as previously described.30 For kinetic analysis of H3K79me2 levels following EPZ-5676 compound addition or removal, exponentially growing MV4-11 cells were seeded in a 75-cm2 culture flask at 2 × 105 cells/mL and incubated in the presence of 1 µM EPZ-5676 for up to 7 days. A total of 1 to 2 × 106 cells were harvested at the appropriate time point and histones were extracted as previously described.30 For the washout component of the experiment, cells were incubated with 1 µM EPZ-5676 for 4 days, washed twice with phosphate-buffered saline, and resuspended in media free of compound. Cells (1–2 × 106) were then harvested daily for 7 days and histones were extracted as previously described.30 To assess inhibition of H3K79me2 in xenograft tumor and bone marrow tissue from rats dosed with EPZ-5676, tissues were harvested and histones extracted as described in supplemental Methods.
Quantitative real-time polymerase chain reaction (qRT-PCR).
To assess inhibition of HOXA9 and MEIS1 messenger RNA (mRNA) expression by EPZ-5676, exponentially growing MV4-11 cells were seeded in 75-cm2 culture flasks at 2 × 105 cells/mL and incubated in the presence of 0.2% DMSO or increasing concentrations of EPZ-5676. On day 4, cells were maintained in log phase culture by reseeding at 5 × 105 cells/mL and compound was replenished. On day 6, cells were harvested, total RNA extracted, and HOXA9 and MEIS1 mRNA levels assessed and normalized to the β2-microglobulin by qRT-PCR as previously described.30 To assess inhibition of HOXA9 and MEIS1 mRNA expression in xenograft tumor tissue from rats dosed with EPZ-5676, tumors were collected and total RNA isolated as described in supplemental Methods.
Cell proliferation assays.
Proliferation assays and IC50 value determinations were performed as described previously.30
Cell-cycle analysis and Annexin staining.
Exponentially growing MV4-11 cells were incubated with 1 μM EPZ-5676 for up to 10 days. Cell treatments and flow cytometric analysis of DNA content and Annexin V staining was performed as previously described.30
Pharmacokinetic studies
A detailed description of the pharmacokinetic studies can be found in supplemental Methods.
Rat MV4-11 xenograft studies
In vivo studies were conducted after review by the appropriate animal care and use committee at Charles River Discovery Research Services (Durham, NC) and Oncodesign S.A. (Dijon, France). A detailed description of the rat MV4-11 xenograft studies is provided in supplemental Methods. Briefly, MV4-11 cells were implanted subcutaneously in female athymic nude rats (rnu/rnu; Harlan). EPZ-5676 was delivered by IV infusion into the femoral vein continuously (24 hours/day), or intermittently (8 hours/day). A control group received infusions of the vehicle, 5% hydroxypropyl-β-cyclodextrin in saline. Animals were weighed and tumors calipered twice weekly until the end of the study. Each test animal was killed when its neoplasm reached the predetermined end-point volume or on the last day of the study, whichever came first. A detailed description of tissue isolation and processing for PBMC whole cell lysates, tumor and bone marrow histones, and tumor RNA is provided in supplemental Methods.
Results
EPZ-5676 is a potent and selective DOT1L inhibitor
EPZ-5676 was the result of structure-guided design and optimization of a series of aminonucleoside compounds that includes the previously described DOT1L inhibitor EPZ004777.30,35 The chemical structure of EPZ-5676 is shown in Figure 1A. The X-ray crystal structure of EPZ-5676 in complex with the human DOT1L methyltransferase domain (Figure 1B; supplemental Figure 1) reveals that EPZ-5676 occupies the S-adenosyl methionine (SAM) binding pocket and induces conformational changes in DOT1L leading to the opening of a hydrophobic pocket beyond the amino acid portion of SAM as described for other members of this series.33,35 This structural rearrangement creates additional interaction surfaces that contribute to high-affinity binding and selectivity of EPZ-5676.
Structure, binding, and inhibitory activity of EPZ-5676. (A) Chemical structure of EPZ-5676. (B) Superposition of DOT1L-EPZ-5676 (green) and DOT1L-SAM (gray; Protein Data Base accession number 3QOW). To accommodate the extended hydrophobic tail of the inhibitor, significant rearrangement of the protein is required, including the loop between β-strands 10 and 11 (L10-11). Details on the interactions between the protein and compound and the induced changes caused by EPZ-5676 binding are shown in supplemental Figure 1. (C) Selectivity profile of EPZ-5676 inhibitory activity against representative members of the lysine (left) and arginine (right) enzyme families. The diameter of the sphere for each enzyme is directly related to the magnitude of inhibition by EPZ-5676. Larger circles correlate to increased potency; gray circles indicate no measurable inhibition up to 10 µM of EPZ-5676. (D) Comparison of EPZ-5676 and EPZ004777 potency, selectivity, and cell-based activity. The enzyme inhibition Ki values for EPZ004777 and EPZ-5676 in DOT1L enzymatic assays are listed (n = 3; mean values ± SD are shown). Residence time for each compound is listed and was calculated as the reciprocal of the enzymatic-ligand dissociation rate as determined by surface plasmon resonance. Also listed are inhibitory activities for both compounds in MV4-11 proliferation assays (n = 3 [EPZ-5676] or n = 2 [EPZ004777]; mean values ± SD are shown), MV4-11 cell H3K79me2 ELISA assays (n = 2; mean values ± SD are shown) and MV4-11 cell HOXA9 and MEIS1 qRT-PCR assays (n = 2; mean values ± SD are shown).
Structure, binding, and inhibitory activity of EPZ-5676. (A) Chemical structure of EPZ-5676. (B) Superposition of DOT1L-EPZ-5676 (green) and DOT1L-SAM (gray; Protein Data Base accession number 3QOW). To accommodate the extended hydrophobic tail of the inhibitor, significant rearrangement of the protein is required, including the loop between β-strands 10 and 11 (L10-11). Details on the interactions between the protein and compound and the induced changes caused by EPZ-5676 binding are shown in supplemental Figure 1. (C) Selectivity profile of EPZ-5676 inhibitory activity against representative members of the lysine (left) and arginine (right) enzyme families. The diameter of the sphere for each enzyme is directly related to the magnitude of inhibition by EPZ-5676. Larger circles correlate to increased potency; gray circles indicate no measurable inhibition up to 10 µM of EPZ-5676. (D) Comparison of EPZ-5676 and EPZ004777 potency, selectivity, and cell-based activity. The enzyme inhibition Ki values for EPZ004777 and EPZ-5676 in DOT1L enzymatic assays are listed (n = 3; mean values ± SD are shown). Residence time for each compound is listed and was calculated as the reciprocal of the enzymatic-ligand dissociation rate as determined by surface plasmon resonance. Also listed are inhibitory activities for both compounds in MV4-11 proliferation assays (n = 3 [EPZ-5676] or n = 2 [EPZ004777]; mean values ± SD are shown), MV4-11 cell H3K79me2 ELISA assays (n = 2; mean values ± SD are shown) and MV4-11 cell HOXA9 and MEIS1 qRT-PCR assays (n = 2; mean values ± SD are shown).
EPZ-5676 is superior to previously described inhibitors of DOT1L, including EPZ004777,30,33,35-38 by a number of biochemical and pharmacologic measures (Figure 1C-D). For example, we tested the ability of EPZ-5676 to inhibit the enzymatic activity of DOT1L and a panel of 15 additional human lysine and arginine methyltransferases. EPZ-5676 inhibits DOT1L enzyme activity with a Ki of ≤0.08 nM and demonstrates a much-extended drug-target residence time relative to EPZ004777 (Figure 1D). As illustrated in the compound potency map, EPZ-5676 has > 37 000-fold selectivity against all of the tested protein methyltransferases (Figure 1C-D; supplemental Table 1).
EPZ-5676 inhibits cellular H3K79 methylation and MLL-fusion target gene expression
DOT1L methylates lysine 79 of histone H3 (H3K79); this activity is required for aberrant expression of MLL-fusion target genes such as HOXA9 and MEIS1.20-23,25,26,28-30,32 Incubation of the MLL-AF4 expressing acute leukemia cell line MV4-11 in the presence of increasing concentrations of EPZ-5676 led to a concentration-dependent decrease in global cellular methylated H3K79 levels as measured by immunoblot analysis of extracted histones with an antibody specific for dimethylated H3K79 (H3K79me2) (Figure 2A).39,40 A quantitative H3K79me2 ELISA assay was developed and used to derive IC50 values for H3K79me2 inhibition by EPZ-5676 of 3 nM and 5 nM in MV4-11 and HL60 (non–MLL-rearranged) cells, respectively (Figure 2B). Consistent with its exquisite biochemical selectivity, EPZ-5676 treatment had no effect on any of the other histone lysine and arginine methylation sites examined (Figure 2C). Cellular H3K79me2 levels declined exponentially with time after EPZ-5676 treatment with a half-life (t1/2) of 1.0 days; thus, 3 to 4 days of treatment are required to effect ≥90% inhibition of this mark (Figure 2D). Prior kinetic studies have demonstrated that the turnover rate of methylated H3K79 (t1/2 ∼1.1 to 1.8 days) is similar to that of histone H3 itself (t1/2 ∼1.3 days).41,42 MV4-11 cells have a doubling time of 24 hours; therefore, these results are consistent with disappearance of H3K79me2 because of dilution of existing histones by newly synthesized unmethylated histones upon cell division. Additional time course experiments in which recovery of H3K79 methylation was monitored following EPZ-5676 removal revealed that 7 days are required for H3K79 methylation to return to pretreatment levels (Figure 2D). This includes an initial 3-day lag phase in which no increase in H3K79me2 levels was observed; this may reflect the extended drug-target residence time43 for EPZ-5676 on DOT1L that was observed in biochemical assays (Figure 1D). Similar lag phases in recovery of intracellular protein phosphorylation have been observed with kinase inhibitors of long residence time.44 These results suggest that reduced global H3K79 methylation can be used as a pharmacodynamic readout for DOT1L inhibitor activity in vivo, but that there is likely to be a lag time of several days between exposure to the inhibitor and full pharmacodynamic response. We also demonstrated that EPZ-5676 incubation caused a concentration-dependent decrease in HOXA9 and MEIS1 mRNA levels consistent with previously described gene expression changes upon small molecule DOT1L inhibition in MLL-rearranged cells (Figure 2E).29-31 These data were used to derive IC50 values for inhibition of HOXA9 and MEIS1 mRNA expression of 67 nM and 53 nM, respectively (Figure 1D). Time course experiments revealed that full depletion of HOXA9 and MEIS1 mRNA took approximately 8 days (Figure 2E). This lag in timing relative to H3K79me2 depletion is consistent with H3K79 demethylation being a prerequisite for decreased expression of MLL-fusion target genes.
Inhibition of histone methylation and MLL-fusion target gene expression by EPZ-5676. (A) Immunoblot analysis of H3K79me2 levels in histones extracted from MV4-11 cells treated for 4 days with increasing concentrations of EPZ-5676. (B) Concentration-dependent inhibition of H3K79 methylation in MV4-11 (left) and HL60 (right) cells following 4-day EPZ-5676 treatment as measured by quantitative ELISA assay for H3K79me2. (C) Immunoblot analysis of histones extracted from MV4-11 cells treated for 4 days with either 1 µM EPZ-5676 or vehicle control and probed with a panel of specific methyl-lysine and methyl-arginine antibodies. Total H3 and H4 antibodies were used as the loading controls. (D) Time course of cellular H3K79me2 depletion in MV4-11 cells incubated in the presence of 1 µM EPZ-5676 (left). The diminution of intracellular H3K79me2 as a function of time after dosing with EPZ-5676 was fit to a simple exponential decay function that included a non-zero limit at infinite time, as described.39 Time course of recovery of H3K79me2 levels in MV4-11 cells upon removal of EPZ-5676 from the culture medium following a 4-day incubation (right). In both plots, H3K79me2 levels are normalized to total H3 and expressed as a percent of DMSO-treated (control) cells at each time point. Recovery of intracellular H3K79me2 levels following compound washout displayed a significant lag phase before semilinear recovery. These data were fit to a modified version of the expolinear function for crop growth as a function of photon interception and leaf area.40 The amount of H3K79me2 at any given time after compound washout (y) was fit as y = ymin +[(v/a)ln{1 + exp(a(t-tlag))}], where ymin is the non-zero limit of H3K79me2 at the start of the washout experiment, v is the velocity of the linear phase of recovery, and a is a constant of proportionality for our purposes. (E) Concentration dependent inhibition of HOXA9 and MEIS1 transcription by EPZ-5676 as measured by qRT-PCR in MV4-11 cells following 6 days of treatment (left; n = 2, mean ± SD are shown). Time course of inhibition of HOXA9 and MEIS1 mRNA expression as measured by qRT-PCR in MV4-11 cells incubated in the presence of 1 µM EPZ-5676 (right). Relative mRNA expression is plotted as a percentage of those at day 0 (n = 2; mean values ± SD are shown).
Inhibition of histone methylation and MLL-fusion target gene expression by EPZ-5676. (A) Immunoblot analysis of H3K79me2 levels in histones extracted from MV4-11 cells treated for 4 days with increasing concentrations of EPZ-5676. (B) Concentration-dependent inhibition of H3K79 methylation in MV4-11 (left) and HL60 (right) cells following 4-day EPZ-5676 treatment as measured by quantitative ELISA assay for H3K79me2. (C) Immunoblot analysis of histones extracted from MV4-11 cells treated for 4 days with either 1 µM EPZ-5676 or vehicle control and probed with a panel of specific methyl-lysine and methyl-arginine antibodies. Total H3 and H4 antibodies were used as the loading controls. (D) Time course of cellular H3K79me2 depletion in MV4-11 cells incubated in the presence of 1 µM EPZ-5676 (left). The diminution of intracellular H3K79me2 as a function of time after dosing with EPZ-5676 was fit to a simple exponential decay function that included a non-zero limit at infinite time, as described.39 Time course of recovery of H3K79me2 levels in MV4-11 cells upon removal of EPZ-5676 from the culture medium following a 4-day incubation (right). In both plots, H3K79me2 levels are normalized to total H3 and expressed as a percent of DMSO-treated (control) cells at each time point. Recovery of intracellular H3K79me2 levels following compound washout displayed a significant lag phase before semilinear recovery. These data were fit to a modified version of the expolinear function for crop growth as a function of photon interception and leaf area.40 The amount of H3K79me2 at any given time after compound washout (y) was fit as y = ymin +[(v/a)ln{1 + exp(a(t-tlag))}], where ymin is the non-zero limit of H3K79me2 at the start of the washout experiment, v is the velocity of the linear phase of recovery, and a is a constant of proportionality for our purposes. (E) Concentration dependent inhibition of HOXA9 and MEIS1 transcription by EPZ-5676 as measured by qRT-PCR in MV4-11 cells following 6 days of treatment (left; n = 2, mean ± SD are shown). Time course of inhibition of HOXA9 and MEIS1 mRNA expression as measured by qRT-PCR in MV4-11 cells incubated in the presence of 1 µM EPZ-5676 (right). Relative mRNA expression is plotted as a percentage of those at day 0 (n = 2; mean values ± SD are shown).
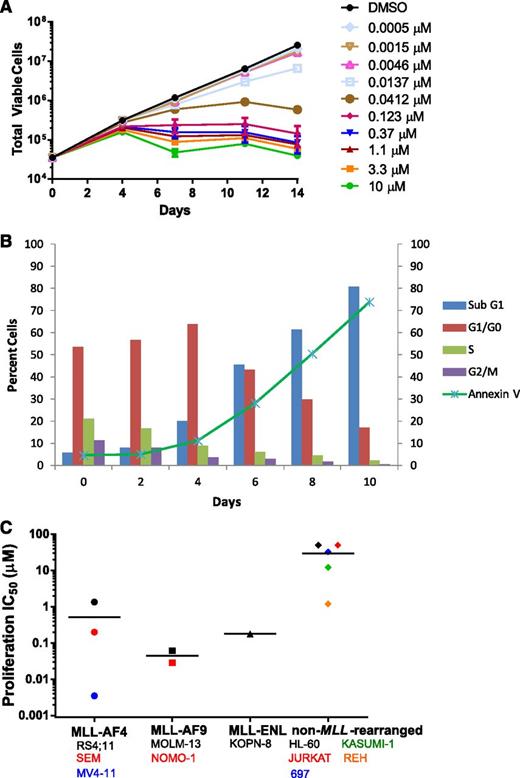
EPZ-5676 selectively inhibits proliferation of MLL-rearranged leukemia cells
EPZ-5676 is a potent inhibitor of MV4-11 proliferation with an IC50 value of 3.5 nM following 14 days of incubation (Figures 1D and 3A,C). Antiproliferative activity was realized after 4 days and was most clear after 7 days of EPZ-5676 treatment (Figure 3A). This delayed effect on proliferation is similar to that previously described for small-molecule DOT1L inhibition and is consistent with a mechanism involving depletion of cellular H3K79 methylation followed by inhibition of MLL-fusion target gene expression and a reversal in the leukemogenic gene expression program.29-31 We examined the mechanism of EPZ-5676–mediated cell killing using flow cytometry for DNA content and Annexin V staining to monitor the effects of EPZ-5676 on cell cycle and apoptosis over 10 days. As shown in Figure 3B, EPZ-5676 treatment led to an increase in the percentage of cells in the G0/G1 phase of the cell cycle and a decrease in the percentage of cells in S-phase cells over the first 4 days. This was followed by an increase in sub-G1 and Annexin V–positive cells indicative of cell death by apoptosis over the following 6 days.
Selective inhibition of MLL-rearranged cell proliferation by EPZ-5676. (A) Inhibition of the proliferation of MV4-11 cells following 14-day treatment with the indicated concentrations of EPZ-5676 (n = 3, mean ± SD are shown). (B) Effect of treatment of MV4-11 cells with 1 μM EPZ-5676 on cell-cycle phase and apoptosis over time as measured by staining for DNA content and Annexin V. (C) EPZ-5676 selectively kills acute leukemia cell lines harboring MLL fusions. IC50 values for inhibition of proliferation are plotted for each cell line following 14 days of exposure to increasing concentrations of EPZ-5676 (n = 2, mean value is shown, apart from MV4-11, where n = 3). Cell lines are grouped according to MLL status and horizontal lines represent the mean IC50 value for each group. IC50 values are also listed along with IC90 values in supplemental Table 2.
Selective inhibition of MLL-rearranged cell proliferation by EPZ-5676. (A) Inhibition of the proliferation of MV4-11 cells following 14-day treatment with the indicated concentrations of EPZ-5676 (n = 3, mean ± SD are shown). (B) Effect of treatment of MV4-11 cells with 1 μM EPZ-5676 on cell-cycle phase and apoptosis over time as measured by staining for DNA content and Annexin V. (C) EPZ-5676 selectively kills acute leukemia cell lines harboring MLL fusions. IC50 values for inhibition of proliferation are plotted for each cell line following 14 days of exposure to increasing concentrations of EPZ-5676 (n = 2, mean value is shown, apart from MV4-11, where n = 3). Cell lines are grouped according to MLL status and horizontal lines represent the mean IC50 value for each group. IC50 values are also listed along with IC90 values in supplemental Table 2.
Ablation of DOT1L histone methyltransferase activity by small-molecule inhibition or genetic approaches has been demonstrated to selectively kill cultured leukemia cells bearing MLL translocations, while having little or no effect on non–MLL-rearranged leukemia cells.21,26,28-33 We tested the effect of EPZ-5676 treatment in 14-day proliferation assays with a panel of 11 acute leukemia cell lines with or without MLL rearrangements. The proliferation IC50 and 90% inhibitory concentration (IC90) values of each cell line are listed in supplemental Table 2 and IC50 values are plotted in Figure 3C. EPZ-5676 demonstrates nanomolar antiproliferative activity against most of the MLL-rearranged cell lines tested, whereas most of the non–MLL-rearranged cell lines gave IC50 readings that were at least 3 orders of magnitude higher. Exceptions were the MLL-AF4 expressing cell line RS4;11, which gave an IC50 of 1.3 μM and the non–MLL-rearranged cell line REH, which gave an IC50 value of 1.2 μM (although an IC90 value for inhibition of proliferation in REH was not achieved [supplemental Table 2]). The reasons for this are not currently understood, but may reflect additional genetic determinants of DOT1L inhibitor sensitivity that have yet to be identified. Overall, these results demonstrate that EPZ-5676 preferentially inhibits the proliferation of leukemia cells expressing MLL fusions.
EPZ-5676 causes complete and sustained regression in a rat xenograft model of MLL-rearranged leukemia
Based on overall activity in the biochemical and cell based assays summarized in Figure 1D, EPZ-5676 represents a significant improvement in potency and selectivity over EPZ004777. We therefore wished to determine the therapeutic potential of EPZ-5676 in vivo. For this, we tested the ability of EPZ-5676 to inhibit tumor growth in rodent subcutaneous (SC) MV4-11 xenograft models. Characterization of the pharmacokinetic properties of EPZ-5676 in mouse and rat (Figure 4) revealed that the compound has low oral bioavailability and high intraperitoneal (IP) bioavailability in mouse and high clearance in both mouse and rat. Pharmacokinetic analysis of EPZ-5676 following IP administration of a dose of 20 mg/kg revealed that plasma levels reached almost 5 μM shortly after injection, but dropped to concentrations below those required to completely block the proliferation of MV4-11 cells in vitro (ie, ≥0.123 μM; Figure 3A) within 2 hours of dosing (Figure 4B). Lack of antitumor activity in mice bearing SC MV4-11 xenografts following IP administration of EPZ-5676 (supplemental Figure 2) led us to believe that extended maintenance of plasma levels above a threshold level may be important for efficacy. We therefore selected continuous IV infusion as the route of administration for subsequent studies. Pilot studies in Sprague-Dawley rats confirmed that this dosing methodology allowed EPZ-5676 plasma levels to be maintained at a constant level over several days (Figure 4D). Because IV infusion for extended periods is technically challenging in mice, we developed a SC MV4-11 xenograft model in immunocompromised rats. Treatment of MV-411 xenograft bearing rats with EPZ-5676 by continuous IV administration for 21 days led to dose-dependent tumor regression, as illustrated in Figure 5A (supplemental Figure 3A). At an intermediate dose of 35 mg/kg, tumor stasis was achieved and sustained for about 7 days after cessation of compound administration. At the higher dose of 70 mg/kg, complete regressions of established SC MV4-11 tumors were achieved. Furthermore, most rats showed little or no regrowth of tumor for more than 30 days beyond the cessation of compound treatment until the study was terminated at day 52 (Figure 5A; supplemental Figure 3A). One group was tested using a modified dosing schedule in which EPZ-5676 was infused daily for 8 hours per day for 21 days at a dose of 67 mg/kg per day. This dose also caused tumor stasis, which was then followed by eventual resumption of growth several days after the end of the last infusion. EPZ-5676 plasma levels during this study averaged 0.6, 0.33, and 4.39 μM in the 70, 35, and 67 mg/kg per day groups, respectively (supplemental Figure 3B). That the intermittent 8-hour daily infusion group showed less efficacy than a similar daily dose delivered continuously is evidence that continuous maintenance of EPZ-5676 plasma levels above a threshold concentration may be a more important driver of efficacy than intermittent high-level exposure. All dose levels were well tolerated by the animals with no significant weight loss observed during the course of treatment (supplemental Figure 3C). We performed a second efficacy study to determine whether potent antitumor activity could be observed with IV infusion periods of <21 days. As in the first study, continuous infusion at 70 mg/kg per day for 21 days caused sustained tumor regressions (Figure 5B; supplemental Figure 4). Reducing the length of infusion to 14 days also caused sustained tumor regressions, although growth in most tumors had resumed before the study was terminated at day 50 (29 days postinfusion). Further reducing the length of continuous infusion to 7 days led to a dramatic reduction in antitumor activity (Figure 5B; supplemental Figure 4). These results show that EPZ-5676 is highly efficacious and achieves optimal activity when delivered by continuous IV infusion for 14 days or longer.
The preclinical pharmacokinetics of EPZ-5676 determined in mouse and rat. (A) Table of EPZ-5676 pharmacokinetic parameters as determined in mouse and rat. All values reported as mean ± standard deviation (SD) with exception of mouse IP and rat IV infusion study where nonserial, sparse blood sampling was used. In this case, all values are means. Pharmacokinetic data analysis was performed using noncompartmental analysis and WinNonlin Phoenix v6.2 software. Calculated pharmacokinetic parameters show high clearance (CL) in both species with low oral bioavailability and high intraperitoneal bioavailability in mouse (the latter calculated using the IV area under the curve [AUC] from CD-1 mouse). Volume of distribution at steady state (VDss) was greater than total body water of 0.7 L/kg in both species. Terminal half-lives (t1/2) ranged from 1 to 5 hours depending on the route of administration, whereas mean residence times (MRT) were short, ranging from 0.35 to 0.84 hours. Cmax, the maximum plasma concentration, is determined at time tmax. AUC0-t and AUC0-inf are the areas under the curve to the last measurable data point and extrapolated to infinity, respectively. Data are shown graphically as (B) concentration vs time profile of mean ± SD (n = 3) plasma concentrations following IV bolus (5 mg/kg, red line) and PO (20 mg/kg, blue line) administrations to CD-1 mouse as well as IP (20 mg/kg, black line) administration to NCr nu/nu mouse (all formulated in 10% ethanol:90% saline). The dotted line represents the EPZ-5676 concentration required to completely block proliferation of MV4-11 cells in vitro (Fig. 2C); (C) concentration vs time profile of mean ± SD (n = 3) plasma concentrations following IV bolus (1 mg/kg formulated in 0.4% hydroxypropyl-β-cyclodextrin in saline) administration to Sprague-Dawley rat; (D) concentration vs time profile of mean ± SD (n = 3) plasma concentrations following IV infusion (4.7 mg/kg per day for 7 days formulated in 10% PEG400:90% saline) administration to Sprague-Dawley rat (elimination phase data after 168 hours not shown for clarity). F, absolute bioavailability.
The preclinical pharmacokinetics of EPZ-5676 determined in mouse and rat. (A) Table of EPZ-5676 pharmacokinetic parameters as determined in mouse and rat. All values reported as mean ± standard deviation (SD) with exception of mouse IP and rat IV infusion study where nonserial, sparse blood sampling was used. In this case, all values are means. Pharmacokinetic data analysis was performed using noncompartmental analysis and WinNonlin Phoenix v6.2 software. Calculated pharmacokinetic parameters show high clearance (CL) in both species with low oral bioavailability and high intraperitoneal bioavailability in mouse (the latter calculated using the IV area under the curve [AUC] from CD-1 mouse). Volume of distribution at steady state (VDss) was greater than total body water of 0.7 L/kg in both species. Terminal half-lives (t1/2) ranged from 1 to 5 hours depending on the route of administration, whereas mean residence times (MRT) were short, ranging from 0.35 to 0.84 hours. Cmax, the maximum plasma concentration, is determined at time tmax. AUC0-t and AUC0-inf are the areas under the curve to the last measurable data point and extrapolated to infinity, respectively. Data are shown graphically as (B) concentration vs time profile of mean ± SD (n = 3) plasma concentrations following IV bolus (5 mg/kg, red line) and PO (20 mg/kg, blue line) administrations to CD-1 mouse as well as IP (20 mg/kg, black line) administration to NCr nu/nu mouse (all formulated in 10% ethanol:90% saline). The dotted line represents the EPZ-5676 concentration required to completely block proliferation of MV4-11 cells in vitro (Fig. 2C); (C) concentration vs time profile of mean ± SD (n = 3) plasma concentrations following IV bolus (1 mg/kg formulated in 0.4% hydroxypropyl-β-cyclodextrin in saline) administration to Sprague-Dawley rat; (D) concentration vs time profile of mean ± SD (n = 3) plasma concentrations following IV infusion (4.7 mg/kg per day for 7 days formulated in 10% PEG400:90% saline) administration to Sprague-Dawley rat (elimination phase data after 168 hours not shown for clarity). F, absolute bioavailability.
EPZ-5676 causes tumor regressions and demonstrates in vivo target inhibition in tumor and surrogate tissue in a rat model of MLL-rearranged leukemia. (A) Effect of EPZ-5676 administration on the growth of MV4-11 xenograft tumors implanted SC in immunocompromised rats. Rats were dosed continuously by IV infusion with vehicle (black line), with EPZ-5676 at 70 mg/kg/day (blue line), 35 mg/kg/day (red line), or for 8 hours daily at 67 mg/kg per day (green line) for 21 days. Median tumor sizes are plotted for each group (n = 10). Tumor growth in individual animals is shown in supplemental Figure 3. Significant P values of P ≤ .0005, calculated using a repeated-measures analysis of variance (ANOVA) and Dunnett posttest, were determined between the vehicle and all 3 treated groups. (B) Effect of additional EPZ-5676 dose and schedules on MV4-11 xenograft tumor growth. Significant tumor growth inhibition was observed in rats treated with 70 mg/kg per day EPZ-5676 by continuous IV infusion for 14 (orange line) or 21 days (blue line) (P = .0004, repeated-measures ANOVA with a Dunnett posttest). Treatment with 70 mg/kg per day continuously for 7 days followed by 14 days at 33.5 mg/kg per day infused for 8 hours daily (green line) led to weak, but statistically significant tumor growth inhibition (P = .04, repeated-measures ANOVA with a Dunnett posttest). A final group dosed for 21 days with 33.5 mg/kg per day infused for 8 hours daily (purple line) showed no tumor growth inhibition when compared with the vehicle control. Median tumor sizes are plotted for each group (vehicle [n = 9], 70 mg/kg per day, 21-day infusion [n = 9], 70 mg/kg per day, 14-day infusion [n = 10], 70 mg/kg per day, 7-day infusion, then 14 days at 33.5 mg/kg per day infused for 8 hours daily [n = 9], 33.5 mg/kg per day, 21 days infused for 8 hours daily [n = 10]). Tumor growth in individual animals is shown in supplemental Figure 4. (C) H3K79me2 levels in tumor tissue, PBMCs, and bone marrow cells harvested from nude rats dosed continuously by IV infusion for 14 days with vehicle or EPZ-5676 at 35 and 70 mg/kg per day. H3K79me2 levels were determined in acid extracted histones by ELISA (tumor, bone marrow) or quantitated immunoblot of whole cell lysates (PBMCs). H3K79me2 levels were normalized to those of total histone H3 in the same sample and are plotted as a percent of the mean H3K79me2 level in tissue from the vehicle-treated (control) group, which is set at 100%. Horizontal lines represent the mean percent H3K79me2 values for each group. (D) Reduced expression of MLL-fusion target genes HOXA9 and MEIS1 measured by qRT-PCR of RNA extracted from tumor tissue harvested from rats dosed continuously by IV infusion for 14 days with EPZ-5676 at 35 and 70 mg/kg per day. HOXA9 or MEIS1 transcript levels are plotted as a percent of the mean transcript level in tumors from the vehicle-treated (control) group, which is set at 100%. Horizontal lines represent the mean percent transcript level in each group.
EPZ-5676 causes tumor regressions and demonstrates in vivo target inhibition in tumor and surrogate tissue in a rat model of MLL-rearranged leukemia. (A) Effect of EPZ-5676 administration on the growth of MV4-11 xenograft tumors implanted SC in immunocompromised rats. Rats were dosed continuously by IV infusion with vehicle (black line), with EPZ-5676 at 70 mg/kg/day (blue line), 35 mg/kg/day (red line), or for 8 hours daily at 67 mg/kg per day (green line) for 21 days. Median tumor sizes are plotted for each group (n = 10). Tumor growth in individual animals is shown in supplemental Figure 3. Significant P values of P ≤ .0005, calculated using a repeated-measures analysis of variance (ANOVA) and Dunnett posttest, were determined between the vehicle and all 3 treated groups. (B) Effect of additional EPZ-5676 dose and schedules on MV4-11 xenograft tumor growth. Significant tumor growth inhibition was observed in rats treated with 70 mg/kg per day EPZ-5676 by continuous IV infusion for 14 (orange line) or 21 days (blue line) (P = .0004, repeated-measures ANOVA with a Dunnett posttest). Treatment with 70 mg/kg per day continuously for 7 days followed by 14 days at 33.5 mg/kg per day infused for 8 hours daily (green line) led to weak, but statistically significant tumor growth inhibition (P = .04, repeated-measures ANOVA with a Dunnett posttest). A final group dosed for 21 days with 33.5 mg/kg per day infused for 8 hours daily (purple line) showed no tumor growth inhibition when compared with the vehicle control. Median tumor sizes are plotted for each group (vehicle [n = 9], 70 mg/kg per day, 21-day infusion [n = 9], 70 mg/kg per day, 14-day infusion [n = 10], 70 mg/kg per day, 7-day infusion, then 14 days at 33.5 mg/kg per day infused for 8 hours daily [n = 9], 33.5 mg/kg per day, 21 days infused for 8 hours daily [n = 10]). Tumor growth in individual animals is shown in supplemental Figure 4. (C) H3K79me2 levels in tumor tissue, PBMCs, and bone marrow cells harvested from nude rats dosed continuously by IV infusion for 14 days with vehicle or EPZ-5676 at 35 and 70 mg/kg per day. H3K79me2 levels were determined in acid extracted histones by ELISA (tumor, bone marrow) or quantitated immunoblot of whole cell lysates (PBMCs). H3K79me2 levels were normalized to those of total histone H3 in the same sample and are plotted as a percent of the mean H3K79me2 level in tissue from the vehicle-treated (control) group, which is set at 100%. Horizontal lines represent the mean percent H3K79me2 values for each group. (D) Reduced expression of MLL-fusion target genes HOXA9 and MEIS1 measured by qRT-PCR of RNA extracted from tumor tissue harvested from rats dosed continuously by IV infusion for 14 days with EPZ-5676 at 35 and 70 mg/kg per day. HOXA9 or MEIS1 transcript levels are plotted as a percent of the mean transcript level in tumors from the vehicle-treated (control) group, which is set at 100%. Horizontal lines represent the mean percent transcript level in each group.
Pharmacodynamic effects of EPZ-5676 in tumor and surrogate tissue
We evaluated H3K79 methylation in MV4-11 SC xenograft tissue harvested from rats dosed by continuous IV infusion for 14 days at 70 and 35 mg/kg per day in a parallel experiment (Figure 5C). H3K79me2 levels were reduced in both EPZ-5676 dose groups relative to vehicle-treated rats. We also found that H3K79me2 levels were reduced in bone marrow cells and PBMCs isolated from the same rats. These results demonstrate in vivo target engagement by EPZ-5676 in tumor tissue and show that H3K79 methylation in surrogate tissues, including readily accessible PBMCs, can be effectively used as a pharmacodynamic assay to track EPZ-5676 activity in vivo. Tumors taken from EPZ-5676–treated rats also showed reduced HOXA9 and MEIS1 mRNA levels (Figure 5D). EPZ-5676 treatment is therefore also able to reduce MLL-fusion target gene expression in vivo.
Discussion
DOT1L has emerged as an attractive target for therapeutic inhibition in MLL-rearranged leukemias. The majority of MLL rearrangements result in the expression of MLL-fusion proteins that gain the ability to recruit DOT1L directly or indirectly to MLL target genes. This leads to inappropriate hypermethylation and increased expression of a panel of target genes including HOXA9 and MEIS1 that promote leukemogenesis. We have shown that EPZ-5676 is a highly potent and selective inhibitor of DOT1L in biochemical and cellular assays. Our crystallographic analysis demonstrates that, in common with previously described DOT1L inhibitors of this series, EPZ-5676 causes conformational changes upon interaction with the SAM binding pocket of DOT1L.33,35 These structural rearrangements create additional interaction surfaces that contribute to high-affinity binding and selectivity of EPZ-5676. The compound demonstrates inhibition of H3K79 methylation and MLL-fusion target gene expression in cultured cells and selective killing of cell lines bearing MLL-AF9, MLL-AF4, and MLL-ENL fusions. The selectivity of EPZ-5676 antiproliferative activity for MLL-rearranged cells is encouraging from the standpoint of developing DOT1L inhibitors as targeted therapeutics and is consistent with previous reports that have used genetic ablation or small molecule tool compounds to demonstrate that unique dependence of MLL-rearranged cells for DOT1L activity.21,26,28-33 Pharmacokinetic characterization of EPZ-5676 demonstrated low oral bioavailability and relatively high clearance in mice and rats. Our preliminary in vivo efficacy studies using IP dosing revealed that continuous maintenance of plasma exposure above a threshold level as likely to be required for efficacy. We therefore turned to IV infusion in a nude rat SC xenograft model of MLL-rearranged leukemia. Using this route of administration, we achieved complete and sustained tumor regressions with EPZ-5676 at doses that were well tolerated with no overt signs of toxicity or significant body weight loss. In the highest dose group, complete regressions were achieved following 21 days of dosing and tumor regrowth was not observed for the duration of the experiment. Shortening the length of dosing to 14 days also caused tumor regression; however, further reducing the duration to 7 days did not show significant efficacy. In addition, we found that intermittent dosing for 8 hours daily provided less efficacy than a similar daily dose administered continuously over a 24-hour period. Therefore, our results suggest that continuous inhibition of DOT1L activity for at least 14 days is required for optimal efficacy. We also evaluated the pharmacodynamic activity of EPZ-5676 in the rat MLL-rearranged xenograft model and demonstrated that H3K79me2 levels and HOXA9 and MEIS1 mRNA levels were reduced in xenograft tissue in animals dosed with EPZ-5676. We also demonstrated inhibition of H3K79 methylation in PBMCs and bone marrow cells. These results establish the feasibility of monitoring in vivo target engagement in surrogate tissues that are readily accessible in a clinical setting. We note that the limitations of consistently maintaining efficacious plasma exposures in mice via intermittent or implanted osmotic minipump strategies (supplemental Figure 2 and data not shown) have precluded evaluation of EPZ-5676 in additional models of interest, such as primary patient-derived leukemia xenografts. We are actively exploring alternative EPZ-5676 delivery routes and formulations in mice with the goal of overcoming this technical obstacle and expanding the repertoire of accessible preclinical models.
EPZ-5676 represents a significant advance over previously described inhibitors in terms of potency, selectivity, drug-target residence time, and the robust in vivo efficacy demonstrated in this report. The profound and sustained inhibition of tumor growth demonstrated by in EPZ-5676 at well-tolerated doses in a rodent model of MLL-fusion leukemia together with preclinical safety and toxicological assessments of EPZ-5676 that support human dosing have prompted us to transition the inhibitor into clinical development. EPZ-5676 is under evaluation as a targeted therapy for acute leukemias bearing MLL rearrangements and is the first reported histone methyltransferase inhibitor to enter human clinical trials.
The data reported in this article have been deposited in the Protein Data Bank (accession number 4HRA).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank P. Pearson, A. A. Avrutskaya, R. J. Mullin, B. Hollister, S. A. Armstrong, and R. Gould for helpful discussions, and J. M. Davis, O. Raguin, and M. Leblanc for executing in vivo studies.
Authorship
Contribution: S.R.D., E.J.O., A.B., L.J., A.R., M.P.S., V.M.R., and R.M.P. designed studies; S.R.D., E.J.O., C.A.T., A.B., C.J.A., C.R.K., and A.R. performed research; S.R.D., E.J.O., R.A.C., and R.M.P. wrote the paper; and all authors analyzed and interpreted data, discussed the results, and commented on the manuscript.
Conflict-of-interest disclosure: All authors are employees of, and/or hold equity in, Epizyme, Inc.
The current affiliation for V.M.R. is Sanofi, Cambridge, MA.
Correspondence: Roy M. Pollock, Epizyme, Inc., 400 Technology Square, 4th Floor, Cambridge, MA 02139; e-mail: rpollock@epizyme.com.
References
Author notes
S.R.D. and E.J.O. contributed equally to this study.

![Figure 1. Structure, binding, and inhibitory activity of EPZ-5676. (A) Chemical structure of EPZ-5676. (B) Superposition of DOT1L-EPZ-5676 (green) and DOT1L-SAM (gray; Protein Data Base accession number 3QOW). To accommodate the extended hydrophobic tail of the inhibitor, significant rearrangement of the protein is required, including the loop between β-strands 10 and 11 (L10-11). Details on the interactions between the protein and compound and the induced changes caused by EPZ-5676 binding are shown in supplemental Figure 1. (C) Selectivity profile of EPZ-5676 inhibitory activity against representative members of the lysine (left) and arginine (right) enzyme families. The diameter of the sphere for each enzyme is directly related to the magnitude of inhibition by EPZ-5676. Larger circles correlate to increased potency; gray circles indicate no measurable inhibition up to 10 µM of EPZ-5676. (D) Comparison of EPZ-5676 and EPZ004777 potency, selectivity, and cell-based activity. The enzyme inhibition Ki values for EPZ004777 and EPZ-5676 in DOT1L enzymatic assays are listed (n = 3; mean values ± SD are shown). Residence time for each compound is listed and was calculated as the reciprocal of the enzymatic-ligand dissociation rate as determined by surface plasmon resonance. Also listed are inhibitory activities for both compounds in MV4-11 proliferation assays (n = 3 [EPZ-5676] or n = 2 [EPZ004777]; mean values ± SD are shown), MV4-11 cell H3K79me2 ELISA assays (n = 2; mean values ± SD are shown) and MV4-11 cell HOXA9 and MEIS1 qRT-PCR assays (n = 2; mean values ± SD are shown).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/6/10.1182_blood-2013-04-497644/4/m_1017f1.jpeg?Expires=1769129892&Signature=x6WzpM4owDIHG0inwMLwqgQ7RupRY~IAxcj13HqIkv~H3q2ikkf2lKCiEcbFiLKKFLULERhLxWrk0UwxwMWBdZPR8B0-IWyuNfqu80DEnQN54hb3yV~ZwACeuQh6a5M9PS2IKES4dhnJuSAouZo6wx9zQrEf4foanZXr-LGVOAv4Xi9kkNxa74QHkBCLoZyYZ6uD4kCAux8VpoRnfJSIrgE9pi7E7dUHb6fSSbp-4ay2psiJgrRxH8kFPc8Rgmv5kseIpjJHoBzxpVbTtliUXp4KHPaiOrsLA1FbhzrY2vy6g9Cxeh0vzJlznCENlWozx53HERE4nSzKoMcY0JczQA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. Inhibition of histone methylation and MLL-fusion target gene expression by EPZ-5676. (A) Immunoblot analysis of H3K79me2 levels in histones extracted from MV4-11 cells treated for 4 days with increasing concentrations of EPZ-5676. (B) Concentration-dependent inhibition of H3K79 methylation in MV4-11 (left) and HL60 (right) cells following 4-day EPZ-5676 treatment as measured by quantitative ELISA assay for H3K79me2. (C) Immunoblot analysis of histones extracted from MV4-11 cells treated for 4 days with either 1 µM EPZ-5676 or vehicle control and probed with a panel of specific methyl-lysine and methyl-arginine antibodies. Total H3 and H4 antibodies were used as the loading controls. (D) Time course of cellular H3K79me2 depletion in MV4-11 cells incubated in the presence of 1 µM EPZ-5676 (left). The diminution of intracellular H3K79me2 as a function of time after dosing with EPZ-5676 was fit to a simple exponential decay function that included a non-zero limit at infinite time, as described.39 Time course of recovery of H3K79me2 levels in MV4-11 cells upon removal of EPZ-5676 from the culture medium following a 4-day incubation (right). In both plots, H3K79me2 levels are normalized to total H3 and expressed as a percent of DMSO-treated (control) cells at each time point. Recovery of intracellular H3K79me2 levels following compound washout displayed a significant lag phase before semilinear recovery. These data were fit to a modified version of the expolinear function for crop growth as a function of photon interception and leaf area.40 The amount of H3K79me2 at any given time after compound washout (y) was fit as y = ymin +[(v/a)ln{1 + exp(a(t-tlag))}], where ymin is the non-zero limit of H3K79me2 at the start of the washout experiment, v is the velocity of the linear phase of recovery, and a is a constant of proportionality for our purposes. (E) Concentration dependent inhibition of HOXA9 and MEIS1 transcription by EPZ-5676 as measured by qRT-PCR in MV4-11 cells following 6 days of treatment (left; n = 2, mean ± SD are shown). Time course of inhibition of HOXA9 and MEIS1 mRNA expression as measured by qRT-PCR in MV4-11 cells incubated in the presence of 1 µM EPZ-5676 (right). Relative mRNA expression is plotted as a percentage of those at day 0 (n = 2; mean values ± SD are shown).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/6/10.1182_blood-2013-04-497644/4/m_1017f2.jpeg?Expires=1769129892&Signature=WVctj5Mk-PaXnVjr~fd2I6E1jPjVGX3cknjbcxRoJF0pMgm4V1I~lJx2r0aMRNtZmQmjgmmr1XqQ7Pn1czOpszbTNc1R5uHbH0r1dD4CkHCKjZTMwzfAOhN48xPjXAIK5t1hjprZ35zJmjh7tzlE6DVFqo4EKEMZZK3s4RgqjTOpU0fStFKoc4yiL-gEcvpokFONAvayGZJw~aCg4Jnl~~zKcUMIE7nosoYzVuurpB4w8XA~lJgSku7iwMqS6OmIvz-bHX0SooN7Yjd4jxJCHvOVF9b85uc5~aYTXLtRh700SffMl8L~5jDNTLHR3OcBKZNcFRz5i-cwpxcGMCMKWw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. The preclinical pharmacokinetics of EPZ-5676 determined in mouse and rat. (A) Table of EPZ-5676 pharmacokinetic parameters as determined in mouse and rat. All values reported as mean ± standard deviation (SD) with exception of mouse IP and rat IV infusion study where nonserial, sparse blood sampling was used. In this case, all values are means. Pharmacokinetic data analysis was performed using noncompartmental analysis and WinNonlin Phoenix v6.2 software. Calculated pharmacokinetic parameters show high clearance (CL) in both species with low oral bioavailability and high intraperitoneal bioavailability in mouse (the latter calculated using the IV area under the curve [AUC] from CD-1 mouse). Volume of distribution at steady state (VDss) was greater than total body water of 0.7 L/kg in both species. Terminal half-lives (t1/2) ranged from 1 to 5 hours depending on the route of administration, whereas mean residence times (MRT) were short, ranging from 0.35 to 0.84 hours. Cmax, the maximum plasma concentration, is determined at time tmax. AUC0-t and AUC0-inf are the areas under the curve to the last measurable data point and extrapolated to infinity, respectively. Data are shown graphically as (B) concentration vs time profile of mean ± SD (n = 3) plasma concentrations following IV bolus (5 mg/kg, red line) and PO (20 mg/kg, blue line) administrations to CD-1 mouse as well as IP (20 mg/kg, black line) administration to NCr nu/nu mouse (all formulated in 10% ethanol:90% saline). The dotted line represents the EPZ-5676 concentration required to completely block proliferation of MV4-11 cells in vitro (Fig. 2C); (C) concentration vs time profile of mean ± SD (n = 3) plasma concentrations following IV bolus (1 mg/kg formulated in 0.4% hydroxypropyl-β-cyclodextrin in saline) administration to Sprague-Dawley rat; (D) concentration vs time profile of mean ± SD (n = 3) plasma concentrations following IV infusion (4.7 mg/kg per day for 7 days formulated in 10% PEG400:90% saline) administration to Sprague-Dawley rat (elimination phase data after 168 hours not shown for clarity). F, absolute bioavailability.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/6/10.1182_blood-2013-04-497644/4/m_1017f4.jpeg?Expires=1769129892&Signature=GcBbVuAvWTsHIbu1z07ZseHT-72c9DBtbumua0SP0OHI037q7CiYFLTC19o9MJ6o87Az57J2~bcjPSDWTazv3HTazOFY839uJg7m2P4XfK-0teeJ-N26opidLhFGXmI5cEEH5sNYuY5YmYkCtFkqiJf0nE9ACo418G9UnaN669udOkns4wdgeqREDQDKmIM2qGp8mmKvVgvzas6M5JKLpx9Q0a4pKbB2xfWeGj47jeCAMxzsJcfX2w0G3ycrq9JrQXRB~5tHZMmtMaiJ2eqXVGUVOXWwfQpEkkhzpaayPvXTa8IvoIK5nbD~5kIzXmV6j0tiY62qkVHYB3WTBvP1Lg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. EPZ-5676 causes tumor regressions and demonstrates in vivo target inhibition in tumor and surrogate tissue in a rat model of MLL-rearranged leukemia. (A) Effect of EPZ-5676 administration on the growth of MV4-11 xenograft tumors implanted SC in immunocompromised rats. Rats were dosed continuously by IV infusion with vehicle (black line), with EPZ-5676 at 70 mg/kg/day (blue line), 35 mg/kg/day (red line), or for 8 hours daily at 67 mg/kg per day (green line) for 21 days. Median tumor sizes are plotted for each group (n = 10). Tumor growth in individual animals is shown in supplemental Figure 3. Significant P values of P ≤ .0005, calculated using a repeated-measures analysis of variance (ANOVA) and Dunnett posttest, were determined between the vehicle and all 3 treated groups. (B) Effect of additional EPZ-5676 dose and schedules on MV4-11 xenograft tumor growth. Significant tumor growth inhibition was observed in rats treated with 70 mg/kg per day EPZ-5676 by continuous IV infusion for 14 (orange line) or 21 days (blue line) (P = .0004, repeated-measures ANOVA with a Dunnett posttest). Treatment with 70 mg/kg per day continuously for 7 days followed by 14 days at 33.5 mg/kg per day infused for 8 hours daily (green line) led to weak, but statistically significant tumor growth inhibition (P = .04, repeated-measures ANOVA with a Dunnett posttest). A final group dosed for 21 days with 33.5 mg/kg per day infused for 8 hours daily (purple line) showed no tumor growth inhibition when compared with the vehicle control. Median tumor sizes are plotted for each group (vehicle [n = 9], 70 mg/kg per day, 21-day infusion [n = 9], 70 mg/kg per day, 14-day infusion [n = 10], 70 mg/kg per day, 7-day infusion, then 14 days at 33.5 mg/kg per day infused for 8 hours daily [n = 9], 33.5 mg/kg per day, 21 days infused for 8 hours daily [n = 10]). Tumor growth in individual animals is shown in supplemental Figure 4. (C) H3K79me2 levels in tumor tissue, PBMCs, and bone marrow cells harvested from nude rats dosed continuously by IV infusion for 14 days with vehicle or EPZ-5676 at 35 and 70 mg/kg per day. H3K79me2 levels were determined in acid extracted histones by ELISA (tumor, bone marrow) or quantitated immunoblot of whole cell lysates (PBMCs). H3K79me2 levels were normalized to those of total histone H3 in the same sample and are plotted as a percent of the mean H3K79me2 level in tissue from the vehicle-treated (control) group, which is set at 100%. Horizontal lines represent the mean percent H3K79me2 values for each group. (D) Reduced expression of MLL-fusion target genes HOXA9 and MEIS1 measured by qRT-PCR of RNA extracted from tumor tissue harvested from rats dosed continuously by IV infusion for 14 days with EPZ-5676 at 35 and 70 mg/kg per day. HOXA9 or MEIS1 transcript levels are plotted as a percent of the mean transcript level in tumors from the vehicle-treated (control) group, which is set at 100%. Horizontal lines represent the mean percent transcript level in each group.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/6/10.1182_blood-2013-04-497644/4/m_1017f5.jpeg?Expires=1769129892&Signature=HM~E2SArxYtU7LBNLQTPJ4FPPb-g3Ry6~QA~sM41SZsD9f1MUo-stvLfrspDpDtZ85Qyd6H0wtcE5JxoAoz1r4ojK63kZJL~Z0cEw-A78A7RaxD~Pfx5hG9Qdl1ZLXfQt6-gSEIQneFvfJoAGSdxVDJbynmyX-Rk2Zn-C2~U47972XZkQa5dBBGPIivlFb3dLenJV5WEjD2TUTpZpKzj8wBRDMhmLVp8jVwrF-9OBZRyTJ~u1iVOADau0Fsx16VCpKoeVDxCuRtyOo7CbUQKK~fkmGyrFNecRTpMhiJ3T5~v4QW9ZXndrnJ52S8qjJ7nGoqXynJt0a8VgKFBwoCOSA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal