Key Points
Two Gcsf ligands function redundantly through the Gcsf receptor to promote myelopoiesis in zebrafish.
Gcsf signaling is required for HSPC emergence and expansion in zebrafish.
Abstract
Granulocyte colony-stimulating factor (Gcsf) drives the proliferation and differentiation of granulocytes, monocytes, and macrophages (mφs) from hematopoietic stem and progenitor cells (HSPCs). Analysis of the zebrafish genome indicates the presence of 2 Gcsf ligands, likely resulting from a duplication event in teleost evolution. Although Gcsfa and Gcsfb share low sequence conservation, they share significant similarity in their predicted ligand/receptor interaction sites and structure. Each ligand displays differential temporal expression patterns during embryogenesis and spatial expression patterns in adult animals. To determine the functions of each ligand, we performed loss- and gain-of-function experiments. Both ligands signal through the Gcsf receptor to expand primitive neutrophils and mφs, as well as definitive granulocytes. To further address their functions, we generated recombinant versions and tested them in clonal progenitor assays. These sensitive in vitro techniques indicated similar functional attributes in supporting HSPC growth and differentiation. Finally, in addition to supporting myeloid differentiation, zebrafish Gcsf is required for the specification and proliferation of hematopoietic stem cells, suggesting that Gcsf represents an ancestral cytokine responsible for the broad support of HSPCs. These findings may inform how hematopoietic cytokines evolved following the diversification of teleosts and mammals from a common ancestor.
Introduction
Granulocyte colony-stimulating factor (Gcsf), also known as colony-stimulating factor 3 (Csf3), is a cytokine responsible for the proliferation, survival, function, and differentiation of neutrophilic granulocytes, monocytes, macrophages (mφs), and their respective progenitors.1-5 Gcsf exerts these actions through binding to its cognate receptor, Gcsfr, activating downstream signaling cascades important for the survival, migration, proliferation, and differentiation of neutrophils during steady-state and emergency hematopoiesis.6 Gcsf is predominantly produced by cells of the monocyte/mφ lineage but is also produced by endothelial cells,7 fibroblasts,8 and mesothelial cells9 under proper stimulatory conditions. Importantly, Gcsfr is present on a variety of hematopoietic cells, including myeloid progenitors, mature neutrophils, monocytes, B cells, and T cells.6
Gcsf−/−1 and Gcsfr−/−2 mice have reduced levels of myeloid progenitors and show defective granulopoiesis resulting in chronic neutropenia. In humans, a number of GCSFR mutations have been characterized in patients with severe congenital neutropenia, and blastic cells containing these mutations can progress to myelodysplastic syndrome and acute myeloid leukemia.10 Importantly, GCSF is used therapeutically to stimulate granulopoiesis in patients with congenital, chemotherapy-induced, and radiation-induced neutropenia to prevent life-threatening infections, as well as for mobilization of hematopoietic stem and progenitor cells (HSPCs) from their niche in the bone marrow for harvesting and transplantation.11
Gcsf was first purified and characterized in mice3,5 and subsequently identified in a number of other nonmammalian vertebrates, including chicken (Gallus gallus),12 Japanese flounder (Paralichthys olivaceus),12 fugu (Takifugu rubipes),12 green spotted pufferfish (Tetraodon nigroviridis),12 and zebrafish (Danio rerio).13 Additionally, Gcsfr is present in goldfish (Carassius auratus L.).14 Gcsf ligands show significant conservation in their structure, binding domains, and synteny across species. Because of a whole-genome duplication event that occurred early in teleost evolution, fugu and green spotted pufferfish have 2 copies of Gcsf that likely arose from a single common ancestral gene. An additional copy of Gcsf that we identified in zebrafish was not detected in these homology-based studies. Although few functional studies have been performed in other teleosts,12,14 Gcsf signaling promotes myelopoiesis and myeloid cell functions13,15 in zebrafish.
Zebrafish possess the same major blood lineages found in mammals, enabling comparative studies of hematopoiesis among vertebrate phyla. Zebrafish, like all other vertebrate animals studied to date, possess sequential waves of blood cell formation during development. Our laboratory and others have demonstrated that hematopoiesis proceeds through 4 independent phases. The first initiates before 24 hours post fertilization (hpf) with the generation of primitive mφs16 and granulocytes17 from cephalic mesoderm. Primitive erythroid cells develop in the intermediate cell mass, which enter circulation ∼26 hpf. These first blood cell types have been termed “primitive” because each lineage arises only transiently during embryogenesis without passaging through a multipotent progenitor. Multipotency is first observed in erythromyeloid progenitors (EMPs), which arise in the posterior blood island (PBI) at 26 to 30 hpf.18 The EMP is a transient definitive precursor responsible for generating adult-type myeloid and erythroid cells in the zebrafish18,19 and mouse20,21 embryo. Finally, hematopoietic stem cells (HSCs) arise from the transdifferentiation of ventral aortic endothelium between 36 and 72 hpf,22-25 generating erythroid, myeloid, and lymphoid cells for the remainder of life. The signals that regulate specification and maturation of each wave are incompletely understood.
In this manuscript, we have determined the roles of Gcsf signaling during development of the zebrafish hematopoietic system and during steady-state hematopoiesis in the adult animal. We report the identification and characterization of the second Gcsf ligand present in the zebrafish genome. Comparison of these 2 ligands elucidated several similarities and some important differences. Both copies of Gcsf stimulate primitive and definitive hematopoiesis by signaling through a single Gcsfr. However, each cytokine shows distinct temporal expression during embryogenesis and distinct spatial patterns in the adult animal, suggesting that the duplication of Gcsf has provided additional mechanisms of transcriptional control over genes with apparently redundant functions. Finally, zebrafish Gcsf is also required for the specification and proliferation of HSCs. This novel finding suggests that Gcsf represents an ancient cytokine whose functions were diversified into new gene families following gene duplication events over evolution.
Materials and methods
MOs and mRNA injection
Four antisense splice-site–targeting morpholinos (MOs) (Table 1; Gene Tools) were resuspended in diethyl pyrocarbonate–treated water and injected at the 1-cell stage of development. In vitro–transcribed messenger RNAs (mRNAs) were generated with mMessage mMachine (Life Technologies); 1 ng of mRNA resuspended in diethyl pyrocarbonate–treated water at a concentration of 1 mg/mL was injected at the 1-cell stage of development. Zebrafish were mated, staged, and raised in accordance with University of California at San Diego Institutional Animal Care and Use Committee guidelines.
MOs used in study
| Name . | Sequence (5′-3′) . | Amount injected . | Volume injected . |
|---|---|---|---|
| Gcsfr-MO13 | 5′-TTTGTCTTTACAGATCCGCCAGTTC-3′ | 8.3 ng | 1 nL |
| Gcsfa-MO | 5′-AAAAACCTCTTGGAACTCACCAGAC-3′ | 8.3 ng | 1 nL |
| Gcsfb-MO | 5′-CAGGTGTTAGTGTGTATTTACCAGT-3′ | 4.1 ng | 1 nL |
| Scrambled-MO | 5′-CCTCTTACCTCAGTTACAATTTATA-3′ | 8.3 ng | 1 nL |
| Name . | Sequence (5′-3′) . | Amount injected . | Volume injected . |
|---|---|---|---|
| Gcsfr-MO13 | 5′-TTTGTCTTTACAGATCCGCCAGTTC-3′ | 8.3 ng | 1 nL |
| Gcsfa-MO | 5′-AAAAACCTCTTGGAACTCACCAGAC-3′ | 8.3 ng | 1 nL |
| Gcsfb-MO | 5′-CAGGTGTTAGTGTGTATTTACCAGT-3′ | 4.1 ng | 1 nL |
| Scrambled-MO | 5′-CCTCTTACCTCAGTTACAATTTATA-3′ | 8.3 ng | 1 nL |
Clonal assays
Unfractionated whole kidney marrow (WKM) was isolated from gata1:DsRed; mpx:GFP fish and plated in methylcellulose as described26 at 2.5 × 104 cells per mL. Complete media26 contained 0.1% carp serum, 10% bovine serum albumin in Iscove's Modified Dulbecco's Media (Stemcell Technologies), 0.1 μg/mL zebrafish Epo, and Gcsfa or Gcsfb. Colonies were enumerated and photographed after 7 days on a fluorescent microscope (Leica DMI-6000; Wetzlar, Germany). Images were processed as described.27
Enumeration of myeloid cells and HSCs
Animals were anesthetized with tricane,28 and fluorescent images collected at developmental time points. Manual counts of fluorescent cells were performed. To image and enumerate HSCs, confocal microscopy was performed.22 Z-sections were taken and manually counted to avoid enumerating multiciliated cells within pronephric tubules.23
Zebrafish stocks and embryos
Zebrafish were mated, staged, and raised as described28 and maintained in accordance with University of California at San Diego Institutional Animal Care and Use Committee guidelines. Various transgenics were used (Table 2).29,30
Transgenic animals used in study
| Transgenic line . | Cell type labeled . | Reference . |
|---|---|---|
| Tg(mpx:eGFP)i114 | Neutrophils | 30 |
| Tg(gata1:DsRed)sd2; Tg(lmo2:eGFP)zf72 | EMPs | 18, 31, 32 |
| Tg(kdrl:HsHRAS-mCherry)s896; Tg(cmyb:eGFP)zf169 | HSCs | 22 |
| Tg(lyz:GFP)nz117 | Neutrophils | 37 |
| Tg(mpeg1:eGFP)gl22 | mφs | 38 |
| Tg(lyz:DsRed)nz50 | Neutrophils | 37 |
| Transgenic line . | Cell type labeled . | Reference . |
|---|---|---|
| Tg(mpx:eGFP)i114 | Neutrophils | 30 |
| Tg(gata1:DsRed)sd2; Tg(lmo2:eGFP)zf72 | EMPs | 18, 31, 32 |
| Tg(kdrl:HsHRAS-mCherry)s896; Tg(cmyb:eGFP)zf169 | HSCs | 22 |
| Tg(lyz:GFP)nz117 | Neutrophils | 37 |
| Tg(mpeg1:eGFP)gl22 | mφs | 38 |
| Tg(lyz:DsRed)nz50 | Neutrophils | 37 |
Flow cytometry and FACS
Embryos were dechorionated at 24 hpf and maintained in E3 with phenylthiourea to prevent pigmentation.28 Each sample contained 5 embryos, digested with Liberase (Roche) according to manufacturer’s instructions. Samples were triturated, rinsed with phosphate-buffered saline (PBS), and filtered. SYTOXRed (Life Technologies) was added to exclude dead cells. Fluorescence-activated cell sorting (FACS) to isolate hematopoietic cell populations was performed as described.31
Quantitative reverse-transcription polymerase chain reaction analysis
RNA was isolated from tissues with RNeasy (Qiagen), and complementary DNA generated with qScript Supermix (Quanta BioSciences). Primers to detect zebrafish ef1α23 and csf3r (gcsfr)32 have been described. The gcsfa and gcsfb primers are listed in Table 3. Relative expression levels of genes were calculated by the following formula: relative expression = 2–(Ct[gene of interest] – Ct[ef1α]).
Primers used for qRT-PCR
| Gene . | Forward primer 5′-3′ . | Reverse primer 5′-3′ . | Product size . | Gene annotation . |
|---|---|---|---|---|
| gcsfa | AACTACATCTGAACCTCCTG (exon 7) | GACTGCTCTTCTGATGTCTG (exon 8) | 165 bp | NM_001145242.1 |
| gcsfb | GGAGCTCTGCGCACCCAACA (exon 2) | GGCAGGGCTCCAGCAGCTTC (exon 4) | 184 bp | EU267077.1 |
| Gene . | Forward primer 5′-3′ . | Reverse primer 5′-3′ . | Product size . | Gene annotation . |
|---|---|---|---|---|
| gcsfa | AACTACATCTGAACCTCCTG (exon 7) | GACTGCTCTTCTGATGTCTG (exon 8) | 165 bp | NM_001145242.1 |
| gcsfb | GGAGCTCTGCGCACCCAACA (exon 2) | GGCAGGGCTCCAGCAGCTTC (exon 4) | 184 bp | EU267077.1 |
Generation of recombinant cytokines
Generation and purification of Gcsfa and Gcsfb are shown in supplemental Figure 1 (supplemental Materials and methods, available on the Blood Web site).
Results
Identification of Gcsfb
Although other teleosts possess 2 copies of Gcsf,12 only 1 copy of Gcsf was annotated in zebrafish.13 To identify other potential Gcsf genes, we performed synteny analyses with Basic Local Alignment Search Tool (National Center for Biotechnology Information). As shown in Figure 1A, Gcsf shows a high degree of synteny across vertebrate phyla, with Stard3 and Psmd3 upstream, and Med24 and Top2A downstream. This synteny is maintained in zebrafish, with the previously described Gcsf ligand (which we refer to as Gcsfa) present on chromosome 12, upstream of Med24 and Top2A.13 The newly discovered Gcsf ligand (Gcsfb) is located on chromosome 19, downstream of Stard3 and Psmd3. Gcsfb is a 558-bp mRNA that encodes a 185-amino-acid protein with 5 exons, similar to reported Gcsf ligands. Whereas Gcsfa and Gcsfb show little sequence similarity (Figure 1B), they share a similar predicted structure (Figure 1C).
Gcsfa and Gcsfb are paralogous genes that arose from a gene duplication event early in teleost evolution. (A) Synteny analysis of Gcsf ligands across species. Data from chicken, green spotted pufferfish, and fugu are adapted from Santos et al.12 (B) Alignment of human, mouse, and zebrafish Gcsfa and Gcsfb with Clustal Omega (EMBL-European Bioinformatics Institute). Predicted α-helical regions are underlined, modeled on the structure of human GCSF. (C) Proposed structure of Gcsfa and Gcsfb, modeled on the structure of human GCSF; α-helical regions of Gcsfa and Gcsfb are underlined in panel B.
Gcsfa and Gcsfb are paralogous genes that arose from a gene duplication event early in teleost evolution. (A) Synteny analysis of Gcsf ligands across species. Data from chicken, green spotted pufferfish, and fugu are adapted from Santos et al.12 (B) Alignment of human, mouse, and zebrafish Gcsfa and Gcsfb with Clustal Omega (EMBL-European Bioinformatics Institute). Predicted α-helical regions are underlined, modeled on the structure of human GCSF. (C) Proposed structure of Gcsfa and Gcsfb, modeled on the structure of human GCSF; α-helical regions of Gcsfa and Gcsfb are underlined in panel B.
Expression of Gcsfr, Gcsfa, and Gcsfb in zebrafish embryos and adults
To determine if Gcsf signaling was important for zebrafish hematopoietic development, we performed reverse-transcription polymerase chain reaction (qRT-PCR) analysis on embryos. gcsfr was expressed as early as 6 hpf, increased at 24 hpf, and maintained at similar levels when assayed up to 72 hpf, indicating that Gcsf signaling is active during both primitive and definitive hematopoiesis (Figure 2A). To investigate if gcsf ligands were expressed differentially during these time windows, we examined expression of gcsfa and gcsfb. gcsfa was expressed at low levels in 6 hpf embryos, with levels increasing over time (Figure 2B). In contrast, expression of gcsfb was more dynamic (Figure 2B). Notably, gcsfb was expressed at higher levels early in development, decreasing at 24 hpf, then returning to higher levels by 60 hpf. To investigate if these ligands showed different spatial expression patterns in the developing embryo, whole mount in situ hybridization (WISH) was performed. Similar to previous studies,13 and likely because of their low expression, WISH analyses were uninformative.
Expression levels of gcsfr, gcsfa, and gcsfb in embryonic and adult zebrafish. (A) qRT-PCR analysis of gcsfr in pooled whole zebrafish embryos at developmental stages listed along x-axis. (B) qRT-PCR analysis of gcsf ligands (gcsfa, black bars; gcsfb, gray bars) in pooled whole zebrafish embryos at developmental stages listed along x-axis. (C) qRT-PCR analysis of gcsfr in adult zebrafish tissues listed along x-axis. Lymphoid, myeloid, and precursor cells are different cell populations isolated by FACS from the adult WKM. (D) qRT-PCR analysis of gcsfa (black bars) and gcsfb (gray bars) in adult zebrafish tissues listed along x-axis. Lymphoid, myeloid, and precursor cells are different cell populations isolated by FACS from the adult WKM. Expression levels are relative to the housekeeping gene ef1α. All embryonic samples are biological triplicate preparations of at least 10 embryos per time point. All adult tissue samples are biological triplicate preparations from individual adult fish. All bars represent the mean of the samples, and error bars represent standard deviation.
Expression levels of gcsfr, gcsfa, and gcsfb in embryonic and adult zebrafish. (A) qRT-PCR analysis of gcsfr in pooled whole zebrafish embryos at developmental stages listed along x-axis. (B) qRT-PCR analysis of gcsf ligands (gcsfa, black bars; gcsfb, gray bars) in pooled whole zebrafish embryos at developmental stages listed along x-axis. (C) qRT-PCR analysis of gcsfr in adult zebrafish tissues listed along x-axis. Lymphoid, myeloid, and precursor cells are different cell populations isolated by FACS from the adult WKM. (D) qRT-PCR analysis of gcsfa (black bars) and gcsfb (gray bars) in adult zebrafish tissues listed along x-axis. Lymphoid, myeloid, and precursor cells are different cell populations isolated by FACS from the adult WKM. Expression levels are relative to the housekeeping gene ef1α. All embryonic samples are biological triplicate preparations of at least 10 embryos per time point. All adult tissue samples are biological triplicate preparations from individual adult fish. All bars represent the mean of the samples, and error bars represent standard deviation.
Because Gcsf signaling is essential for the continual production of myeloid cells from progenitors, we assessed where gcsfr and its ligands were expressed in adult fish. We found that gcsfr was expressed highly in the kidney, the main site of hematopoiesis in adult teleosts33 (Figure 2C), and fractionation of WKM by light scatter characteristics31 indicated that gcsfr was highly expressed in mature myeloid cells. Additionally, gcsfr was expressed in the “precursor” population, likely because of the presence of myeloid progenitors in this scatter fraction,26,27 including putative common myeloid progenitors and granulocyte-monocyte progenitors.34 gcsfr was also detected in the spleen and skin, likely because of the presence of neutrophils. Examination of gcsfa and gcsfb showed differential expression among various tissues (Figure 2D). gcsfb was expressed highly in gills, skin, and testes. gcsfa was also present in these organs, but highly expressed in the heart. Both gcsfa and gcsfb were expressed in the kidney, but gcsfb was expressed at much higher levels in the lymphoid, myeloid, and precursor scatter fractions. Because of the differences in the expression patterns of these Gcsf ligands, we hypothesized that each likely played differential roles in hematopoietic development and homeostasis.
Gcsf signaling promotes primitive myelopoiesis
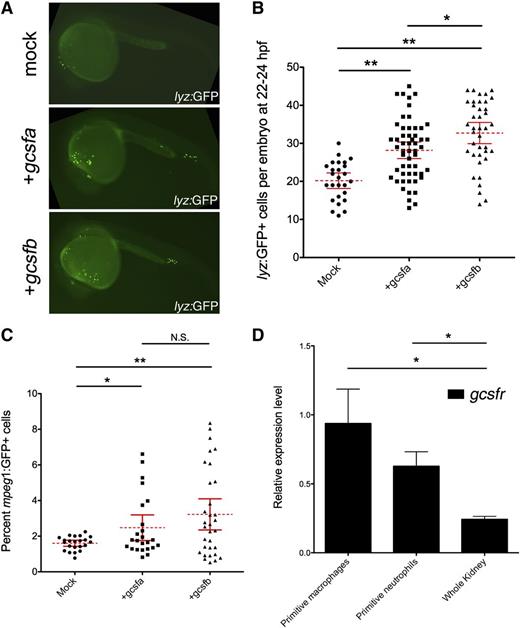
To examine if each ligand showed differential effects on primitive myelopoiesis, we first performed gain-of-function experiments, injecting in vitro–transcribed mRNA into lyz:GFP transgenic embryos whereby green fluorescent protein (GFP) marks primitive mφs35 and neutrophils36 at 22 to 24 hpf. Both gcsfa and gcsfb significantly expanded lyz:GFP+ cells when compared with mock-injected embryos (Figure 3A-B). To examine if gcsfs specifically expanded primitive mφs, we used mφ-specific mpeg1:GFP transgenics.36 Because the fluorescence from these fish is weak, we analyzed embryos by flow cytometry. We observed that gcsfa and gcsfb significantly expanded the numbers of primitive mφs in developing embryos (Figure 3C). In agreement with these data, we observed that FACS-isolated subsets of primitive mφs and neutrophils both expressed high levels of gcsfr (Figure 3D).
Gcsfa and Gcsfb ligands both expand primitive neutrophils and mφs in the zebrafish embryo. (A) Fluorescence images of 24-hpf lyz:GFP embryos injected at the 1-cell stage of development with PBS (mock) and either gcsfa or gcsfb mRNA. Fluorescent images taken on a Leica DMI-6000 inverted fluorescent scope with a Hamamatsu Photonics Orca 3CCD color digital camera (Hamamatsu, Japan) at ×50 and processed by Volocity (Perkin Elmer, MA) and Photoshop (Adobe Systems, San Jose, CA) software. (B) Numbers of lyz:GFP+ cells at 22 to 24 hpf (y-axis) after injection of scrambled MO (mock, circles) and in vitro transcribed gcsfa (squares) or gcsfb (triangles) mRNA. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .02; **P < .0001. (C) Percentage of mpeg1:GFP+ cells at 22 to 24 hpf (y-axis) after injection of PBS (mock, circles) or in vitro transcribed gcsfa (squares) or gcsfb (triangles) mRNA. Each data point corresponds to 5 embryos pooled together before digestion and flow cytometry. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .02; **P < .001. N.S., no significance. (D) Primitive mφs and neutrophils express the gcsf receptor. qRT-PCR analysis of gcsfr in FACS-isolated primitive mφs (mpeg1:GFP+ cells at 20-24 hpf), primitive neutrophils (mpx:GFP+ and lyz:GFP+ cells at 20-24 hpf), and adult zebrafish kidney (Whole Kidney). Levels are relative to the housekeeping gene ef1α. All samples are at least biological duplicate preparations. Bars represent the mean, and error bars represent standard error of the mean (SEM). *P < .05.
Gcsfa and Gcsfb ligands both expand primitive neutrophils and mφs in the zebrafish embryo. (A) Fluorescence images of 24-hpf lyz:GFP embryos injected at the 1-cell stage of development with PBS (mock) and either gcsfa or gcsfb mRNA. Fluorescent images taken on a Leica DMI-6000 inverted fluorescent scope with a Hamamatsu Photonics Orca 3CCD color digital camera (Hamamatsu, Japan) at ×50 and processed by Volocity (Perkin Elmer, MA) and Photoshop (Adobe Systems, San Jose, CA) software. (B) Numbers of lyz:GFP+ cells at 22 to 24 hpf (y-axis) after injection of scrambled MO (mock, circles) and in vitro transcribed gcsfa (squares) or gcsfb (triangles) mRNA. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .02; **P < .0001. (C) Percentage of mpeg1:GFP+ cells at 22 to 24 hpf (y-axis) after injection of PBS (mock, circles) or in vitro transcribed gcsfa (squares) or gcsfb (triangles) mRNA. Each data point corresponds to 5 embryos pooled together before digestion and flow cytometry. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .02; **P < .001. N.S., no significance. (D) Primitive mφs and neutrophils express the gcsf receptor. qRT-PCR analysis of gcsfr in FACS-isolated primitive mφs (mpeg1:GFP+ cells at 20-24 hpf), primitive neutrophils (mpx:GFP+ and lyz:GFP+ cells at 20-24 hpf), and adult zebrafish kidney (Whole Kidney). Levels are relative to the housekeeping gene ef1α. All samples are at least biological duplicate preparations. Bars represent the mean, and error bars represent standard error of the mean (SEM). *P < .05.
To examine if Gcsf ligands signaled through Gcsfr to promote primitive hematopoiesis, we used an MO designed against gcsfr.13 Injection of gcsfr-MO caused a reduction in lyz:GFP+ cells when compared with embryos injected with a control MO (supplemental Figure 2). To determine if Gcsfa and Gcsfb were signaling through Gcsfr, we attempted to rescue the knockdown of myeloid cells by coinjecting individual ligands with gcsfr-MO. In agreement with previous studies,13 we found that Gcsfa was signaling through Gcsfr based on the inability of excess ligand to rescue the knockdown phenotype (supplemental Figure 2). We obtained similar results with Gcsfb, suggesting that it also signals through the Gcsfr. Taken together, these data indicate that both ligands function in a redundant manner, signaling through Gcsfr to expand primitive myeloid cells in zebrafish.
Gcsf signaling promotes definitive myelopoiesis
To investigate if Gcsf ligands were capable of expanding myeloid cells originating from definitive, multilineage precursors, we examined later developmental time points. We performed these studies at 72 hpf, a time whereby the majority of blood cells are derived from definitive HSPCs. We observed that gcsfa and gcsfb significantly expanded the number of mpx:GFP+ myeloid cells present at 72 hpf (Figure 4A-B). To ensure that this was not transgene specific, we also performed these experiments in lyz:DsRed transgenic animals, recapitulating the neutrophil expansion (data not shown). We also observed a significant expansion of mpeg1:GFP+ mφs in injected embryos (Figure 4C). FACS-isolated neutrophils and mφs both expressed gcsfr (Figure 4D). Finally, we observed that Gcsfa and Gcsfb failed to rescue the knockdown of neutrophils induced by gcsfr-MO (supplemental Figure 3), indicating that the ligands were signaling through Gcsfr at later time points in myeloid development.
Gcsfa and Gcsfb expand definitive neutrophils and mφs in the zebrafish embryo. (A) Fluorescence images of 72 hpf mpx:GFP transgenic zebrafish injected with PBS (mock) and either in vitro–transcribed gcsfa or gcsfb mRNA. Fluorescent images taken on a Leica DMI-6000 inverted fluorescent scope with a Hamamatsu Photonics Orca 3CCD color digital camera at ×50 and processed by Volocity (Perkin Elmer) and Photoshop (Adobe Systems) software. Images shown are 2 images stitched together. (B) Numbers of mpx:GFP+ cells at 72 hpf (y-axis) after injection of PBS (mock, circles) and gcsfa (squares) or gcsfb (triangles) mRNA at the single-cell stage of development. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .0001. (C) Percentage of mpeg1:GFP+ cells at 72 hpf (y-axis) after injection of PBS (mock, circles) and gcsfa (squares) or gcsfb (triangles) mRNA. Each data point corresponds to 5 embryos pooled together before digestion and flow cytometry. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .003; **P < .0006; ***P < .0001. (D) At 72 hpf, mφs and neutrophils express gcsfr. qRT-PCR analysis of gcsfr in FACS-isolated macrophages (mpeg1:GFP+ cells at 72 hpf), neutrophils (mpx:GFP+ cells at 72 hpf), and adult zebrafish kidney (Whole Kidney) shown for reference. Levels are relative to the housekeeping gene ef1α. All samples are at least biological duplicate preparations. Bars represent the mean, and error bars represent SEM. *P < .0001.
Gcsfa and Gcsfb expand definitive neutrophils and mφs in the zebrafish embryo. (A) Fluorescence images of 72 hpf mpx:GFP transgenic zebrafish injected with PBS (mock) and either in vitro–transcribed gcsfa or gcsfb mRNA. Fluorescent images taken on a Leica DMI-6000 inverted fluorescent scope with a Hamamatsu Photonics Orca 3CCD color digital camera at ×50 and processed by Volocity (Perkin Elmer) and Photoshop (Adobe Systems) software. Images shown are 2 images stitched together. (B) Numbers of mpx:GFP+ cells at 72 hpf (y-axis) after injection of PBS (mock, circles) and gcsfa (squares) or gcsfb (triangles) mRNA at the single-cell stage of development. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .0001. (C) Percentage of mpeg1:GFP+ cells at 72 hpf (y-axis) after injection of PBS (mock, circles) and gcsfa (squares) or gcsfb (triangles) mRNA. Each data point corresponds to 5 embryos pooled together before digestion and flow cytometry. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .003; **P < .0006; ***P < .0001. (D) At 72 hpf, mφs and neutrophils express gcsfr. qRT-PCR analysis of gcsfr in FACS-isolated macrophages (mpeg1:GFP+ cells at 72 hpf), neutrophils (mpx:GFP+ cells at 72 hpf), and adult zebrafish kidney (Whole Kidney) shown for reference. Levels are relative to the housekeeping gene ef1α. All samples are at least biological duplicate preparations. Bars represent the mean, and error bars represent SEM. *P < .0001.
Gcsf signaling expands HSCs
To investigate if Gcsf signaling regulated the specification and expansion of embryonic HSPCs, we again performed gain-of-function experiments and analyzed the number of HSCs and EMPs present. Surprisingly, both ligands caused expansion of runx1+ HSCs at 24 hpf when analyzed by WISH (Figure 5A), and quantitation of runx1+ cells along the dorsal aorta and in the PBI indicated significant expansion (Figure 5B). Knockdown of Gcsf signaling resulted in a significant reduction of runx1+ cells that exogenous ligand could not rescue (supplemental Figure 4). Further analysis of cells expressing the HSC marker cmyb along the dorsal aorta and caudal hematopoietic tissue (CHT) at 36 hpf confirmed that both ligands were expanding HSCs (Figure 5C-D). We next analyzed the effect of increased Gcsf signaling on EMPs, transient definitive progenitors that lack lymphoid potential. Whereas enforced Gcsf expression significantly expanded HSCs, neither ligand showed significant effects on lmo2:GFP+; gata1:DsRed+ EMPs when analyzed by flow cytometry (Figure 5E). To examine if these 2 progenitor subtypes could respond to Gcsf signaling, we examined their gcsfr expression. Although FACS-isolated HSCs expressed significant levels of gcsfr, EMPs expressed significantly less gcsfr than HSCs (Figure 5F), potentially explaining their inability to respond to Gcsf signaling.
Gcsfa and Gcsfb expand HSCs but not EMPs in the zebrafish embryo. (A) WISH of 24 hpf embryos for runx1 after injection of PBS (mock) or in vitro–transcribed gcsfa or gcsfb mRNA at the single-cell stage of development. Bright field images taken on a Leica M165C upright dissecting scope with a Leica DFC295 color digital camera at ×6 and processed by Photoshop (Adobe Systems) software. (B) Numbers of runx1+ cells along the dorsal aorta and PBI region quantitated from 3 independent experiments as in panel A. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .0001. (C) WISH of 36 hpf embryos for cmyb after injection of PBS (mock) or in vitro–transcribed gcsfa or gcsfb mRNA at the single-cell stage of development. Bright field images taken on a Leica M165C upright dissecting scope with a Leica DFC295 color digital camera at ×8 and processed by Photoshop (Adobe Systems) software. (D) Numbers of cmyb+ cells along the dorsal aorta and CHT region quantitated from 3 independent experiments as in panel C. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .02; **P < .0001. (E) Percentage of lmo2:GFP+; gata1:DsRed+ EMPs at 30 to 32 hpf (y-axis) after injection of PBS (mock, circles) or in vitro–transcribed gcsfa (squares) or gcsfb (triangles) mRNA at the single-cell stage of development. Each data point corresponds to 5 embryos pooled together before digestion and flow cytometry. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. (F) qRT-PCR analysis of gcsfr in FACS-isolated HSCs (cmyb:GFP+; flk1:DsRed+ cells at 48 hpf), EMPs (lmo2:GFP+; gata1:DsRed+ cells at 30 hpf), and adult zebrafish kidney (Whole Kidney). Levels are relative to the housekeeping gene ef1α. All samples are at least biological duplicate preparations. *P < .05; **P < .0003.
Gcsfa and Gcsfb expand HSCs but not EMPs in the zebrafish embryo. (A) WISH of 24 hpf embryos for runx1 after injection of PBS (mock) or in vitro–transcribed gcsfa or gcsfb mRNA at the single-cell stage of development. Bright field images taken on a Leica M165C upright dissecting scope with a Leica DFC295 color digital camera at ×6 and processed by Photoshop (Adobe Systems) software. (B) Numbers of runx1+ cells along the dorsal aorta and PBI region quantitated from 3 independent experiments as in panel A. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .0001. (C) WISH of 36 hpf embryos for cmyb after injection of PBS (mock) or in vitro–transcribed gcsfa or gcsfb mRNA at the single-cell stage of development. Bright field images taken on a Leica M165C upright dissecting scope with a Leica DFC295 color digital camera at ×8 and processed by Photoshop (Adobe Systems) software. (D) Numbers of cmyb+ cells along the dorsal aorta and CHT region quantitated from 3 independent experiments as in panel C. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .02; **P < .0001. (E) Percentage of lmo2:GFP+; gata1:DsRed+ EMPs at 30 to 32 hpf (y-axis) after injection of PBS (mock, circles) or in vitro–transcribed gcsfa (squares) or gcsfb (triangles) mRNA at the single-cell stage of development. Each data point corresponds to 5 embryos pooled together before digestion and flow cytometry. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. (F) qRT-PCR analysis of gcsfr in FACS-isolated HSCs (cmyb:GFP+; flk1:DsRed+ cells at 48 hpf), EMPs (lmo2:GFP+; gata1:DsRed+ cells at 30 hpf), and adult zebrafish kidney (Whole Kidney). Levels are relative to the housekeeping gene ef1α. All samples are at least biological duplicate preparations. *P < .05; **P < .0003.
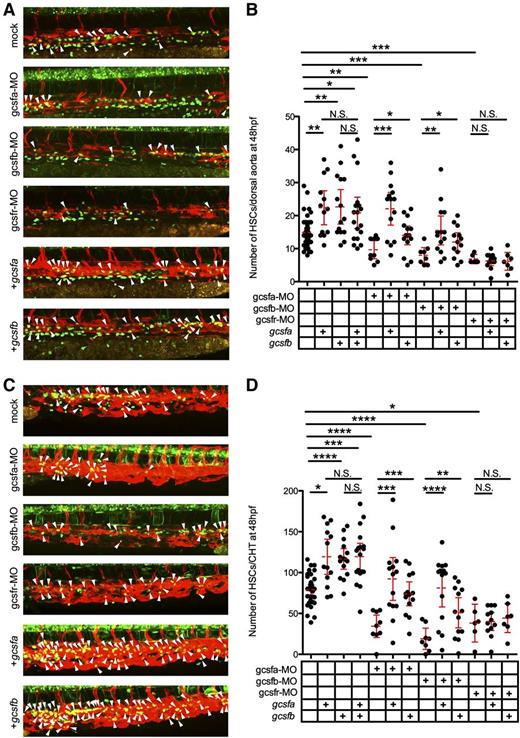
To investigate if Gcsf ligands were directly responsible for the expansion of HSCs, we visualized the emergence of individual cmyb:GFP+; flk1:mCherry+ HSCs along the aortic floor at 48 hpf.22 In agreement with our previous data, we observed that gcsf ligand injections significantly increased HSCs along the dorsal aorta (Figure 6A-B). We also investigated the CHT region of these embryos because this area has been suggested to be the first site of HSC expansion following emergence from aortic endothelium.25 The CHT also showed significant increases in HSCs (Figure 6C-D). To investigate if Gcsf signaling was responsible for this expansion, we again used gcsfr-MO, which caused significant decreases in HSCs (Figure 6B,D). Injection of gcsfa and gcsfb mRNA failed to rescue this reduction, indicating that both ligands were signaling through the Gcsfr to expand HSCs. To further investigate, we generated ligand-specific MOs to see if the depletion of either ligand would have similar effects. The knockdown of either gcsfa or gcsfb (supplemental Figure 5) significantly reduced the amount of myeloid cells at 72 hpf (supplemental Figure 3B) and HSCs in the aorta (Figure 6A-B) and CHT (Figure 6C-D) at 48 hpf, indicating that both ligands play a role in the emergence and expansion of myeloid cells and HSCs in the embryo. We were able to rescue the depletion of Gcsfa by injecting either gcsfa or gcsfb mRNA, indicating not only that was this effect directly attributable to Gcsf signaling, but also that these ligands function redundantly (supplemental Figure 3B; Figure 6B,D). The same results were achieved when Gcsfb was depleted and rescued (supplemental Figure 3B; Figure 6B,D). Confirmation that these ligands function redundantly vs synergistically is demonstrated by the finding that adding both ligands failed to cause a statistically significant increase in HSCs (Figure 6B,D). Taken together, these data indicate that embryonic HSCs are responsive to Gcsf signaling, and that this signaling is essential for their expansion.
Gcsfa and Gcsfb expand HSCs in the zebrafish embryo. (A) Maximum projection of multiple z-stack images of the dorsal aorta region of 48 hpf cmyb:GFP+; flk1:mCherry+ transgenic animals after injection of PBS (mock, top), gcsfa-MO, gcsfb-MO, and gcsfr-MO, in vitro–transcribed gcsfa, and in vitro transcribed gcsfb at the single-cell stage of development. White arrowheads (yellow cells) denote double-positive HSCs located between the dorsal aorta and cardinal vein. Fluorescence images taken on a Leica TCS SP5 inverted confocal system at ×250 and processed by Volocity (Perkin Elmer) and Photoshop (Adobe Systems) software. (B) Numbers of HSCs (cmyb:GFP+; flk1:mCherry+) were enumerated from individual confocal z-stacks from the dorsal aorta region of 48 hpf cmyb:GFP+; flk1:mCherry+ transgenic animals as shown in panel A. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .05; **P < .01; ***P < .0002. (C) Maximum projection of multiple z-stack images of the CHT region of embryos shown in panel A. White arrowheads (yellow cells) denote double-positive HSCs located in contact with the vascular plexus of the CHT. Fluorescence images taken on a Leica TCS SP5 inverted confocal system at ×250 and processed by Volocity (Perkin Elmer) and Photoshop (Adobe Systems) software. (D) Numbers of HSCs (cmyb:GFP+; flk1:mCherry+) were enumerated from individual confocal z-stacks from the CHT region of 48 hpf cmyb:GFP+; flk1:mCherry+ transgenic animals as shown in panel C. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .003; **P < .006; ***P < .0004; ****P < .0001.
Gcsfa and Gcsfb expand HSCs in the zebrafish embryo. (A) Maximum projection of multiple z-stack images of the dorsal aorta region of 48 hpf cmyb:GFP+; flk1:mCherry+ transgenic animals after injection of PBS (mock, top), gcsfa-MO, gcsfb-MO, and gcsfr-MO, in vitro–transcribed gcsfa, and in vitro transcribed gcsfb at the single-cell stage of development. White arrowheads (yellow cells) denote double-positive HSCs located between the dorsal aorta and cardinal vein. Fluorescence images taken on a Leica TCS SP5 inverted confocal system at ×250 and processed by Volocity (Perkin Elmer) and Photoshop (Adobe Systems) software. (B) Numbers of HSCs (cmyb:GFP+; flk1:mCherry+) were enumerated from individual confocal z-stacks from the dorsal aorta region of 48 hpf cmyb:GFP+; flk1:mCherry+ transgenic animals as shown in panel A. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .05; **P < .01; ***P < .0002. (C) Maximum projection of multiple z-stack images of the CHT region of embryos shown in panel A. White arrowheads (yellow cells) denote double-positive HSCs located in contact with the vascular plexus of the CHT. Fluorescence images taken on a Leica TCS SP5 inverted confocal system at ×250 and processed by Volocity (Perkin Elmer) and Photoshop (Adobe Systems) software. (D) Numbers of HSCs (cmyb:GFP+; flk1:mCherry+) were enumerated from individual confocal z-stacks from the CHT region of 48 hpf cmyb:GFP+; flk1:mCherry+ transgenic animals as shown in panel C. Mean (dashed red line) with 95% confidence interval (red error bars) and level of statistical significance. *P < .003; **P < .006; ***P < .0004; ****P < .0001.
Gcsfa is more efficient than Gcsfb at differentiating adult HSPCs
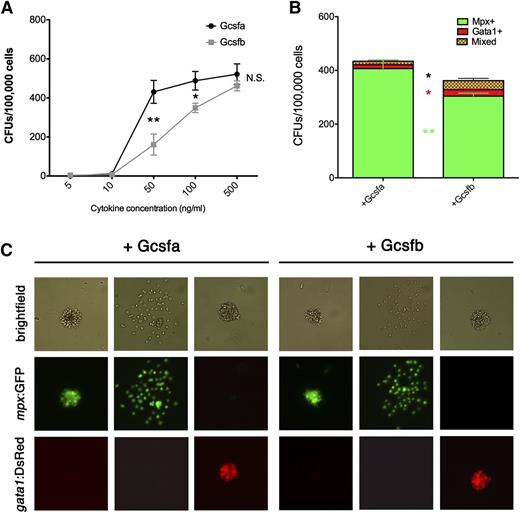
To determine if each Gcsf ligand had redundant effects on adult HSPC differentiation, we produced recombinant versions of Gcsfa and Gcsfb ligands (supplemental Figure 1), performing in vitro assays to quantitatively and functionally assess their proliferative and differentiation activity. First, we used these recombinant ligands to validate that Gcsfa and Gcsfb were responsible for HSPC proliferation, adding ligands to WKM labeled with the membrane dye PKH-26, which indicated that HSPCs were expanded (supplemental Figure 6). To analyze erythromyeloid differentiation, we plated WKM from mpx:GFP+; gata1:DsRed+ adult fish, adding carp serum, zebrafish Epo, and increasing concentrations of Gcsf ligands. Gcsfb encouraged slightly more colony formation at 5 and 10 ng/mL, but between 10 and 100 ng/mL, Gcsfa encouraged significantly more colony formation (Figure 7A). At 500 ng/mL, both Gcsf ligands encouraged similar numbers of colony growth. To assess whether there were functional differences in the maturation of these colonies, we enumerated myeloid colonies marked by the myeloid-specific mpx:GFP transgene. Additionally, we enumerated erythroid colonies marked by the erythroid-specific gata1:DsRed transgene to determine if Gcsf ligands skewed the differentiation of EMPs. Finally, we observed and enumerated “mixed” colonies that contained erythroid and myeloid cells that putatively arose from multilineage EMP cells (Figure 7B). Gcsfa and Gcsfb both stimulated GFP+ ruffled myeloid colonies (composed mainly of neutrophils),26 as well as spread GFP+ colonies (composed of neutrophils, monocytes, and mφs)26 (Figure 7C). There were no differences in colony size, indicating that the 2 ligands induced no measurable difference in progenitor proliferation capacity. Neither Gcsfa nor Gcsfb prevented or skewed the differentiation of erythroid colonies (Figure 7B-C). To confirm that Gcsfa and Gcsfb had differential biological activities, we stably transfected Baf3 murine cells with zebrafish Gcsfr and found that both proteins stimulated the survival of these cells in a ligand-dependent manner (supplemental Figure 7). Importantly, these studies indicated that Gcsfa stimulated survival at lower concentrations than Gcsfb, indicating higher biological activity. Overall, these data confirm that Gcsfa and Gcsfb possess redundant roles in the differentiation of myelomonocytic cells, although Gcsfa has higher colony forming unit (CFU)-promoting capabilities and biological activity.
Gcsfa is more efficient than Gcsfb at expanding erythromyeloid HSPCs cells in the adult zebrafish. (A) Numbers of CFUs (combined erythroid, myeloid, and mixed) per 100 000 cells plated from unfractionated WKM grown with Gcsfa (black line, circles) and Gcsfb (gray line, squares) cytokine concentrations as listed along the x-axis. Each point represents the mean of biological triplicate experiments, and error bars represent the SEM of those triplicate experiments. Statistical significance represents the difference between Gcsfa and Gcsfb effects. *P < .08; **P < .03. (B) Breakdown of different CFU types per 100 000 cells plated from unfractionated WKM grown with 100 ng/mL of Gcsfa and Gcsfb in the presence of Epo. Green bars represent Mpx:GFP+ ruffled and spread colonies (Mpx+), red bars represent Gata1:DsRed+ compact colonies (Gata1+), and green/red checkered bars represent colonies with Mpx:GFP+ and Gata1:DsRED+ cells both present (Mixed). Bars represent the mean of biological duplicate experiments, and error bars represent the SEM of those experiments. Statistical significance represents the difference between Gcsfa and Gcsfb effects. *P < .09; **P < .04. (C) Images of representative colonies derived from unfractionated WKM after stimulation with 100 ng/mL of Gcsfa (left half of panel) or Gcsfb (right half of panel). All cultures also had carp serum, 10% bovine serum albumin, and Epo added. Brightfield (top row), mpx:GFP (middle row), and gata1:DsRed (bottom row) images are shown to illustrate representative colonies seen with these different growth conditions. Mixed colonies are not shown but were present in both cultures. Brightfield and fluorescent images taken on a Leica DMI-6000 inverted fluorescent scope with a Hamamatsu Photonics Orca 3CCD color digital camera at ×400 and processed by Volocity (Perkin Elmer) and Photoshop (Adobe Systems) software.
Gcsfa is more efficient than Gcsfb at expanding erythromyeloid HSPCs cells in the adult zebrafish. (A) Numbers of CFUs (combined erythroid, myeloid, and mixed) per 100 000 cells plated from unfractionated WKM grown with Gcsfa (black line, circles) and Gcsfb (gray line, squares) cytokine concentrations as listed along the x-axis. Each point represents the mean of biological triplicate experiments, and error bars represent the SEM of those triplicate experiments. Statistical significance represents the difference between Gcsfa and Gcsfb effects. *P < .08; **P < .03. (B) Breakdown of different CFU types per 100 000 cells plated from unfractionated WKM grown with 100 ng/mL of Gcsfa and Gcsfb in the presence of Epo. Green bars represent Mpx:GFP+ ruffled and spread colonies (Mpx+), red bars represent Gata1:DsRed+ compact colonies (Gata1+), and green/red checkered bars represent colonies with Mpx:GFP+ and Gata1:DsRED+ cells both present (Mixed). Bars represent the mean of biological duplicate experiments, and error bars represent the SEM of those experiments. Statistical significance represents the difference between Gcsfa and Gcsfb effects. *P < .09; **P < .04. (C) Images of representative colonies derived from unfractionated WKM after stimulation with 100 ng/mL of Gcsfa (left half of panel) or Gcsfb (right half of panel). All cultures also had carp serum, 10% bovine serum albumin, and Epo added. Brightfield (top row), mpx:GFP (middle row), and gata1:DsRed (bottom row) images are shown to illustrate representative colonies seen with these different growth conditions. Mixed colonies are not shown but were present in both cultures. Brightfield and fluorescent images taken on a Leica DMI-6000 inverted fluorescent scope with a Hamamatsu Photonics Orca 3CCD color digital camera at ×400 and processed by Volocity (Perkin Elmer) and Photoshop (Adobe Systems) software.
Discussion
Zebrafish are an increasingly popular model for understanding vertebrate hematopoiesis. Although functional assays in the zebrafish have been designed to investigate the differentiation and proliferation of embryonic18,19 and adult HSPCs,26,27 the specific roles of cytokine signaling in these processes have yet to be defined. To refine and further develop these assays, we set out to more fully characterize cytokine signaling important in hematopoietic development and maintenance.
Here we report and characterize for the first time the second copy of Gcsf in the zebrafish genome. Although duplicate copies of Gcsf had been identified in other teleosts,12 only 1 copy had been described in the zebrafish.13 Because multiple copies of other myeloid cytokines had been described in zebrafish,37 we began an extensive synteny analysis of the genome to look for Gcsf paralogues. We discovered a duplicate copy of Gcsf, which we have designated Gcsfb, that shares extensive upstream genomic synteny with human chromosome 17 and murine chromosome 11. The previously described copy of Gcsf shares extensive downstream synteny with human and mouse, implying that the chromosomal regions harboring these 2 zebrafish paralogues share a common ancestral origin. Additionally, this synteny suggests that the zebrafish Gcsf paralogues resulted from a chromosome/genome duplication event.
Interestingly, zebrafish Gcsfa and Gcsfb seem to possess only partially orthologous functions with their mammalian counterparts. As in mammals, both zebrafish paralogues stimulate granulocytic differentiation,3,4 along with the differentiation of monocytes/mφs.38 However, Gcsf signaling appears to play a broader role in zebrafish hematopoiesis. Our studies demonstrate that Gcsf signaling is required for HSC specification and expansion, roles that have not been described for mammalian Gcsf. One possible explanation is that, during vertebrate evolution, the Gcsf signaling pathway initially functioned to broadly support many levels of hematopoiesis. Following the radiation of mammals, other cytokines likely evolved to take on more specialized roles following gene duplication events.39,40
Supporting this idea, zebrafish lack several members of the class I cytokine signaling pathway, which regulate hematopoiesis and immune cell function.41 Extensive searches in the zebrafish genome indicate that zebrafish lack both the ligands and receptors for the interleukin-3 (IL-3) subfamily, which includes IL-5, Gmcsf, and IL-3. In mammals, these cytokines encourage the production and maintenance of myelomonocytic lineages, including neutrophils, monocytes/mφs, and eosinophils. That each of these lineages exists and functions in teleosts without these cytokines indicates that the ancestral requirements for these specialized cytokines were less complex. Our findings, in addition to phylogenetic analyses, suggest that Gcsf possesses many functions that became diversified over evolution through modification of the Gcsf paradigm. Phylogenetic predictions indicate that identified teleost Gcsfs form a single evolutionary clade outside other related class I cytokine families such as IL-6, leukemia inhibitory factor, and oncostatin M, suggesting that all of these cytokines are orthologous and arose from a common ancestral source.12 Synteny analyses have identified the highly homologous IL-3, IL-4, IL-5, IL-13, and Gmcsf genes clustered on chicken chromosome 13, murine chromosome 11, and human chromosome 5, implying that each of these genes likely arose from a duplication event from a common gene ancestor.40 Supporting this postulate, many of these cytokines have overlapping roles in hematopoiesis that can be compensated for by other family members. For instance, IL-3, originally named multi-CSF for its ability to stimulate the proliferation and differentiation of multiple hematopoietic cell types in vitro,42,43 is dispensable for normal hematopoiesis in vivo.44 Thus, the diversification of class I cytokines appears to have resulted in specialization to fine-tune cytokine signaling present in our common ancestors.
Prior to this report, Gcsf had never been shown to directly stimulate HSC specification or proliferation, although it had been implicated in the production and differentiation of myeloid progenitors.1,2,45 Gcsfr−/− mice have similar numbers of long-term culture-initiating cells,45 and although radiolabeled Gcsf46 and biotinylated GCSF47 bind to the surface of HSCs, they have very few Gcsf receptors relative to more mature myeloid progenitors46 and GCSFR mRNA levels are barely detectable on sorted HSCs.48
Another difference between teleosts and mammals is the duplication of gcsf. We found that gcsfa and gcsfb are differentially expressed temporally during development and spatially in adult fish. We observed that gcsfa was expressed at low levels early in development but increased over time, whereas gcsfb was initially expressed at high levels and decreased later in development. Although we hypothesized that the dynamic expression of each Gcsf ligand may reflect different roles during development, overexpression of either showed redundant functions. The possibility exists that these ligands are expressed in different locations of the embryo and have distinct, specific roles in different tissues, even though WISH analysis was inconclusive, likely because of the low expression of these ligands in the embryo. Interestingly, gcsfb was expressed highly in the kidney, the main site of hematopoiesis in the zebrafish, as well as the testes, skin, and gills. gcsfa was also present in all of these tissues, but at lower levels. The only 2 tissues where gcsfa levels were significantly higher in the adult zebrafish were the heart and spleen. However, both of these ligands stimulated granulocytic and monocyte/mφ differentiation from WKM in vitro, indicating that even though gcsfa was not highly expressed in the kidney, it still retains the ability to differentiate myeloid progenitors.
These studies are the first to compare the functional ability of recombinant zebrafish cytokines to differentiate HSPCs. Although Gcsfa stimulated myeloid progenitors in vitro,26 we were interested to see if Gcsfb would have the same functional effect. Interestingly, we found that the 2 ligands had redundant functions in vitro, both generating myeloid colonies. We found that the 2 Gcsf ligands stimulated colony formation at different concentrations; at low concentrations, Gcsfb encouraged slightly more CFUs, but between 10 and 100 ng/mL, Gcsfa stimulated significantly more colonies. Additionally, Gcsfa stimulated the survival of Baf3 cells transfected with zebrafish Gcsfr, indicating that it possessed higher biological activity. Further biochemical studies are required to investigate the reasons behind the differential activities of these 2 ligands.
In conclusion, we have identified Gcsf as a broadly required cytokine in the formation and function of the zebrafish hematopoietic system. Unlike its mammalian counterpart, Gcsf signaling is required for HSC specification. In contrast, Gcsf signaling is dispensable for the formation and function of EMPs, the first multipotent progenitors generated in the embryo. Similar to its role in mammals, Gcsf is important in the support of myelomonocytic cells, whether derived from the first primitive waves of the embryo or from definitive precursors. The importance of the Gcsf signaling axis in hematopoietic development is highlighted by the observation that reduction of individual Gcsf ligands caused an increase in the other functionally redundant ligand. It will be of interest in future studies to determine if the signals elaborated by the zebrafish Gcsfr are conserved with its mammalian counterparts. Nevertheless, our findings suggest that Gcsf represents an ancient cytokine whose functions were diversified into new gene families following gene duplication events over evolution.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Jiří Brynda for structural predictions of Gcsfa and Gcsfb, Roger Rainville and Lisa Phelps for animal care, and Karen Ong for laboratory management.
This work was supported by grants from the National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (K01-DK087814-01A1) (D.L.S.), 305/10/0953 projects and Fullbright Scholars award (P.B.), California Institute of Regenerative Medicine New Faculty Award (1-00575-1), American Heart Association Innovative Science Award (#12PILT12860010), and National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (R01-DK074482) (D.T.).
Authorship
Contribution: D.L.S., O.S., P.B., R.P.L., R.E.-P., and C.A.C. performed research; D.L.S., P.B., O.S., and D.T. designed the research; D.L.S. and D.T. wrote the manuscript; and P.B. and L.I.Z. provided critical reagents for the work, as well as suggestions on experimental design.
Conflict-of-interest disclosure: L.I.Z. is a founder and stockholder of Fate Inc. and a scientific advisor for Stemgent. The remaining authors declare no competing financial interests.
Correspondence: David Traver, Department of Cellular and Molecular Medicine, University of California at San Diego, La Jolla, CA, 92093-0380; e-mail: dtraver@ucsd.edu.
References
Author notes
D.L.S. and O.S. contributed equally to this study.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal