Key Points
Human inherited IL-10 receptor deficiency is associated with a very high risk of non-EBV–related diffuse large B-cell lymphoma.
IL-10 signaling may be involved in the immune control of germinal center B-cell lymphoma.
Abstract
Monogenic interleukin-10 (IL-10) and IL-10 receptor (IL-10R) deficiencies cause very early onset severe inflammatory bowel disease. Here, we report that 5 patients with an IL-10R1 (n = 1) or IL-10R2 (n = 4) deficiency developed B-cell non-Hodgkin lymphoma between the ages of 5 and 6 years (which was recurrent in 1 patient). These lymphomas had some of the characteristics of diffuse large B-cell lymphomas and contained monoclonal, Epstein-Barr virus–negative germinal center B cells. The tumors displayed a remarkably homogeneous signature, with original activation of the nuclear factor κB pathway and a decrease in intratumor T-cell infiltration. Hence, IL-10R deficiency is associated with a high risk of developing B-cell lymphoma. Our results revealed an unexpected role of the IL-10R pathway in lymphomagenesis.
Introduction
Inflammatory bowel disease (IBD) encompasses a heterogeneous group of diseases that are characterized by chronic intestinal inflammation with a complex etiology.1-3 Very-early-onset IBDs (VEO-IBDs) are particularly severe, treatment-resistant conditions.4,5 Loss-of-function mutations in one or another of the interleukin-10 (IL-10) receptor’s two chains (IL-10R1 and IL-10R2) or in IL-10 itself are detected in about 20% of VEO-IBD patients.6-10 Studies of IL-10/IL-10R knockout mice have shown that this cytokine is a key checkpoint for maintaining immune homeostasis toward intestinal microbiota.11,12
Chronic intestinal inflammation is a known risk factor for the development of malignancies.13 Infection with Epstein-Barr virus (EBV) and long-term administration of immunosuppressive medication are also associated with a slight increase in the risk of lymphoma in young children with colitis14 and in adults with IBD.15 A Mendelian predisposition to B-cell lymphoma16 has been observed in some primary immunodeficiencies related to DNA repair (such as ataxia-telangiectasia17 ) and dominant-negative STAT3 mutations18,19 but not previously in the monogenic form of IBD. Here, we report on the occurrence and recurrence of diffuse large B-cell lymphomas (DLBCLs) in 5 children with IL-10R1 (n = 1) or IL-10R2 deficiencies (n = 4). None of the DLBCLs was related to EBV infection. Our data strongly suggest the existence of a direct relationship between IL-10R deficiency and the development of B-cell lymphomas.
Patients and methods
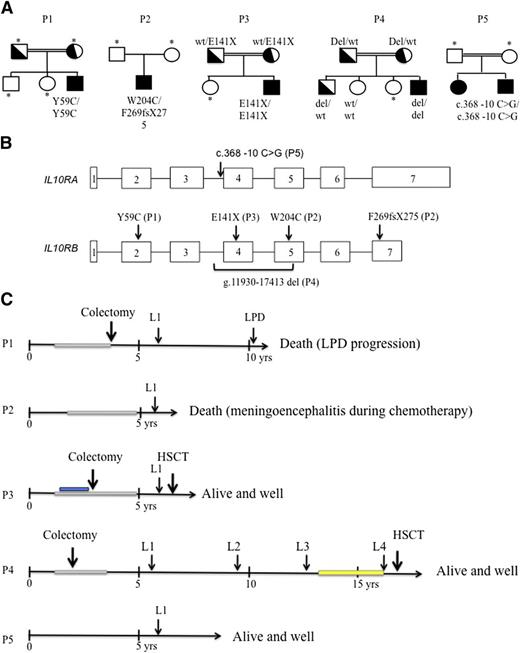
Of 18 children being monitored for VEO-IBD at Necker Children’s Hospital (Paris, France), 7 were diagnosed with a defect in the IL-10 pathway (IL-10R1, n = 1; IL-10R2, n = 6). Similarly, IL-10 pathway defects were diagnosed in 18 children (IL-10R1, n = 7; IL-10R2, n = 8; IL-10, n = 3) of 60 with VEO-IBD treated at Munich Children’s Hospital (Kotlarz et al9 and personal data). Four of the 14 patients with IL-10R2 deficiency and 1 of the 8 patients with IL-10R1 deficiency developed lymphomas. The clinical characteristics of the 5 patients with VEO-IBD who subsequently developed lymphoma (P1 through P5) are summarized in supplemental Table 1. The first signs of colitis (bloody diarrhea) were observed between 2 and 12 weeks of age. The patients subsequently developed recurrent stomatitis and perineal inflammation. In P1, P2, P3, and P5, several episodes of perianal abscesses were observed. All of the patients except P5 had recurrent flare-ups of cutaneous folliculitis, which had also started during the first year of life. Immunophenotype, T-cell function, immunoglobulin levels, and antibody functions of these patients were normal. In view of the persistent colitis, four patients were treated with steroids and various immunosuppressive drugs (azathioprine, n = 4; cyclosporine, n = 2; mycophenolate mofetil and anti–tumor necrosis factor α monoclonal antibodies [infliximab]) at between 1.5 and 5 years of age. P5 did not receive any immunosuppressive drugs prior to lymphoma onset. In view of the poor observed long-term remission, colectomy was performed in P1, P3, and P4 at between 3 and 4 years of age (Figure 1). Furthermore, P4 had brachysyndactyly of both hands. All participants or their parents/guardians gave their written informed consent to participation in accordance with the Declaration of Helsinki. The study was approved by the Comité Consultatif Pour la Recherche Biologique.
Genetic characteristics, time course of lymphoma development, and treatment in patients with IL-10R deficiency. (A) Pedigrees of the 5 patients with IL-10R1– and IL-10R2–deficient patients. Consanguinity (double horizontal bars), affected individuals (black boxes and circles), carriers (half-filled boxes and circles), and subjects not available for participation in the study (asterisks) are indicated. P1 carried a homozygous missense mutation in exon 2 (p.Y59C) of the IL-10RB gene. This mutation was absent from genome databases (including HGMD, Ensembl, and 1000 Genomes). P2 was a heterozygous composite with a frameshift mutation in exon 7 (F269fsX275) and a missense mutation in exon 5 (p.W204C) of IL-10RB. P3 carried an homozygous nonsense mutation in exon 3 of IL-10RB (p. E141X, as previously reported by Begue et al8 ). P4 displayed homozygous deletion (g.11930-17413 del) in IL-10RB. P5 carried a homozygous c.368-10 C>G mutation in intron 3 of IL-10RA. (B) Positions of the IL-10RA and IL-10RB mutations within the gene sequence. (C) Time course of lymphoma development and treatment. Immunosuppressive treatment with azathioprine is represented as a gray rectangle, and administration of an anti–tumor necrosis factor antibody is shown in blue. Maintenance therapy with 6-mercaptopurine and methotrexate was given in P4 after remission of L3 and is represented by a yellow rectangle. P1 (IL-10R2) died as a result of the progression of EBV-positive Hodgkin-like lymphoproliferative disease (LPD) (despite chemotherapy); P2 (IL-10R2) died of meningoencephalitis (of undetermined etiology) that occurred during chemotherapy; P3 (IL-10R2) was in remission from lymphoma when he received a transplant from a mismatched family donor and is now alive and well 12 months after HSCT; P4 (IL-10R2) was in remission from L4 when he received a transplant from a geno-identical sibling. He is alive and well 18 months after HSCT. P5 (IL-10R1) is in remission from lymphoma and is alive and well 3.4 years after completion of chemotherapy (HSCT is pending). wt, wild-type; yrs, years; occurrence shown by arrows (↓).
Genetic characteristics, time course of lymphoma development, and treatment in patients with IL-10R deficiency. (A) Pedigrees of the 5 patients with IL-10R1– and IL-10R2–deficient patients. Consanguinity (double horizontal bars), affected individuals (black boxes and circles), carriers (half-filled boxes and circles), and subjects not available for participation in the study (asterisks) are indicated. P1 carried a homozygous missense mutation in exon 2 (p.Y59C) of the IL-10RB gene. This mutation was absent from genome databases (including HGMD, Ensembl, and 1000 Genomes). P2 was a heterozygous composite with a frameshift mutation in exon 7 (F269fsX275) and a missense mutation in exon 5 (p.W204C) of IL-10RB. P3 carried an homozygous nonsense mutation in exon 3 of IL-10RB (p. E141X, as previously reported by Begue et al8 ). P4 displayed homozygous deletion (g.11930-17413 del) in IL-10RB. P5 carried a homozygous c.368-10 C>G mutation in intron 3 of IL-10RA. (B) Positions of the IL-10RA and IL-10RB mutations within the gene sequence. (C) Time course of lymphoma development and treatment. Immunosuppressive treatment with azathioprine is represented as a gray rectangle, and administration of an anti–tumor necrosis factor antibody is shown in blue. Maintenance therapy with 6-mercaptopurine and methotrexate was given in P4 after remission of L3 and is represented by a yellow rectangle. P1 (IL-10R2) died as a result of the progression of EBV-positive Hodgkin-like lymphoproliferative disease (LPD) (despite chemotherapy); P2 (IL-10R2) died of meningoencephalitis (of undetermined etiology) that occurred during chemotherapy; P3 (IL-10R2) was in remission from lymphoma when he received a transplant from a mismatched family donor and is now alive and well 12 months after HSCT; P4 (IL-10R2) was in remission from L4 when he received a transplant from a geno-identical sibling. He is alive and well 18 months after HSCT. P5 (IL-10R1) is in remission from lymphoma and is alive and well 3.4 years after completion of chemotherapy (HSCT is pending). wt, wild-type; yrs, years; occurrence shown by arrows (↓).
Details of the IL10RA and IL10RB gene mutations found in these 5 patients and the functional validation of these mutations (ie, the absence of IL-10R expression or an in vitro response to IL-10) are given in Figure 1 and supplemental Data. We further characterized these lymphomas by performing immunohistochemical studies, genome-wide array-based comparative genomic hybridization (aCGH) (n = 4), cytogenetic tests, fluorescence in situ hybridization (FISH) analysis (n = 4), and gene expression profiling (n = 3) on tumor samples. Furthermore, whole-exome sequencing of tumor DNA and counterpart genomic DNA was performed in 3 tumors from 2 patients. The methods are described in the supplemental Data. The study was conducted in accordance with the Declaration of Helsinki.
Results
Characteristics of lymphomas in 5 IL-10R–deficient patients
P1 through P5 all developed high-grade B-cell non-Hodgkin lymphoma (NHL) at between 5.5 and 6.5 years of age (Figure 1). Three further lymphomas occurred in P4 at 9.5, 12.1, and 16.5 years of age (referred to as P4-L2, P4-L3, and P4-L4, respectively) (Figure 1). The B-cell lymphomas were found at various locations within abdominal lymph nodes (n = 7), thoracic lymph nodes (n = 3), spleen (n = 5), liver (n = 3), and bone (n = 2) (Table 1). Lymphoproliferative mucosal lesions were not documented. All lymphomas were treated with chemotherapy, which was combined in all but one case (P1-L1) with administration of an anti-CD20 monoclonal antibody (Table 1). P1 was in long-term remission when an ultimately fatal EBV-related lymphoproliferative disease occurred 4.5 years after chemotherapy. P2 died during chemotherapy as a result of meningoencephalitis of unknown etiology. P3 underwent hematopoietic stem cell transplantation (HSCT) from a mismatched family donor 12 months after the completion of effective chemotherapy and is currently (12 months post-HSCT) in remission. Two months after remission of his fourth lymphoma, P4 underwent HSCT from a geno-identical sibling and is currently (18 months post-HSCT) in remission. P5 is currently in remission (3.3 years after the completion of chemotherapy), and HSCT is being considered. Data on immunoglobulin H (IgH) sequences, cytogenetic test results, and aCGH data on tumor cells from P4 confirmed that the first 3 lymphomas (at least) were distinct and had occurred sequentially.
Characteristics of lymphomas, treatment, and outcome in 5 IL10R deficient patients
| Patient . | Lymphoma . | Age at onset (years) . | IS at lymphoma onset . | Organ involvement . | Treatment . | Outcome (post HSCT) . |
|---|---|---|---|---|---|---|
| P1 | P1-L1 | 5.8 | 0 | Liver, spleen, retroperitoneal and intra-abdominal LN, bone | COP, COPADM × 2, CYM × 2 | Remission of lymphoma, EBV-related LPD at 10.3 y, died of progression |
| P2 | P2-L1 | 5.3 | Azathioprine | Liver, bone | COP, R-COPADM | Died during treatment |
| P3 | P3-L1 | 6.5 | Azathioprine | Liver, intra-abdominal LN | COP, R-COPADM × 2, R-CYM × 2, HSCT | Remission, alive (+12 mo) |
| P4 | P4-L1 | 5.5 | 0 | Spleen, pelvis, and inguinal LN | COP, COPADM × 2, CYM × 2 | |
| P4-L2 | 9.7 | 0 | Spleen, mediastinal, and intra-abdominal LN | COP, R-COPADM × 2, R-CYM × 2 | ||
| P4-L3 | 12.1 | 0 | Spleen, mediastinal, and intra-abdominal LN | COP, R-COPADM × 2, R-CYM × 2 | ||
| P4-L4 | 16.5 | 6MP and MTX | Spleen, mediastinal, and intra-abdominal LN | R-ICE × 4, HSCT | Remission, alive (+18 mo) | |
| P5 | P5-L1 | 6.5 | 0 | Abdominal LN | Up-front R prephase,* (2A4-2B4) × 4 | Remission, alive |
| Patient . | Lymphoma . | Age at onset (years) . | IS at lymphoma onset . | Organ involvement . | Treatment . | Outcome (post HSCT) . |
|---|---|---|---|---|---|---|
| P1 | P1-L1 | 5.8 | 0 | Liver, spleen, retroperitoneal and intra-abdominal LN, bone | COP, COPADM × 2, CYM × 2 | Remission of lymphoma, EBV-related LPD at 10.3 y, died of progression |
| P2 | P2-L1 | 5.3 | Azathioprine | Liver, bone | COP, R-COPADM | Died during treatment |
| P3 | P3-L1 | 6.5 | Azathioprine | Liver, intra-abdominal LN | COP, R-COPADM × 2, R-CYM × 2, HSCT | Remission, alive (+12 mo) |
| P4 | P4-L1 | 5.5 | 0 | Spleen, pelvis, and inguinal LN | COP, COPADM × 2, CYM × 2 | |
| P4-L2 | 9.7 | 0 | Spleen, mediastinal, and intra-abdominal LN | COP, R-COPADM × 2, R-CYM × 2 | ||
| P4-L3 | 12.1 | 0 | Spleen, mediastinal, and intra-abdominal LN | COP, R-COPADM × 2, R-CYM × 2 | ||
| P4-L4 | 16.5 | 6MP and MTX | Spleen, mediastinal, and intra-abdominal LN | R-ICE × 4, HSCT | Remission, alive (+18 mo) | |
| P5 | P5-L1 | 6.5 | 0 | Abdominal LN | Up-front R prephase,* (2A4-2B4) × 4 | Remission, alive |
2A4, dexamethasone, vincristine, cytarabine, etoposide, methotrexate, ifosfamide; 2B4, dexamethasone, vincristine, doxorubicin, methotrexate, cyclophosphamide;6MP, 6 mercaptopurine; BM, bone marrow; CHOP, cyclophosphamide, doxorubicin, vincristine, prednisone; COP, cyclophosphamide, vincristine, prednisone; COPADM, cyclophosphamide, vincristine, prednisone, doxorubicin, methotrexate; CYM, cytarabine, methotrexate; ICE, ifosfamide, carboplatin, etoposide; IS, immunosuppressive therapy; LN, lymph node; LPD, lymphoproliferative disease; MTX, methotrexate; R, rituximab.
Prephase, dexamethasone and cyclophosphamide.
Immunohistochemical characteristics of the B-cell lymphomas
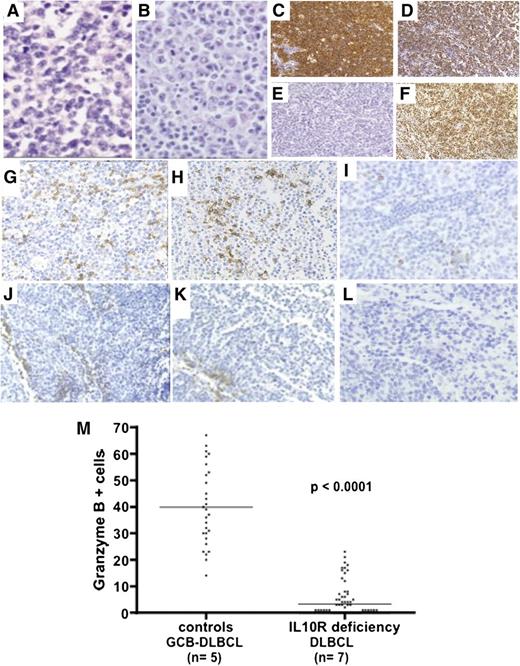
Immunohistochemical studies were performed on lymph node, liver, and spleen biopsies (Table 2 and Figure 2). All the lymphomas had common histologic features, with destruction of the normal architecture by diffuse proliferation of monomorphic, large, lymphoid cells (Figure 2A-B). There were many mitotic figures and apoptotic bodies. Immunohistochemical staining revealed that the large, atypical cells were CD20+ B cells (Figure 2C). Almost all the B cells (80% to 90%) stained positive for the proliferation-associated nuclear antigen Ki67 (Figure 2D). FISH assays based on an EBV-encoded RNA (EBER) probe were negative (Figure 2E). All lymphomas were BCL6+ (Figure 2F) and 4 were CD10+, suggesting a germinal center origin (Table 2). In 3 BCL6+/CD10– tumors, MUM1/IRF4 was absent (n = 1) or heterogeneous and was positive in <30% of cells (n = 2). Thus, these B-cell lymphomas shared many of the features of DLBCLs. CD3 staining revealed a predominantly perivascular distribution, with very few lymphoma-infiltrating T cells; these data contrasted with the detection of both perivascular and infiltrating T cells in 5 control germinal-center–like B-cell (GCB) DLBCLs (Figure 2). Granzyme B staining was negative or scattered in all the tested IL-10R–deficient samples (DLBCLs, n = 7) and was positive in 5 lymphoma-infiltrating T-cell control GCB DLBCLs (Figure 2).
Immunopathological features and IgH clonality of lymphomas in IL-10R–deficient patients
| Patient . | Lymphoma . | Biopsy . | Classification . | CD20 . | BCL2 . | CD10 . | BCL6 . | CD30 . | KI67 . | MUM-1 . | EBER . | c-REL . | IgH . | MYC . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | L1 | Liver | DLBCL | +++ | N/D | N/D | N/D | N/D | N/D | N/D | – | N/D | N/D | − |
| P2 | L1 | Liver | DLBCL | +++ | +/− | – | + | – | +++ | – | – | C++/N++ | Monoclonal | − |
| P3 | L1 | LN | DLBCL | +++ | – | + | + | – | +++ | – | – | C++/N+ | Monoclonal | − |
| P4 | L1 | Spleen | DLBCL | +++ | + | – | + | + | +++ | +/− | – | C++/N++ | Monoclonal | − |
| L2 | LN | DLBCL | +++ | – | + | + | + | +++ | + | – | C++/N+/− | Monoclonal | − | |
| L3 | LN | DLBCL | +++ | – | – | + | + | +++ | +/− | – | C++/N+ | Monoclonal | − | |
| L4 | LN | DLBCL | +++ | +/− | + | + | + | +++ | + | – | C++/N+ | N/D | − | |
| P5 | L1 | LN | DLBCL | +++ | + | + | + | – | +++ | +/− | – | C++/N+ | Monoclonal | N/D |
| Patient . | Lymphoma . | Biopsy . | Classification . | CD20 . | BCL2 . | CD10 . | BCL6 . | CD30 . | KI67 . | MUM-1 . | EBER . | c-REL . | IgH . | MYC . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| P1 | L1 | Liver | DLBCL | +++ | N/D | N/D | N/D | N/D | N/D | N/D | – | N/D | N/D | − |
| P2 | L1 | Liver | DLBCL | +++ | +/− | – | + | – | +++ | – | – | C++/N++ | Monoclonal | − |
| P3 | L1 | LN | DLBCL | +++ | – | + | + | – | +++ | – | – | C++/N+ | Monoclonal | − |
| P4 | L1 | Spleen | DLBCL | +++ | + | – | + | + | +++ | +/− | – | C++/N++ | Monoclonal | − |
| L2 | LN | DLBCL | +++ | – | + | + | + | +++ | + | – | C++/N+/− | Monoclonal | − | |
| L3 | LN | DLBCL | +++ | – | – | + | + | +++ | +/− | – | C++/N+ | Monoclonal | − | |
| L4 | LN | DLBCL | +++ | +/− | + | + | + | +++ | + | – | C++/N+ | N/D | − | |
| P5 | L1 | LN | DLBCL | +++ | + | + | + | – | +++ | +/− | – | C++/N+ | Monoclonal | N/D |
Lymphomas were classified according to the World Health Organization.
–, negative staining; +/–, heterogeneous staining; +++, strongly positive staining; C, cytoplasm; LN, lymph node; MYC, rearrangement of MYC locus detected by FISH; N, nuclear; N/D, not done.
Histochemical characteristics of lymphomas. (A-B) Hematoxylin-eosin staining (original magnification, ×400) of P3-L1 (intra-abdominal lymph node biopsy) and P4-L2 (inguinal lymph node biopsy), showing diffuse, massive infiltration by monomorphic large lymphoid cells. (C-F) Immunohistochemical studies on P3-L1 biopsy showing (C) CD20 staining, (D) Ki67 staining, (E) FISH for EBV-encoded RNA, and (F) BCL6 staining. (G-L) CD3, CD8, and granzyme B immunohistochemical studies: in control GCB DLBCL biopsy, (G) CD3 and (H) CD8 T cells were detected at perivascular and intratumor sites. (I) Granzyme B was detected in many T cells. In the IL-10R–deficient DLBCL (P4-L3) biopsy, (J-K) CD3/CD8 T cells were found in the perivascular region, whereas (L) no granzyme B was detected within the tumor (×200 magnification). (M) Counts of granzyme-B-positive cells in 5 control GCB DLBCLs and 7 lymphomas from IL-10R–deficient patients are shown. Counts per optic field (×400 magnification). Horizontal bars represent mean values.
Histochemical characteristics of lymphomas. (A-B) Hematoxylin-eosin staining (original magnification, ×400) of P3-L1 (intra-abdominal lymph node biopsy) and P4-L2 (inguinal lymph node biopsy), showing diffuse, massive infiltration by monomorphic large lymphoid cells. (C-F) Immunohistochemical studies on P3-L1 biopsy showing (C) CD20 staining, (D) Ki67 staining, (E) FISH for EBV-encoded RNA, and (F) BCL6 staining. (G-L) CD3, CD8, and granzyme B immunohistochemical studies: in control GCB DLBCL biopsy, (G) CD3 and (H) CD8 T cells were detected at perivascular and intratumor sites. (I) Granzyme B was detected in many T cells. In the IL-10R–deficient DLBCL (P4-L3) biopsy, (J-K) CD3/CD8 T cells were found in the perivascular region, whereas (L) no granzyme B was detected within the tumor (×200 magnification). (M) Counts of granzyme-B-positive cells in 5 control GCB DLBCLs and 7 lymphomas from IL-10R–deficient patients are shown. Counts per optic field (×400 magnification). Horizontal bars represent mean values.
Clonality
All tested specimens (6 of 6) displayed monoclonal rearrangements of the IgH gene locus (Table 2 and data not shown). Somatic mutations in IgV regions were detected in 2 of 3 cases studied (supplemental Table 2).
Tumor gene expression profiling
In an attempt to better characterize the lymphomas, gene expression profiles were determined for tumors P3-L1, P4-L2, and P5-L1 and compared with those in well-defined B-cell lymphoma entities and normal GCB cells from tonsils. Microarrays signatures from these various entities were downloaded from the open-access Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/; identifier, GSE 39503) and compared with samples from our patients in an unsupervised analysis. Remarkably, lymphomas P3-L1, P4-L2, and P5-L1 displayed similar profiles (R > 0.650) and clustered in the branch containing all the DLBCL (despite the latter’s known heterogeneity) and follicular lymphoma samples (R > 0.125) (Figure 3A). This clustering fits with the initial diagnosis and the immunohistochemical phenotype of the tumors. In view of the heterogeneity of DLBCLs, we sought to determine whether the patients’ lymphomas were primarily activated B-cell–like or GCB-like.16,20 In fact, they all clustered in the GCB branch, confirming a major enrichment in the GCB signature (Figure 3B) characterized by markers of germinal center differentiation (MME, BCL6, BCL7, LMO2, CD21 (CR2), CD22, CD27, CD86, XBP1, LRMP, SERPINA9, FAM3C, RGS13, LCK, and IRS1; P < .0001). Furthermore, the 3 lymphomas had additional functional pathways in common with GCB DLBCLs, such as upregulation of the neurotrophin signaling pathway (P < .0003), unsaturated fatty acid biosynthesis (P < .0005), the MAPK-signaling pathway (P < .003), and downregulation of the BCR signaling pathway (P < .02).
Gene expression profiles. (A) Unsupervised, hierarchical clustering of IL-10R lymphoma samples and other lymphoma samples. The dendrogram is based on expression of the most variable genes. Genes whose median expression was above background and whose standard deviation across all samples was >2 in at least 1 lymphoma entity were used for unsupervised hierarchical clustering (n = 3364). Samples from P3-L1, P4-L2, and P5L1 are shown in white and are indicated by arrows. (B) Hierarchical clustering of IL-10R lymphoma samples and activated B-cell–like (ABC) (light pink) and GCB (dark pink) DLBCL samples. Samples from P3-L1, P4-L2, and P5L1 are shown in white and are indicated by arrows. A list of 227 genes used to discriminate between ABC and GCB DLBCL was extracted via a nonparametric test implemented in the LIMMA software package (P < .05). Gene lists were submitted to the Cluster program for calculation of the Pearson correlation coefficient (as a similarity metric) and centroid linkage clustering. The results were visualized with TreeView software. BL, Burkitt’s lymphoma; CB, centroblasts; CC, centrocytes; cHL, classic Hodgkin’s lymphoma; FL, follicular lymphoma; MCL, mantle cell lymphoma; NPLHL, nodular predominance lymphocytic Hodgkin lymphoma; PMBL, primary mediastinal B-cell lymphoma; THRBL, T-cell/histiocyte-rich large B-cell lymphoma; Ton, tonsils.
Gene expression profiles. (A) Unsupervised, hierarchical clustering of IL-10R lymphoma samples and other lymphoma samples. The dendrogram is based on expression of the most variable genes. Genes whose median expression was above background and whose standard deviation across all samples was >2 in at least 1 lymphoma entity were used for unsupervised hierarchical clustering (n = 3364). Samples from P3-L1, P4-L2, and P5L1 are shown in white and are indicated by arrows. (B) Hierarchical clustering of IL-10R lymphoma samples and activated B-cell–like (ABC) (light pink) and GCB (dark pink) DLBCL samples. Samples from P3-L1, P4-L2, and P5L1 are shown in white and are indicated by arrows. A list of 227 genes used to discriminate between ABC and GCB DLBCL was extracted via a nonparametric test implemented in the LIMMA software package (P < .05). Gene lists were submitted to the Cluster program for calculation of the Pearson correlation coefficient (as a similarity metric) and centroid linkage clustering. The results were visualized with TreeView software. BL, Burkitt’s lymphoma; CB, centroblasts; CC, centrocytes; cHL, classic Hodgkin’s lymphoma; FL, follicular lymphoma; MCL, mantle cell lymphoma; NPLHL, nodular predominance lymphocytic Hodgkin lymphoma; PMBL, primary mediastinal B-cell lymphoma; THRBL, T-cell/histiocyte-rich large B-cell lymphoma; Ton, tonsils.
Although GCB DLBCLs were the closest malignant entities to lymphomas P3-L1, P4-L2, and P5-L1, a gene set enrichment analysis and a pathway analysis revealed that the latter also shared some features distinct from GCB lymphomas (supplemental Figure 2 and supplemental Table 3) and nonmalignant, proliferative tonsil B cells (supplemental Table 4 and supplemental Figure 3). When compared with GCB DLBCLs, lymphomas P3-L1, P4-L2, and P5-L1 showed greater expression of the spliceosome pathway (P < 2 × 10−12) and ubiquitin-mediated proteolysis (P < 2 × 10−11). These characteristics fit with the observed active cell cycling and frozen GC phenotype. The activation of many pathways involved in cancer (such as NOTCH, mTOR, ERBB2, and nuclear factor κB [NF-κB]) (P < 2 × 10−5) is consistent with the high proliferation rate observed in these lymphomas. Strikingly, expression of molecules related to cytotoxicity and dendritic cell function was low and thus agreed with the immunohistologic data on T cells. A comparison with the nonmalignant, proliferating B cells found in secondary lymphoid organs (supplemental Figure 3 and supplemental Table 4) confirmed the monoclonality of the lymphomas (relative to the polyclonal response observed in tonsils; P < 4 × 10−6) and, most importantly, revealed surprisingly low levels of AICDA, BACH2, and AFF3, 3 molecules that are usually upregulated in GCBs. Last, it is noteworthy that the axon guidance pathway was also enriched (P < .2 × 10−5).
Overall, this signature shows that the lymphomas had high proliferative and survival capacities. The low numbers of antitumor immune cells in the patients’ samples (as assessed by immunohistochemical staining and relative to conventional DLBCLs) was confirmed by the results of a transcriptome analysis.
Amplification and expression of the cellular homolog of the reticuloendotheliosis viral oncogene (c-REL) and constitutive activation of NF-kB
A cytogenetic analysis of the tumor cells revealed multiple chromosome rearrangements (supplemental Table 5), and an aCGH analysis was performed on genomic DNA samples from 4 tumors (P3-L1, P4-L1, P4-L2, and P5-L1). The presence of high copy number variations in the aCGH analysis of tumor cells agreed in full or in part with the results from a standard karyotype. Of the many genomic variations detected, the only recurrent variation was a gain of the 2p16 chromosomal region (supplemental Table 6 and Figure 4A). The minimum common amplified region defined by the breakpoint junctions of the P4-L1 sample spanned 0.25 Mb and included the PAPOLG, FLJ6341, REL, and PUS10 genes. P4-L2 notably exhibited full chromosome 2 trisomy and gain of the 2p16 chromosomal region (thus resulting in 2p16 tetraploidy). In P3-L1, gain of the 2p16 locus was confirmed by a FISH analysis with a probe that flanks the REL locus (Figure 4B).
Duplication of 2p16 in four lymphomas. (A) Schematic representation of chromosome 2, with the various duplicated regions spanning the 2p16 chromosomal band in samples P3-L1, P4-L1, P4-L2, and P5L1. The expanded region is indicated by a large blue column. (B) Involvement of the REL locus in IL-10R deficiency-associated lymphomas, as detected by FISH. The FISH analysis (using an RP11-1118K19 probe for the REL locus [red signals] and a chromosome 2 centromere probe [green signals]) revealed duplication of REL in metaphase spread and interphase nuclei from P3-L1.
Duplication of 2p16 in four lymphomas. (A) Schematic representation of chromosome 2, with the various duplicated regions spanning the 2p16 chromosomal band in samples P3-L1, P4-L1, P4-L2, and P5L1. The expanded region is indicated by a large blue column. (B) Involvement of the REL locus in IL-10R deficiency-associated lymphomas, as detected by FISH. The FISH analysis (using an RP11-1118K19 probe for the REL locus [red signals] and a chromosome 2 centromere probe [green signals]) revealed duplication of REL in metaphase spread and interphase nuclei from P3-L1.
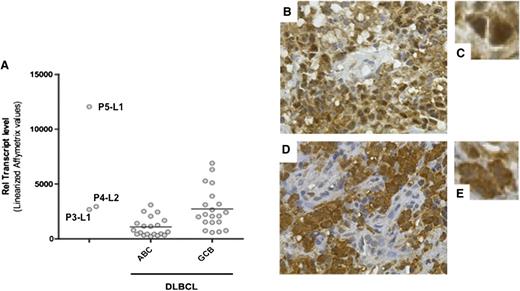
Given that REL locus amplification was found in all four tested cases, we assayed for expression of the cellular homolog c-REL. The expression levels of c-REL messenger RNA in P3-L1, P4-L2, and (above all) P5-L1 were similar to or greater than those found in control GCB DLBCL samples (Figure 5A). The expression and cellular localization of c-REL were thus examined by immunohistochemistry in all specimens but P1-L1. Strikingly, all tested DLBCL specimens stained positive for c-REL in the nucleus (Figure 5B-D and data not shown). Only P4-L2 exhibited mixed, predominantly cytoplasmic c-REL staining. A gene set enrichment analysis of P3-L1, P4-L2, and P5-L1 confirmed the upregulation of transcripts associated with REL functions (such as the cell cycle/proliferation, apoptosis/survival, adhesion/architecture, and innate immune cell functions). The most activated NF-κB–related pathways in the 3 lymphomas studied were Toll-like receptor signaling (P = 9.7 × 10−22) and apoptosis (P = 5.09 × 10−17) (supplemental Table 7). The observed overexpression of directly c-REL–responsive genes (such as CCL5, CXCL9, CCL19, TNFRSF17, TNFRSF9, BCL2A1, and BIRC3) suggested constitutive activation.
Expression of c-REL messenger RNA and protein. (A) c-REL transcript levels in tumor cells from P3-L1, P4-L2, and P5L1 in comparison with ABC DLBCL and GCB DLBCL controls. The horizontal bar represents the mean value. (B-E) Immunohistochemical staining of c-REL on tumor biopsies (B,D: original magnification, ×400; C,E: detail from B and D, respectively). (B-C) A P4-L4 sample showing nuclear expression of c-REL in tumor cells and the absence of staining in endothelial cells. (D-E) A P4-L1 sample showing c-REL expression in both the nucleus and cytoplasm of tumor cells.
Expression of c-REL messenger RNA and protein. (A) c-REL transcript levels in tumor cells from P3-L1, P4-L2, and P5L1 in comparison with ABC DLBCL and GCB DLBCL controls. The horizontal bar represents the mean value. (B-E) Immunohistochemical staining of c-REL on tumor biopsies (B,D: original magnification, ×400; C,E: detail from B and D, respectively). (B-C) A P4-L4 sample showing nuclear expression of c-REL in tumor cells and the absence of staining in endothelial cells. (D-E) A P4-L1 sample showing c-REL expression in both the nucleus and cytoplasm of tumor cells.
To directly assess the status of NF-κB activation, electrophoretic mobility shift assay analysis was performed on protein extracts prepared from P3-L1 and P5-L1. A strong constitutive NF-κB DNA binding activity composed of two major complexes was observed in both samples (supplemental Figure 1). The subunit composition of the NF-κB DNA-binding complexes was determined by supershift analysis. Antibody directed against RelA and p50 supershifted complex I almost completely. Complex II was effectively supershifted with anti-RelB and p50 antibodies (supplemental Figure 1). Altogether, these results suggest that both the canonical (ie, RelA) and noncanonical (ie, RelB) NF-κB activation pathways were constitutively activated in P3-L1 and P5-L1.
To further characterize the key disease events leading to lymphomagenesis in our patients, we performed whole-exome sequencing of P3-L1, P4-L1, and P4-L2 DNA and germline DNA in peripheral blood mononuclear cells from P3 and P4. We detected 26, 22, and 9 somatic mutations in 24, 21, and 9 genes, respectively, in the three tumors (supplemental Table 8). The numbers of somatic mutations found in these lymphomas are in agreement with data on DLBCL.21-23 Some of the somatic mutations previously associated with DLBCL were found in the patients’ lymphomas, including a mutation in MYD88 (p.S219C) (P3-L1).24 It is noteworthy that a somatic nonsense mutation in a master NF-κB regulator gene NFKBIA (p. Q68X) was identified in P4-L1, as previously described in classical Hodgkin lymphoma.25 Remarkably, we did not find any somatic mutations in genes involved in histone and chromatin modifications, in contrast to events described in DLBCL.26,27 Overall, the atypical but homogeneous gene expression profiles, the amplification of 2p16 in all tumors tested, the unconventional localization of c-REL, the constitutive activation of both canonical and noncanonical NF-κB pathways, and the presence of somatic mutations potentially conferring intrinsic activation of NF-κB suggested that the lymphomas in IL-10R–deficient patients represent a distinct/intermediate subtype of DLBCL.
Discussion
Here, we reported the occurrence of B-cell lymphomas in 5 IL-10R–deficient patients. Strikingly, one patient (P4) developed at least 3 distinct lymphomas. This hitherto unrecognized association between IL-10/IL-10R deficiency and lymphoma raises a number of questions. EBV-negative B-cell lymphomas were monoclonal. They contained highly proliferative, monomorphic, large B cells and showed several of the immunohistologic and molecular characteristics of DLBCLs. A gene expression analysis confirmed the origin of the lymphoma germinal center28,29 and revealed a strikingly homogeneous phenotype that was similar (but not identical) to that of GCB DLBCL. A key finding was the amplification of the 2p16 chromosomal region in four tested tumors. This feature has been reported previously in 17% to 26% of GCB DLBCLs16,30 and in Hodgkin lymphoma.31-33 The minimal amplified region contained REL but not the closely located oncogene BCL11A. In fact, REL encodes c-REL, a member of the NF-κB transcription factor complex involved in driving cell survival and proliferation.34 It has been shown that mutations in REL lead to constitutive NF-κB activation in B-cell lymphomas,29,35 with the detection of NF-κB transcription factors (including c-REL) in the nucleus,30,36,37 as observed here for c-REL. This characteristic also distinguishes the patients’ lymphomas from typical GCB DLBCLs.35,38 The observed pattern of gene overexpression is consistent with aberrant triggering of the NF-κB pathway and is further suggested by the observation of somatic mutations that potentially lead to gain of function of the NF-κB pathway in 2 of the 3 lymphomas tested. In addition, electrophoretic mobility shift assay combined with supershift analysis performed on 2 samples showed constitutive activation of both canonical and noncanonical NF-κB pathways. This pattern in unusual for GCB DLBCL.
The occurrence of 1 or more B-cell lymphomas in 5 patients with IL-10R deficiency cannot be viewed as a chance event, given that the incidence of NHL in childhood is 1 × 10−5 per year and that DLBCLs account for only 15% of childhood NHLs.39 Furthermore, the characteristics shared by the lymphomas of all the patients argue strongly against random occurrence. This increased risk of lymphomas might conceivably have been related to the immunosuppressive therapy that 4 of the 5 patients were receiving as treatment for IBD. In adult IBD, a fivefold increase in the risk of lymphoproliferative disorders has been observed in thiopurine-exposed patients.15,40 The CESAME study of almost 20 000 IBD patients reported 23 lymphoproliferative disorders (12 of which were EBV-related).15 However, none of the 53 children with idiopathic VEO-IBD in our series (who were also exposed to thiopurine-based immunosuppression for a comparable period) developed lymphomas.8 Likewise, patients with monogenic IBDs (such as chronic granulomatous disease or X-linked inhibitor of apoptosis protein deficiency) do not develop lymphoma more frequently than would be expected.41,42 Thus, in the context of IL-10R deficiency, azathioprine therapy is probably at most a weak risk factor for lymphomagenesis.
Our data therefore point to a direct relationship between the occurrence of lymphoma and a deficiency in the IL-10 pathway. Taking into account all reported patients with IL-10 (n = 5), IL-10R1 (n = 11), or IL-10R2 (n = 19) deficiencies,6-10,43 the frequency of developing lymphoma is estimated to be 36% (5 of 14) at the age of 7 years (in the absence of a previous HSCT). It is noteworthy that lymphomagenesis has not been reported in mice with impaired IL-10–mediated pathways, despite the onset of IBD,11,44 suggesting either the existence of marked differences between mice and humans or insufficient follow-up of the mice in an adequate environment. A role for the IL-10 pathway in human lymphomagenesis has been suggested by several studies showing a significant association between genetic variants in IL-10 and IL-10RB on one hand and NHL (and particularly DLBCL) on the other.45,46 Since disruption of the IL-10/IL-10R axis in both humans and mice leads to severe inflammatory enterocolitis,12 one can hypothesize that chronic inflammation may create a favorable milieu for B-cell lymphomagenesis, perhaps through protracted B-cell activation.47 However, few of the patients’ nodal lymphomas were in the gut, no mucosa-associated lymphomas were detected, and an activated B-cell lymphoma phenotype was not observed. Furthermore, VEO-IBD was not found to be associated with lymphoma when the IL-10/IL-10R pathway was unaffected. We thus conclude either that gut inflammation is not a main driver of lymphomagenesis or that a distinct pattern of inflammation (related to IL-10/IL-10R deficiency) is involved in lymphomagenesis. Future research will have to evaluate these possibilities.
Hence, two mutually nonexclusive models may account for the occurrence of B-cell lymphomas in the absence of a fully competent IL-10 signaling pathway. Given that IL-10 controls the proliferation of nonactivated B cells,48 defective signaling may lead to an increase in DNA replication and, in turn, acquisition of rearrangement events and somatic mutations. An alternative mechanism might involve a local T-cell immunodeficiency caused by impairment of the IL-10/IL-10R pathway. Surprisingly, in view of the immunosuppressive role of IL-10, recent studies of murine models have found that this cytokine promotes CD8 T-cell infiltration into tumors, local interferon-γ secretion, cytotoxicity, antigen presentation, and thus tumor suppression.49 Furthermore, IL-10 can directly induce cytotoxicity in intratumor CD8 T cells. Overall, these experimental data tend to confer a role of IL-10 in immunosurveillance. This hypothesis is attractive but remains to be tested. The scarcity of intratumor infiltrative granzyme-B–positive T cells in lymphomas from IL-10R–deficient patients (relative to control DLBCLs) shown in immunohistochemical analyses and confirmed at transcriptomic level is consistent with the presence of a deficiency in an IL-10–triggered local antitumor immunity pathway. It is noteworthy that patients with heterozygous STAT3 mutations (resulting in partial loss of function) are also prone to the development of B-cell lymphomas18,19 (albeit to a lesser extent). The observation that IL-10 signaling is STAT3 dependent supports the hypothesis in which the IL-10R/STAT3 pathway is involved in controlling lymphomas. An additional role of the IL-10R2 signaling pathway could be suggested by the fact that the IL-10R2 chain is common to the IL-22, IL-26, and interferon λ (IL-28A, IL-28B, and IL-29) receptors50 However, the occurrence of B-cell lymphoma with the very same characteristics in a patient with IL-10R1 deficiency makes this hypothesis very unlikely.
In conclusion, our results indicate that IL-10R deficiency predisposes to the development of a subtype of DLBCLs with germinal center origin characterized by original constitutive activation of the NF-κB pathway and a defective local T-cell immune response. Further work will be needed to fully elucidate the IL-10R pathway’s protective effect. Meanwhile, our data support early HSCT in patients with an impaired IL-10 pathway; this procedure is able to cure IBD and may well prevent the occurrence or recurrence of lymphoma.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and their families for their cooperation; Dr V. Minard, Dr C. Patte, Dr F. Doz, Dr D. Orbach, and Dr G. Couillaut for excellent care of the patients; Dr N. Cerf Bensussan for critical reading of the manuscript; C. Daussy and J. Pasquet for excellent technical assistance; C. Wouters, C. Janssens, and T. Roeskam for assistance in immunohistochemical staining; T. Molina for discussion and critical review of the manuscript; T. Petrella for providing patient samples; and the staff at the Necker Hospital’s “tumorotheque” biological resource center for analytical assistance.
This work was funded by grants from the Institut National de la Santé et de la Recherche Médicale, the Fondation pour la Recherche Médicale (B. Nadel), the Agence Nationale pour la Recherche (F.R.-L.), The German Research Foundation (SFB1054) (C.K.), and an Advanced Senior Grant from the European Research Council (PID Immune 249816) (A.F.).
Authorship
Contribution: B. Neven designed the research, collected and analyzed data, and participated in drafting the paper and clinical care; E.M. performed and interpreted microarray studies and participated in drafting the manuscript; J.B., D.C., and N.B. performed pathology and immunohistochemical studies and participated in drafting the manuscript; S.K., J.M.-P. and I.R.-W. performed cytogenetic and microarray studies and participated in drafting the manuscript; F.R.-L. and C.P. participated in genetic testing, functional analysis, and critical review of the manuscript; V.A. and F.D. performed clonality and somatic mutations in studies of IgV regions and participated in critical review of the manuscript; C.B. and P. N. performed whole-exome sequencing; F.S., P.F., S.B., O.G., J.-L.C., and O.H. participated in clinical care and critical review of the manuscript; D.K., D.M., and C.K. performed genetic and functional studies and participated in clinical care of patients and critical review of the manuscript; K.B. performed analysis of NF-κB activation status; V.B. designed the NF-κB activation study and participated in data analyses and drafting the manuscript; B. Nadel designed the research and participated in data analyses and drafting the manuscript; F.R. participated in data analyses, drafting the paper, and clinical care; and A.F. designed the research and participated in data analyses, drafting the manuscript, and clinical care.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alain Fischer, Hôpital Necker-Enfant Malades, Assistance Publique des Hôpitaux de Paris, 149 rue de Sevres, 75015 Paris, France; e-mail: alain.fischer@nck.aphp.fr.
References
Author notes
E.M., J.B., and S.K. contributed equally to this work.
B.N. and F.R. contributed equally to this work.


![Figure 4. Duplication of 2p16 in four lymphomas. (A) Schematic representation of chromosome 2, with the various duplicated regions spanning the 2p16 chromosomal band in samples P3-L1, P4-L1, P4-L2, and P5L1. The expanded region is indicated by a large blue column. (B) Involvement of the REL locus in IL-10R deficiency-associated lymphomas, as detected by FISH. The FISH analysis (using an RP11-1118K19 probe for the REL locus [red signals] and a chromosome 2 centromere probe [green signals]) revealed duplication of REL in metaphase spread and interphase nuclei from P3-L1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/23/10.1182_blood-2013-06-508267/4/m_3713f4.jpeg?Expires=1765889245&Signature=5G3g9mHLJgwXLLJBPniCD8BB0NjrTvSd3FFWDu~K6DO3MilV2mVAPh1d8hxCqouL7l4FDRskKo-Fe6rJV109fDX-jWw0DgLCehWyAUTsSvZ1Yb7i-WGb5l1D4HcF~RBmyUzZcW14obEUsqIpUhmGKB8IANsOQC5v8T8bHttb8Z67zH~MI8Kh1XSkCTs880zsChqFOuz4AqLzMiBVZ1HmFjkl-dBqxIcqKGQWUUqF8OOY2iQJYlXBZDAqSH8JsHqekU5efHyB-6Mu9A78NjnLWTWaJe053na8NF0Zh80wMQaFlWvIGceZdfXSgZROFjOqDrve77b1gVcNyNY8ggWlOg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal