Abstract
A growing number of recurrent gene mutations have been identified in AML through novel sequencing techniques. To uncover the functional consequences of these mutations and identify novel therapeutic targets, adequate model systems are needed. Such models should recapitulate the mutational landscape and genetic heterogeneity of AML as closely as possible. In this respect, analyses of primary tumor cells are superior to established permanent cell lines. Xenografts of primary AML blasts in immunodeficient mice thus may represent a valuable platform for functional analyses on a wide spectrum of AML subtypes. However, it has not been determined whether the mutational architecture of AML xenografts faithfully represents the human disease they originate from.
Fresh bone marrow or peripheral blood samples were obtained from adult AML patients. One to ten million cells were injected intravenously or intrafemorally into recipient NOD-scid IL2R-gammanull(NSG) mice. Engraftment was monitored by serial immunophenotyping using anti-human CD33 and anti-human CD45 antibodies. Mice showing >35% human cells in the peripheral blood or signs of disease were sacrificed, and xenotransplanted cells re-isolated from femurs and spleens. Using genomic DNA from xenografts and paired primary human specimens, we profiled mutations in 44 genes recurrently mutated in human hematologic malignancies by targeted, deep amplicon resequencing (Agilent Haloplex / Illumina MiSeq).
To date, 4 pairs of primary AML specimens and matched xenografts have been genetically characterized by targeted resequencing. We obtained between 480k and 1100k paired-end reads (2x250bp) per sample, resulting in >30x coverage for >98.7% of the target sequence. The median coverage for individual target regions ranged from 116-fold (CEBPA) to ∼5200-fold (FLT3).
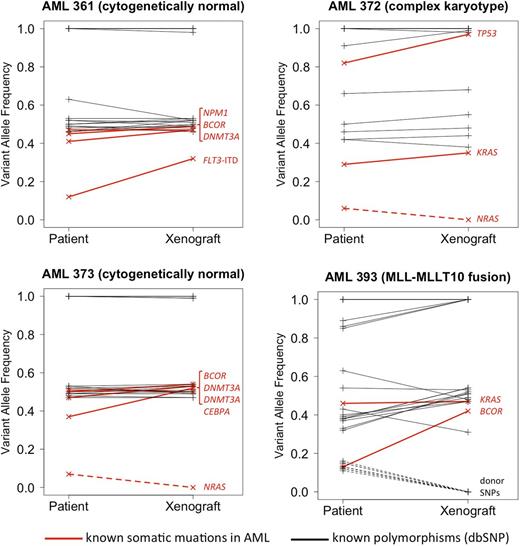
In each patient specimen, 2 to 5 known AML-associated driver mutations were identified in our panel of 44 genes. The Figure shows variant allele frequencies (VAFs) for mutations and known germline polymorphisms (SNPs; dbSNP database v137), in the patient samples and matched xenografts. All mutations that were present in the primary patient samples with a VAF of >10% were also found in the matched xenograft. None of the xenografts acquired new mutations that were undetectable in the original patient specimen. However, each sample pair showed evidence for clonal diversity and clonal evolution. In patient AML361, NPM1, DNMT3A and BCOR mutations were detected at a VAF slightly below .5, consistent with heterozygous mutations present in most cells in the primary specimen. Additionally, a FLT3-internal tandem duplication (ITD) was present at a lower VAF, likely representing a subclonal mutation. In the corresponding xenograft, the FLT3-ITD was observed in a significantly larger fraction of cells, suggesting that the FLT3-mutated clone had a relative growth advantage. Similarly, patient AML393 carried a subclone with mutated BCOR that became the dominant clone in the xenograft. Conversely, in patients AML372 and AML373 we found subclonal NRAS mutations (VAF, 6% and 7%, respectively) that were undetectable in the matched xenografts, indicating that the NRAS-mutated subclones did not engraft. We are currently studying parallel lines of xenografts generated from the same patient, followed by serial re-passaging in NSG mice, to better characterize growth patterns of such patient-derived subclones in our model. Updated results will be presented at the meeting.
Genotypes for known germline SNPs were fully concordant between the human specimens and matched xenografts in 3/4 pairs. The fourth patient (AML393), who had AML relapse after an allogeneic stem cell transplant, showed several SNPs with a low VAF that were not identified in the corresponding xenograft. These SNPs most likely originate from residual donor hematopoiesis in the patient, indicating that non-malignant cells of donor origin did not engraft in the NSG mouse.
Xenografts of primary AML blasts in NSG mice recapitulate the patterns of gene mutations observed in AML patients, and thus provide an opportunity to study the biology of various genetic AML subgroups. Deep, targeted amplicon resequencing can sensitively detect subclonal driver mutations, and in conjunction with the xenograft model can be used to study clonal diversity and clonal evolution in AML.
Greif:Illumina: Honoraria.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal