Abstract
Multiple Myeloma (MM) is a heterogeneous disease with highly variable survival. Gene expression profiling (GEP) classifiers, such as the EMC-92, can consistently distinguish high risk patients from standard risk patients. Other prognostic factors for MM include the international staging system (ISS) and FISH. Here we present a comparison of prognostic factors and introduce a novel stratification based on EMC-92 and ISS.
Scores were calculated for the GEP classifiers EMC-92, UAMS-70, UAMS-17, UAMS-80 and MRC-IX-6 for the following five studies: HOVON-65/GMMG-HD4 (n=328; GSE19784), MRC-IX (n=247; GSE15695), UAMS-TT2 (n=345; GSE2658), UAMS-TT3 (n=238; E-TABM-1138 and GSE2658) and APEX (n=264; GSE9782; for details, see Kuiper R, et al. Leukemia (2012) 26: 2406–2413). FISH data were available for the HOVON-65/GMMG-HD4 trial and the MRC-IX trial. ISS values were available for all datasets except UAMS-TT2. Univariate associations between markers and overall survival (OS) were investigated in a Cox regression analysis, using Bonferroni multiple testing correction. For pair wise analysis of markers, the significance in the increase of partial likelihood was calculated. In order to find the strongest combination (defined as the highest partial likelihood) of GEP-ISS, we compared these pair-wise on the same data. Training sets of classifiers were excluded for those analyses in which that specific classifier was tested. All survival models have been stratified for study. The calculations were done in R using the package survival.
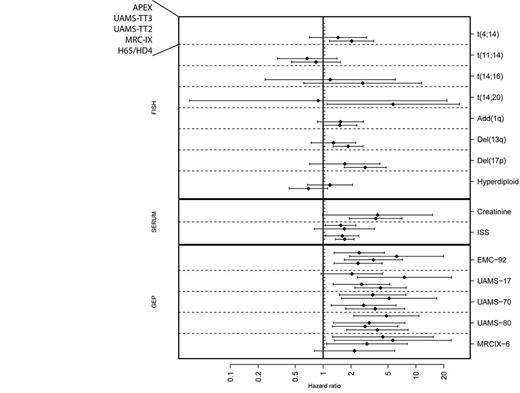
Prognostic value of FISH, GEP and serum markers was determined in relation to overall survival (Figure 1). GEP classifiers generally performed much better than FISH markers. Of 6 FISH markers with known adverse risk, del(17p), t(4;14), t(14;20) and del(13q) demonstrated a significant association only in one of two data sets with available FISH (HOVON-65/GMMG-HD4). GEP classifiers, on the other hand, are much more robust. Classifiers EMC-92, UAMS-70 and UAMS-80 significantly identify a high-risk population in all evaluated data sets, whereas the UAMS-17 and the MRC-IX-6 classifiers predict high-risk patients in three of four datasets. As expected, ISS staging demonstrated stable and significant hazard ratios in most studies (three out of four). Indeed, when evaluating a merged data set, both ISS and all evaluated GEP classifiers are strong prognostic factors independent of each other. Markers with additive value to each other include all combinations of GEP classifiers as well as the combination of GEP classifiers together with ISS. Tested in independent sets, the EMC-92 classifier combined with ISS is the best combination, as compared to other classifier-ISS combinations tested on the same independent data sets. The strongest risk stratification in 3 groups was achieved by splitting the EMC-92 standard risk patients into low risk, based on ISS stage I, and intermediate risk, based on ISS stage II and III. This stratification retains the original EMC-92 high-risk group, and is robust in all cohorts. The proportions of patients defined as low, intermediate and high risk for this combined EMC-92-ISS classifier are 31% / 47% / 22 % (HOVON-65/GMMG-HD4), 19% / 61% / 20 % (MRC-IX), 46% / 39% / 15 % (UAMS-TT3) and 32% / 55% / 13 % (APEX). Variability in low risk proportion is caused by the variable incidence of ISS stage I.
Univariate associations to OS per data set per marker. Hazard ratios and 95% confidence intervals are denoted by diamond shape and whiskers, respectively. Markers which cross the center line are not significant. Panels separated by dashed lines contain statistic values per marker for all data sets. The order of the data sets is given on the top left, i.e. from top to bottom APEX, UAMS-TT3, UAMS-TT2, MRC-IX and HOVON65/GMMG-HD4 per panel. Generally, FISH markers (top) are less robust than GEP markers (bottom).
Univariate associations to OS per data set per marker. Hazard ratios and 95% confidence intervals are denoted by diamond shape and whiskers, respectively. Markers which cross the center line are not significant. Panels separated by dashed lines contain statistic values per marker for all data sets. The order of the data sets is given on the top left, i.e. from top to bottom APEX, UAMS-TT3, UAMS-TT2, MRC-IX and HOVON65/GMMG-HD4 per panel. Generally, FISH markers (top) are less robust than GEP markers (bottom).
We conclude that GEP is the strongest predictor for survival in multiple myeloma and far more robust than FISH. Adding ISS to EMC-92 results in the strongest combination of any of the GEP classifier-ISS combinations. Stratification in low risk, intermediate and high risk may result in improved treatment and ultimately in longer survival of MM patients.
This research was supported by the Center for Translational Molecular Medicine (BioCHIP project)
van Vliet:Skyline Diagnostics: Employment. Mulligan:Millennium Pharmaceuticals: Employment. Morgan:Celgene: Consultancy, Honoraria, Membership on an entity’s Board of Directors or advisory committees; Millenium: Consultancy, Honoraria, Membership on an entity’s Board of Directors or advisory committees; Novartis: Consultancy, Honoraria, Membership on an entity’s Board of Directors or advisory committees; Merck: Consultancy, Honoraria, Membership on an entity’s Board of Directors or advisory committees; Johnson and Johnson: Consultancy, Honoraria, Membership on an entity’s Board of Directors or advisory committees. Goldschmidt:Celgene: Consultancy, Honoraria, Research Funding; Janssen: Consultancy, Honoraria, Research Funding; Novartis: Consultancy, Honoraria, Research Funding. Lokhorst:Genmab A/S: Consultancy, Research Funding; Celgene: Honoraria; Johnson-Cilag: Honoraria; Mudipharma: Honoraria. van Beers:Skyline Diagnostics: Employment. Sonneveld:Janssen-Cilag: Honoraria; Celgene: Honoraria; Onyx: Honoraria; Janssen-Cilag: Research Funding; Millenium: Research Funding; Onyx: Research Funding; Celgene: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal