Abstract
Critical pathway alterations contribute to heterogeneous biology and clinical course in chronic lymphocytic leukemia (CLL). The role of particular mutations for drug sensitivity has been demonstrated for CLL and other entities. Here we systematically investigated heterogeneity of drug response and genetic lesions in primary CLL. The comprehensive identification of pathway dependencies will lead to a better understanding of hematologic malignancies and this may be exploited by development of rational-driven treatment approaches.
Primary CLL cells were characterized through an ex vivo high-throughput drug screening platform in 384-well format. The drug library consisted of 67 compounds (inhibitors targeting critical pathways in CLL and other cancers as well as compounds with known activity against CLL cells (including ABT-263, CAL-101, dasatinib, fludarabine, ibrutinib)) at different concentrations. Heat inactivated human serum was supplemented to mimic micro-environmental conditions. CLL samples represented untreated CLL, sensitive or refractory disease as well as major genetic subgroups. Cell viability was assessed by quantification of ATP (CellTiterGlo®) 48 and 72 hours after drug application. To understand heterogeneous pathway dependencies, drug sensitivity was regressed on genetic profiles. Genetic characterization was performed by FISH, targeted sequencing of recurrent aberrations (BRAF, MYD88, NOTCH1, SF3B1, TP53) as well as whole exome sequencing.
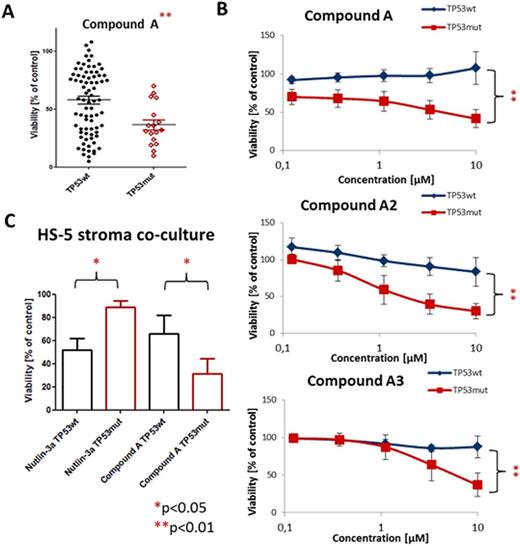
We report results from 97 primary CLL samples screened ex vivo. Validity of the approach was demonstrated by the response profiles of p53-interfering compounds: Nutlin-3a and fludarabine induced cell death in cells with wild-type (n=80) but considerably lessin CLL with mutated p53(n=17; Nutlin-3a 10µM: 51±18vs80±18;fludarabine 10µM: 40±36vs 67±23 [% viability of untreated control], both p<0.001). Furthermore inhibitors with related targets (e.g. drugs targeting the B-cell receptor (BCR) pathway: CAL-101, dasatinib, ibrutinib, R406) showed similar response profiles across patients, suggesting that prediction of drug effects can be based on functional and genetic properties of tumors. Patients can be divided into two major subgroups according to their sensitivity towards BCR and related kinase inhibition (e.g. Akt, Btk, Lyn, PI3K, Syk). A number of genotype-specific screening hits for single genetic aberrations were obtained. We identified compounds with preferential activity in groups with high risk aberrations (e.g. TP53 mutation, NOTCH1 mutation, 11q deletion; Fig. 1A) suggesting synthetic lethal interactions. Validation of the top hit with increased response of TP53-mutated CLL (compound A 10µM wt vs mutTP53: 58±31 vs 37±18 [% viability of untreated control]; Fig. 1A) confirmed genotype-specific cytotoxicity across concentration ranges (0.1-10µM; Fig 1B). Other compounds inhibiting the same target support our observation (compounds A2 and A3; Fig. 1B). The effect was reproducible in co-culture of CLL cells with HS-5 stroma cells (66±16 vs 32±13 [% viability of untreated control], p<0.05; Fig 1C).
Our data provide a first comprehensive map of genetically determined drug sensitivity in CLL. The work offers a novel functional classification of CLL based on drug sensitivity and identifies synthetic lethal interaction for TP53 mutant CLL.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal