Key Points
BCR-ABL1-like signature and IKZF1 deletions are clinically important to identify high-risk acute lymphoblastic patients.
Abstract
Most relapses in childhood B-cell precursor acute lymphoblastic leukemia (BCP-ALL) are not predicted using current prognostic features. Here, we determined the co-occurrence and independent prognostic relevance of 3 recently identified prognostic features: BCR-ABL1-like gene signature, deletions in IKZF1, and high CRLF2 messenger RNA expression (CRLF2-high). These features were determined in 4 trials representing 1128 children with ALL: DCOG ALL-8, ALL9, ALL10, and Cooperative ALL (COALL)-97/03. BCR-ABL1-like, IKZF1-deleted, and CRLF2-high cases constitute 33.7% of BCR-ABL1–negative, MLL wild-type BCP-ALL cases, of which BCR-ABL1-like and IKZF1 deletion (co)occurred most frequently. Higher cumulative incidence of relapse was found for BCR-ABL1-like and IKZF1-deleted, but not CRLF2-high, cases relative to remaining BCP-ALL cases, reflecting the observations in each of the cohorts analyzed separately. No relapses occurred among cases with CRLF2-high as single feature, whereas 62.9% of all relapses in BCR-ABL1–negative, MLL wild-type BCP-ALL occurred in cases with BCR-ABL1-like signature and/or IKZF1 deletion. Both the BCR-ABL1-like signature and IKZF1 deletions were prognostic features independent of conventional prognostic markers in a multivariate model, and both remained prognostic among cases with intermediate minimal residual disease. The BCR-ABL1-like signature and an IKZF1 deletion, but not CRLF2-high, are prognostic factors and are clinically of importance to identify high-risk patients who require more intensive and/or alternative therapies.
Introduction
The 5-year event-free survival (EFS) rate of childhood acute lymphoblastic leukemia (ALL) currently exceeds 80%.1 This is attributed to risk-adjusted treatment, which implements risk factors such as an early treatment response and cytogenetic abnormalities in leukemic cells, as well as the implementation of rationally designed phases in the treatment backbone of ALL including remission induction, central nervous system prophylaxis, and intensification and maintenance phases of treatment. More than half of the children in whom contemporary therapies failed were initially classified as non–high risk,2 highlighting the need for improved prognostic markers.
The IKAROS transcription factor, encoded by IKZF1, was recently associated with an unfavorable prognosis in childhood B-cell precursor (BCP) ALL.3-6 A monoallelic (often partial) deletion in IKZF1 results in a loss of its tumor suppressor function.7,8 EBF1, MSH2, and MCL1 were demonstrated as target genes for IKAROS.9 These genes contribute to the development of B cells (EBF1), DNA repair (MSH2), and survival/antiapoptosis (MCL1). Deletions in IKZF1 aborted the activation of these genes, which may facilitate B-cell leukemogenesis.9 Ten percent to 15% of BCP-ALL cases have an IKZF1 deletion, which therefore represents the most frequently observed genetic marker for an unfavorable outcome identified in children.3-6
The BCR-ABL1-like gene expression signature is a second recently identified unfavorable prognostic marker in childhood BCP-ALL. BCR-ABL1-like ALL was identified based on a gene expression signature of its leukemic cells, which is similar to that of BCR-ABL1–positive ALL, although these leukemic cells do not harbor the BCR-ABL1 translocation.5,10-12 Approximately 15% of BCP-ALL cases have BCR-ABL1-like ALL, which is associated with a 5-year EFS of <60%, similar to that observed in BCR-ABL1–positive BCP-ALL.5,10-12 More than 80% of BCR-ABL1-like ALL cases have abnormalities in genes involved in B-cell development, amongst others. IKZF1 deletions in ∼40%.5,10-12
A third recently identified adverse marker is linked to increased expression of cytokine receptor-like factor 2 (CRLF2).13-16 CRLF2 and interleukin-7 receptor α (IL7RA) form the thymic stromal-derived lymphopoietin receptor, which promotes B-cell progenitor proliferation by activating the Janus kinase (Jak) 2–signal transducer and activator of transcription 5 pathway.16 The high expression of CRLF2 in BCP-ALL arises either via a cryptic deletion, which juxtaposes CRLF2 to the P2RY8 promoter (P2RY8-CRLF2), or from the positioning of CRLF2 under control of the immunoglobulin heavy chain locus (IGH@-CRLF2) enhancer element.16,17 In addition, CRLF2 activity can be increased by gain-of-function mutations either in CRLF2 itself18 or in its partner gene, IL7RA.19 High CRLF2 messenger RNA (mRNA) expression (CRLF2-high) occurs in 5% to 14% of childhood BCP-ALL cases, and its incidence is remarkably high in Down syndrome ALL (50% to 55%).14,17,20
The presence of the aforementioned features was determined in several studies that used different inclusion and outcome criteria and a variety of techniques. Therefore, the relationship between these features, as well as their prognostic independence within a given patient population, remains unclear. Here, we studied these 3 molecular features in 1128 cases from 4 independent cohorts of children with newly diagnosed ALL. This study shows that both the BCR-ABL1-like signature and IKZF1 deletions are strong and independent prognostic features, whereas CRLF2-high is not uniformly predictive for an unfavorable outcome.
Methods
Detailed methods are described in the supplemental Appendix (see the Blood Web site).
Patients and leukemic cell samples
This study comprised 1128 children with newly diagnosed ALL in 3 consecutive Dutch Childhood Oncology Group trials (DCOG ALL-8, ALL-9, and ALL-10)21 and 2 German Cooperative ALL trials (COALL 06-97 and 07-03)22 that were combined for analysis and are referred to hereafter as COALL-97/03. Patients were stratified into low, intermediate, and high risk according to stratification markers in the treatment protocol (DCOG ALL-8,21 ALL-9,21 ALL10 [Supplemental Table 1], COALL 06-97,22 and 07-0322 ). In accordance with the Declaration of Helsinki, written informed consent was obtained from parents or guardians, and institutional review boards approved the use of excess diagnostic material for research purposes. Minimal residual disease (MRD) of DCOG ALL-10 patients was diagnosed by Sanquin (Amsterdam) and Erasmus University Medical Center (MC) Rotterdam.23 Cases were screened for hyperdiploidy (>50 chromosomes or DNA index ≥1.16), ETV6-RUNX1 and BCR-ABL1 fusion products, and rearrangements of TCF3 and MLL by routine diagnostic procedures; complementary data were generated by the Erasmus MC laboratory. Cases negative for these cytogenetic markers were assigned to the B-other group (supplemental Figure 1).
BCR-ABL1-like signature
New BCR-ABL1-like cases were identified using gene expression profiling of leukemic cells with Affymetrix U133 plus 2.0 microarrays. The same set of 110 gene probes and clustering procedure were used to identify novel BCR-ABL1-like cases in a newly arrayed cohort of 572 BCP-ALL cases using the previous study cohort of 107 DCOG cases as a reference (supplemental Figure 2; supplemental Table 2; Gene Expression Omnibus accession number GSE13351).10 Newly arrayed cases that hierarchically clustered together with previously identified BCR-ABL1-like and BCR-ABL1–positive cases were identified as new BCR-ABL1-like cases if proved negative for the BCR-ABL1 translocation (supplemental Figure 2; supplemental Data).10
IKZF1 deletions and mutations
IKZF1 deletions identified using the SALSA P335 ALL-IKZF1 Multiplex Ligation-dependent Probe Amplification (MLPA) assay (MRC-Holland) were confirmed using the P202 MLPA assay, a previously described in-house IKZF1 MLPA probe set,4 or comparative genomic hybridization analysis (SurePrint G3 180K array; Agilent).10 The presence of inactivating mutations in IKZF1 exons 4, 5, 6, and 8 was measured using Sanger sequencing in BCR-ABL1-like cases.4,5
Abnormalities in CRLF2 by genetic lesions and gene expression
P2RY8-CRLF2 and IGH@-CRLF2 (detected by interphase fluorescence in situ hybridization)16 are linked to high CRLF2 mRNA expression levels as determined by Affymetrix gene expression microarrays (supplemental Figure 3).24 Those cases with a signal intensity of the Affymetrix probe set 208303_s_at above the 90th percentile of the total BCP-ALL group were classified as CRLF2-high.
Statistics
Categorical and continuous variables were analyzed using nonparametric statistical tests. The cumulative incidence of relapse (CIR) with death as a competing event was calculated as described by Fine and Gray,25 considering relapse and nonresponse to induction chemotherapy as events. EFS rates were determined using Cox’s univariate and multivariate proportional hazard analyses with relapse, nonresponse, and death by leukemia considered as events. Differences with a P value < .05 were considered significant.
Results
A total of 1128 newly diagnosed children with BCP-ALL (n = 971) and T-lineage ALL (T-ALL; n = 157) were analyzed for the BCR-ABL1-like signature, IKZF1 deletions, and/or high CRLF2 expression.
The DCOG ALL-10 study subset was highly representative of the entire cohort of eligible cases, whereas the other 3 subsets had higher white blood cell (WBC) counts than the cases in each of the trial cohorts (supplemental Table 3). To prevent bias from known high-risk factors in the outcome analyses, CIR and Cox’s EFS analyses were analyzed in BCP-ALL patients after excluding BCR-ABL1–positive and MLL-rearranged cases (supplemental Figure 1).
Frequency and cumulative relapse incidences of BCP-ALL with BCR-ABL1-like expression signature
Ninety-four (16%) new BCR-ABL1-like cases were identified among the total of 572 BCP-ALL with gene expression data (Table 1; supplemental Figure 2). BCR-ABL1-like cases were identified in 52% (30/58) of DCOG ALL-10 B-other cases (ie, negative for hyperdiploidy, ETV6-RUNX1 fusion, TCF3 rearrangement, MLL rearrangement, or BCR-ABL1 translocation). Similar frequencies were observed in the other cohorts (50% ALL-8 [16/32], 64% ALL-9 [25/39], and 49% COALL-97/03 [23/47]). None of the T-ALL patients were found to carry the BCR-ABL1-like signature. BCR-ABL1-like cases were more often assigned to intermediate- and high-risk treatment than other BCP-ALL cases as a consequence of higher WBC count or age at diagnosis (supplemental Tables 6 and 7).
Frequency of new prognostic features in subtypes of pediatric ALL
| Subgroup . | BCR-ABL1-like . | . | IKZF1-deleted . | CRLF2-high . | |||
|---|---|---|---|---|---|---|---|
| n . | % . | Subtype . | n . | % . | n . | % . | |
| Hyperdiploid | 0/130 | 0 | 32/208 | 15 | 25/130 | 19 | |
| ETV6-RUNX1 positive | 0/162 | 0 | 7/225 | 3 | 0/162 | 0 | |
| TCF3 rearranged | 0/19 | 0 | 1/19 | 5 | 0/19 | 0 | |
| MLL rearranged | 0/12 | 0 | 2/27 | 7 | 1/12 | 8 | |
| B-other* | 94/176 | 53 | BCR-ABL1-like† | 37/92 | 40 | 15/94 | 16 |
| Remaining B-other | 16/80 | 20 | 10/82 | 12 | |||
| BCR-ABL1 positive | 0/24 | 0 | 16/23 | 70 | 0/24 | 0 | |
| Unknown BCP-ALL‡ | 0/49 | 0 | 43/233 | 18 | 5/49 | 10 | |
| Total BCP-ALL | 94/572 | 16 | 154/907 | 17 | 56/572 | 10 | |
| T-ALL | 0/80 | 0 | 5/157 | 3 | 1/80 | 1 | |
| Subgroup . | BCR-ABL1-like . | . | IKZF1-deleted . | CRLF2-high . | |||
|---|---|---|---|---|---|---|---|
| n . | % . | Subtype . | n . | % . | n . | % . | |
| Hyperdiploid | 0/130 | 0 | 32/208 | 15 | 25/130 | 19 | |
| ETV6-RUNX1 positive | 0/162 | 0 | 7/225 | 3 | 0/162 | 0 | |
| TCF3 rearranged | 0/19 | 0 | 1/19 | 5 | 0/19 | 0 | |
| MLL rearranged | 0/12 | 0 | 2/27 | 7 | 1/12 | 8 | |
| B-other* | 94/176 | 53 | BCR-ABL1-like† | 37/92 | 40 | 15/94 | 16 |
| Remaining B-other | 16/80 | 20 | 10/82 | 12 | |||
| BCR-ABL1 positive | 0/24 | 0 | 16/23 | 70 | 0/24 | 0 | |
| Unknown BCP-ALL‡ | 0/49 | 0 | 43/233 | 18 | 5/49 | 10 | |
| Total BCP-ALL | 94/572 | 16 | 154/907 | 17 | 56/572 | 10 | |
| T-ALL | 0/80 | 0 | 5/157 | 3 | 1/80 | 1 | |
B-other cases are negative for hyperdiploidy, ETV6-RUNX1, TCF3 rearrangement, MLL rearrangement, and BCR-ABL1 translocation.
BCR-ABL1-like cases are negatively tested for BCR-ABL1 translocation; BCR-ABL1–positive cases are allocated to the BCR-ABL1–positive group.
Unknown BCP-ALL cases are negative for BCR-ABL1 translocation, MLL rearrangement, and BCR-ABL1-like signature, but other cytogenetic lesions were not determined.
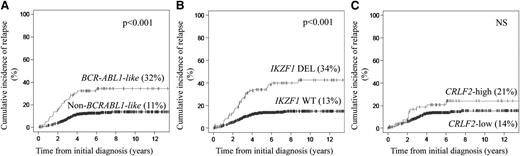
The 94 BCR-ABL1-like ALL had a 5-year CIR of 32% (95% confidence interval [CI] of 27% to 37%) compared with 11% (95% CI of 9% to 12%) for the 442 BCR-ABL1–negative/MLL wild-type BCP-ALL cases (P < .001; Figure 1A; supplemental Table 8). Similar significant differences were measured within each of the 4 study cohorts (supplemental Table 9; supplemental Figure 5). BCR-ABL1-like patients who were treated in the standard-risk (P = .05) and medium-risk (P = .06) arms of the most recent DCOG ALL-10 study exhibited a trend toward an unfavorable outcome (supplemental Figure 9).
CIR for the BCR-ABL1-like expression signature, IKZF1-deleted, and high CRLF2 mRNA expression in newly diagnosed children with BCR-ABL1–negative, MLL wild-type BCP-ALL. CIR with death as a competing event was calculated using the method of Fine and Gray25 in a pooled analysis of all 4 study cohorts and plotted against the time from initial diagnosis. For each feature, CIR at the 5-year follow-up is given in parentheses. (A) CIR of 94 BCR-ABL1-like and 442 non–BCR-ABL1-like BCP-ALL cases. (B) CIR of 136 IKZF1-deleted (DEL) and 721 IKZF1 wild-type (WT) cases. (C) Comparison between 55 cases with high CRLF2 expression and 481 cases with low CRLF2 expression.
CIR for the BCR-ABL1-like expression signature, IKZF1-deleted, and high CRLF2 mRNA expression in newly diagnosed children with BCR-ABL1–negative, MLL wild-type BCP-ALL. CIR with death as a competing event was calculated using the method of Fine and Gray25 in a pooled analysis of all 4 study cohorts and plotted against the time from initial diagnosis. For each feature, CIR at the 5-year follow-up is given in parentheses. (A) CIR of 94 BCR-ABL1-like and 442 non–BCR-ABL1-like BCP-ALL cases. (B) CIR of 136 IKZF1-deleted (DEL) and 721 IKZF1 wild-type (WT) cases. (C) Comparison between 55 cases with high CRLF2 expression and 481 cases with low CRLF2 expression.
Frequency and cumulative relapse incidences of IKZF1-deleted BCP-ALL
IKZF1 deletions were identified in 16% (136/857) of BCR-ABL1–negative and MLL wild-type BCP-ALL cases (Table 1). Whole-gene deletions encompassing IKZF1 exons 1 to 8 accounted for 40.4% of these deleted cases. In the remaining cases, partial deletions of exons 4 to 7 (26.5% of cases), exons 2 to 7 (9.6%), exons 4 to 8 (7.3%), exons 2 to 3 (8.1%), exons 2 to 8 (4.4%), and other exons (3.7%; supplemental Figure 4) were detected. Whole-gene or partial deletions in the protein-coding exons 2 to 8 were considered to compose 1 biological group, as reported previously.4-6
The highest incidence of IKZF1 deletions occurred in BCR-ABL1–positive (70%; 16/23) and BCR-ABL1-like (40%; 37/92) cases (Table 1). IKZF1 deletions were also identified in other BCP-ALL types, including hyperdiploid (15% of cases), TCF3-rearranged (5%), ETV6-RUNX1–positive (3%), MLL-rearranged (7%), and remaining B-other ALL cases (20%). Three percent of T-ALL cases (5/157) contained an IKZF1 deletion (Table 1).
The 136 IKZF1-deleted BCP-ALL cases had a 5-year CIR of 34% (95% CI of 30% to 38%) compared with 13% (95% CI of 12% to 15%) in the 721 nondeleted BCR-ABL1–negative/MLL wild-type cases (P < .001; Figure 1B; supplemental Table 8). Each treatment protocol exhibited similar prognostic differences, except for DCOG ALL-8–treated cases (supplemental Table 9; supplemental Figure 6). A deletion in IKZF1 was primarily predictive of poor outcome in patients treated in the medium-risk arm (P = .008) of the DCOG ALL-10 protocol (supplemental Figure 9). The most discriminating prognostic values for IKZF1 deletions were observed in hyperdiploid (P = .04) and non–BCR-ABL1-like B-other (P = .02) cases, whereas no prognostic difference was observed for IKZF1 deletions in the BCR-ABL1-like and ETV6-RUNX1–positive genetic subtypes in the DCOG ALL-10 study (Figure 2). Inactivating mutations in exons 4 to 6 and 8 were not identified in BCR-ABL1-like, IKZF1 wild-type cases.
CIR for IKZF1 status among subtypes of ALL. CIR with death as a competing event was calculated for the DCOG ALL-10 study cases with (A) high hyperdiploid ALL (>50 chromosomes) including 21 IKZF1-deleted and 87 wild-type cases; (B) ETV6-RUNX1–positive ALL including 4 IKZF1-deleted and 100 wild-type cases; (C) non–BCR-ABL1-like B-other ALL including 5 IKZF1-deleted and 22 wild-type cases; and (D) BCR-ABL1-like ALL including 10 IKZF1-deleted and 19 wild-type cases. For each feature, the CIR at 5 years of follow-up is indicated in parentheses.
CIR for IKZF1 status among subtypes of ALL. CIR with death as a competing event was calculated for the DCOG ALL-10 study cases with (A) high hyperdiploid ALL (>50 chromosomes) including 21 IKZF1-deleted and 87 wild-type cases; (B) ETV6-RUNX1–positive ALL including 4 IKZF1-deleted and 100 wild-type cases; (C) non–BCR-ABL1-like B-other ALL including 5 IKZF1-deleted and 22 wild-type cases; and (D) BCR-ABL1-like ALL including 10 IKZF1-deleted and 19 wild-type cases. For each feature, the CIR at 5 years of follow-up is indicated in parentheses.
Frequency and cumulative relapse incidences of BCP-ALL with high CRLF2 mRNA expression levels
High CRLF2 mRNA expression, irrespective of CRLF2-rearrangement status, was found in hyperdiploid (19% of cases), BCR-ABL1-like (16%), MLL-rearranged (8%), and B-other (12%) cases; whereas no CRLF2-high cases were observed in ETV6-RUNX1–positive, BCR-ABL1–positive, or TCF3-rearranged ALL (Table 1). High CRLF2 mRNA expression was observed in 1% (1/80) of T-ALL cases.
No statistically significant difference in CIR was found for CRLF2 mRNA expression status using the 90th percentile cutoff value in a pooled analysis of all patients or in the individual treatment cohorts, except for COALL-97/03 (P = .007; Figure 1C; supplemental Figure 7; supplemental Tables 8 and 9). The CIR analyses did not reveal a significant difference if the 92.5 and the 95th percentiles were used as cutoff values for high CRLF2 mRNA expression, nor did the CIR differ between cases with and without genetic CRLF2 aberration (IGH@-CRLF2 and P2RY8-CRLF2; supplemental Figure 8). In addition, CRLF2-high (90th percentile cutoff) had no prognostic value within the subset of BCR-ABL1-like cases (supplemental Figure 12).
Univariate and multivariate EFS analyses of BCP-ALL with BCR-ABL1-like signature, IKZF1 deletion, and/or CRLF2-high expression
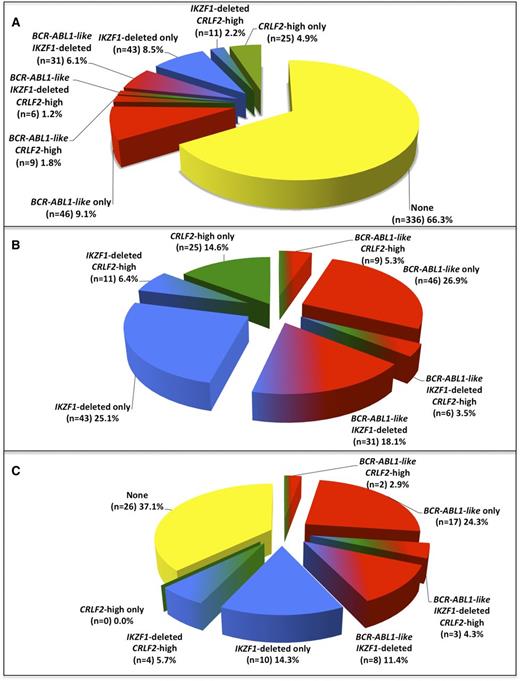
The results of the CIR analyses of BCR-ABL1-like, IKZF1-deleted, and CRLF2-high cases among the cohort of 507 BCR-ABL1–negative/MLL wild-type BCP-ALL patients for which all 3 features were determined (supplemental Table 4; supplemental Figure 10) were similar to those obtained by separate analyses of larger sample sets of BCR-ABL1-like signature (n = 536), IKZF1 status (n = 857), and CRLF2 expression (n = 536; Figure 1). The 3 molecular features together composed 33.7% of BCP-ALL cases (Figure 3A), of which the BCR-ABL1-like signature and IKZF1 deletions were the most prominent both as single lesions and in combination (Figure 3B; supplemental Table 5A). No relapses were predicted by the 14.6% of cases with high CRLF2 expression alone. In contrast, 62.9% (44/70) of all relapses in BCP-ALL (negative for BCR-ABL1 and MLL wild type) occurred in cases with the BCR-ABL1-like signature and/or an IKZF1 deletion. Of these 44 relapsed cases, 30 had a BCR-ABL1-like signature and 25 an IKZF1 deletion, and thus 11 cases had both a BCR-ABL1-like signature and an IKZF1 deletion (Figure 3C; supplemental Table 5B).
Frequency of BCR-ABL1-like, IKZF1-deleted, and CRLF2-high mRNA expression features among children with BCR-ABL1–negative, MLL wild-type BCP-ALL. (A) Distribution of cases with BCR-ABL1-like, IKZF1-deleted, and/or CRLF2-high mRNA expression in 507 children with BCR-ABL1–negative, MLL wild-type BCP-ALL. See also supplemental Table 5A. (B) Pie chart depicting the distribution of patients carrying 1 or more of the aforementioned features. Collectively, these 3 features constitute 33.7% of BCR-ABL1–negative, MLL wild-type BCP-ALL cases as shown in panel A. See also supplemental Table 5A. (C) Pie chart depicting the percentages of relapsed patients carrying 1, 2, or 3 features. In total, 62.9% of all relapsed cases were associated with BCR-ABL1-like and/or IKZF1-deleted features. See also supplemental Table 5B.
Frequency of BCR-ABL1-like, IKZF1-deleted, and CRLF2-high mRNA expression features among children with BCR-ABL1–negative, MLL wild-type BCP-ALL. (A) Distribution of cases with BCR-ABL1-like, IKZF1-deleted, and/or CRLF2-high mRNA expression in 507 children with BCR-ABL1–negative, MLL wild-type BCP-ALL. See also supplemental Table 5A. (B) Pie chart depicting the distribution of patients carrying 1 or more of the aforementioned features. Collectively, these 3 features constitute 33.7% of BCR-ABL1–negative, MLL wild-type BCP-ALL cases as shown in panel A. See also supplemental Table 5A. (C) Pie chart depicting the percentages of relapsed patients carrying 1, 2, or 3 features. In total, 62.9% of all relapsed cases were associated with BCR-ABL1-like and/or IKZF1-deleted features. See also supplemental Table 5B.
Univariate analysis of EFS of the 507 patients stratified according to treatment revealed that the BCR-ABL1-like signature (hazard ratio [HR] 3.5, 95% CI 2.2-5.7, P < .001), IKZF1 deletions (HR 2.8, 95% CI 1.7-4.6, P < .001) and WBC count of ≥50 cells per nL (HR 2.3, 95% CI 1.4-3.8, P = .001) were associated with an unfavorable prognosis; whereas high CRLF2 expression, age, and gender were not (supplemental Table 10A). Multivariate analysis stratified according to treatment protocol suggested that the BCR-ABL1-like signature, a high WBC count, and male gender had independent prognostic value (HR 3.1, 95% CI 1.8-5.2, P < .001; HR 1.8, 95% CI 1.1-3.1, P = .02; HR 1.8, 95% CI 1.1-3.0, P = .02, respectively). In a similar model applied to IKZF1 status, deletions in IKZF1 (HR 2.5, 95% CI 1.5-4.2, P = .001), high WBC count (HR 2.0, 95% CI 1.4-3.4, P = .007), and male gender (HR 1.7, 95% CI 1.1-2.9, P = .03) were significant. High CRLF2 expression did not have prognostic value in this multivariate approach (supplemental Table 10B).
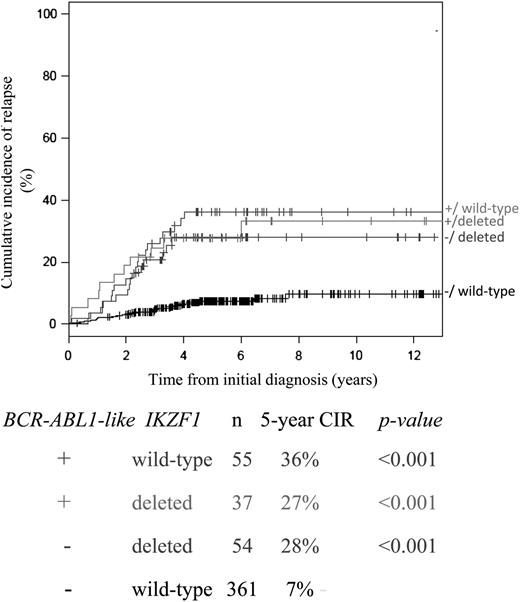
Interestingly, among the BCR-ABL1-like cases, CIR was not affected by the IKZF1 deletion status. Conversely, CIR among the IKZF1-deleted cases was not affected by the BCR-ABL1-like signature (Figure 4). A multivariate interaction model including both BCR-ABL1-like signature and IKZF1 status showed that the HRs for BCR-ABL1-like ALL alone (HR 5.3, 95% CI 2.9-9.7), IKZF1-deleted cases alone (HR 4.4, 95% CI 2.3-8.4), or combined BCR-ABL1-like signature and IKZF1-deleted cases (HR 3.7, 95% CI 2.0-8.4) were similarly adverse for each of these categories compared with cases without the BCR-ABL1-like signature and nondeleted IKZF1 (P < .001, treatment-stratified analysis; supplemental Table 10C).
Interaction between BCR-ABL1-like and IKZF1 status for predicting relapse. CIR with death as a competing event for cases with or without a deletion in IKZF1 among BCR-ABL1-like and non–BCR-ABL1-like ALL cases. The box below the graphs indicates the 5-year CIR, and P values compared with the reference group of non–BCR-ABL1-like and IKZF1 wild-type cases (black line). BCR-ABL1–positive and MLL-rearranged cases were excluded from this analysis.
Interaction between BCR-ABL1-like and IKZF1 status for predicting relapse. CIR with death as a competing event for cases with or without a deletion in IKZF1 among BCR-ABL1-like and non–BCR-ABL1-like ALL cases. The box below the graphs indicates the 5-year CIR, and P values compared with the reference group of non–BCR-ABL1-like and IKZF1 wild-type cases (black line). BCR-ABL1–positive and MLL-rearranged cases were excluded from this analysis.
Relationship between MRD status and BCR-ABL1-like ALL, IKZF1-deleted cases, and CRLF2-high cases in DCOG ALL-10 study
MRD levels were prospectively measured for eligible DCOG ALL-10 cases and used to stratify patient risk (see supplemental Appendix). All 3 MRD categories contained BCR-ABL1-like, IKZF1-deleted, and/or CRLF2-high cases, but the distribution was skewed toward the MRD-intermediate and MRD-high categories for both BCR-ABL1-like (P = .001) and IKZF1-deleted (P = .03) cases relative to control BCP-ALL cases (supplemental Table 6). More precisely, 17.9% (5/28), 6.7% (2/30), and 5.6% (1/18) of the BCR-ABL1-like, IKZF1-deleted, and CRLF2-high BCP-ALL cases, respectively, were assigned to the MRD-high category (supplemental Table 11), collectively, representing 5 distinct BCP-ALL cases. Remarkably, BCR-ABL1-like was the common denominator of these 5 MRD-high cases, although this finding is preliminary (supplemental Table 11). Both the BCR-ABL1-like signature (HR 3.7, 95% CI 1.2-11.5, P = .026) and IKZF1-deleted cases (HR 2.7, 95% CI 1.0-7.0, P = .043), but not CRLF2-high cases, remained predictive of a poor clinical outcome among the MRD-intermediate BCP-ALL cases (supplemental Table 12).
Discussion
Recently, new molecular features were discovered that can predict an unfavorable outcome in childhood BCP-ALL and include a BCR-ABL1-like gene expression signature,10,12 a (partial) deletion in IKZF1,3-6 and high expression of CRLF2.13-16 These factors are predictive among cases that do not exhibit known poor-prognostic features and therefore have potential clinical value. However, to date, these leukemia cell characteristics have been assessed separately in discrete patient cohorts with different inclusion criteria, limiting an analysis of their independent prognostic relevance. Moreover, variable technical approaches and cutoff values for positivity have hampered comparisons between published reports.
This is the first study to analyze the prognostic value and interaction between these 3 recently identified molecular features using standardized conditions and uniform inclusion criteria for newly diagnosed childhood ALL patients enrolled in 4 different treatment protocols. We show that 16%, 17%, and 10% of BCP-ALL cases have the BCR-ABL1-like signature, IKZF1 deletions, and high CRLF2 mRNA expression, respectively. These frequencies are consistent with previous observations in single-marker studies in BCP-ALL.4-6,10,12,15
A relatively high frequency of IKZF1 deletions was found in BCR-ABL1-like cases identified in the present study (40%) and those described by the St. Jude Children’s Research Hospital and Children’s Oncology Group (80% to 90%), although the technique for the identification of BCR-ABL1-like cases was different.11,12 The St. Jude Children’s Research Hospital and Children’s Oncology Group studies identified the BCR-ABL1-like (or Ph-like) cases by using gene expression profiling and gene set enrichment analysis,5 a recognition of outliers by sampling ends clustering approach (so-called R8 cluster),11,12 or a prediction analysis for microarrays classifier model.11,26 These approaches used a dynamic, nonfixed classifier, which differed per study. In the present study, we used a fixed model using our previously identified 110 gene probes and the same identical hierarchical clustering method as in our discovery study.10 A second difference is that the St. Jude Children’s Research Hospital and Children’s Oncology Group studies analyzed patients enrolled in high-risk protocols (eg, P9906 and AALL0232), whereas in the present study the BCR-ABL1-like cases were identified in both standard-, medium-, and high-risk arms. In the present more population-based study, the BCR-ABL1-like signature, IKZF1 deletions, and CRLF2-high cases together constitute 33.7% of BCR-ABL1–negative, MLL wild-type BCP-ALL cases, of which concomitant BCR-ABL1-like signature and IKZF1 deletion was the most frequent (Figure 3).
The BCR-ABL1-like and IKZF1-deleted features were predictive of an unfavorable prognosis in most studied treatment protocols, including the MRD-based DCOG ALL-10 study. This indicates that BCR-ABL1-like and IKZF1-deleted cases are not yet recognized by other means and that no sufficient risk-adapted treatment is provided in contemporary regimens. Correspondingly, multivariate analysis revealed that the adverse prognoses of BCR-ABL1-like signature and IKZF1-deleted cases are independent of the conventional risk factors and remained predictive for an unfavorable outcome in the group of patients with intermediate MRD levels. This highlights the value of both features as strong independent risk factors. Importantly, both BCR-ABL1-like and IKZF1-deleted cases were identified across all stratification arms and were either exclusively (BCR-ABL1-like) or primarily (IKZF1 deletion) present in the B-other category, for which new prognostic markers are urgently needed (Table 1).
The biology underlying BCR-ABL1-like ALL is largely unknown, but there is a relatively high frequency of deletions in B-cell development genes (eg, IKZF1, EBF1, PAX5, and VPREB1) and in cell cycle genes (CDKN2A/2B), whereas mutations and translocations are mainly observed in genes associated with cytokine signaling (eg, CRLF2, IL7R, and EPOR) and kinase pathways (eg, ABL1 and JAK2).5,10-12,26,27 The genomic deletions are most often recurrent (eg, IKZF1 deletion), whereas the mutations and translocations most often affect single genes and different fusion partners. Exceptions are recurrent lesions found for CRLF2 and JAK-family genes and to a lesser extent for the EBF1-PDGFRB translocation in an independent validation cohort.11,27 In the present cohort of BCP-ALL cases, we infrequently found mutations and translocations affecting JAK2 (<2%). These lesions were mainly found in non–BCR-ABL1-like cases with high CRLF2 expression and represent only a minority of our CRLF2-high cases (J. Marchante, M.L.D.B, unpublished observation). This low frequency of JAK2 abnormalities, therefore, cannot explain the poor prognosis ascribed to the BCR-ABL1-like signature or an IKZF1 deletion in the present study. The ABL1-translocated cells were shown to be sensitive to the ABL1 kinase domain inhibitors imatinib and dasatinib, whereas JAK2-affected leukemic cells were sensitive to the JAK-inhibitor ruxolitinib.11,28 Although no common genomic denominator has yet been identified for BCR-ABL1-like ALL, screening of patients for the presence of ABL1 and JAK-family lesions may identify BCR-ABL1-like patients who may benefit from implementing targeted tyrosine kinase inhibitors in their treatment.
IKZF1 deletions were observed in every ALL subtype, including at low frequency in T-ALL, and primarily showed unfavorable prognostic value in hyperdiploid and non–BCR-ABL1-like B-other cases (Figure 2; supplemental Figure 11). Together, this indicates that a broad range of ALL types are affected by aberrant IKAROS function. IKZF1 deletions contribute to uncontrolled proliferation in BCR-ABL1–positive ALL,7,8 most likely by interfering with c-Myb and Bmi1-regulated transcription of B-cell differentiation genes.29,30 Interestingly, IKZF1 deletions had no additive adverse effect on the prognosis of BCR-ABL1-like cases (Figure 4). Moreover, the unfavorable prognosis of BCR-ABL1-like cases without IKZF1 deletion could not be explained by the presence of inactivating mutations in IKZF1. The role of aberrant IKAROS in BCR-ABL1-like and other ALL subtypes is yet unclear, and the current findings underscore the importance of elucidating these mechanisms. One intriguing observation is that expression of the immunoglobulin joining chain (IGJ) gene is deregulated in both IKZF1-deleted and BCR-ABL1-like ALL.10,12 The immunoglobulin joining chain is involved in immunoglobulin maturation,31-33 whereas IKAROS controls the processing of immature B-cell receptors into immunoglobulins.34,35 Whether IKZF1 deletions functionally affect this maturation process clearly warrants further investigation.
In this study, high CRLF2 mRNA expression was the weakest prognostic feature, being predictive of an unfavorable outcome in COALL-97/03 but not in the 3 DCOG-trials. The group with high CRLF2 mRNA expression included all cases with genetic CRLF2 aberrations (supplemental Figure 3), which is in correspondence with previous observations.15 We cannot rule out that high CRLF2 expression will show some prognostic relevance when larger sample sizes are used. We therefore do not want to state that deregulated CRLF2 completely lacks prognostic relevance in pediatric BCP-ALL, but we want to emphasize that the discriminative prognostic value of high CRLF2 mRNA expression is lower than that of the BCR-ABL1-like signature and IKZF1 deletion. The prognostic and clinical value of CRLF2 expression levels in childhood BCP-ALL is controversial and appears to be dependent on the treatment protocol: predictive for adverse outcome in US and German (eg, COALL97/03 in the present study),15,24,36,37 but not Dutch (DCOG ALL-8, ALL-9, and ALL-10) and British treatment protocols.14,37 Other possible explanations of this discrepancy include differences in methodology (eg, mRNA expression vs genomic rearrangement) and inclusion criteria (eg, inclusion of cases with Down syndrome or cases of Hispanic/Latino ethnicity)17,24 and the possibility that the treatment and/or stratification criteria could mitigate any adverse effects of high CRLF2 expression. Moreover, we observed that CRLF2-high cases that lacked IKZF1 deletions and/or the BCR-ABL1-like signature did not relapse, suggesting that the prognostic value attributed to this factor may arise from the co-occurrence of IKZF1 deletions and/or the BCR-ABL1-like signature.13-16
We conclude that the BCR-ABL1-like signature and IKZF1 deletions in BCP-ALL cells are strong independent prognostic features that can identify patients at high risk for treatment failure, even among cases with intermediate MRD levels. In contrast, high CRLF2 mRNA expression lacks such discriminative prognostic power in our studies. These findings highlight the important clinical value of these 2 molecular features for identifying patients at high risk for treatment failure who might otherwise remain unrecognized. The implementation of BCR-ABL1-like signature as new risk factor in DCOG trials is being considered, and to this aim, a feasibility study for gene expression profiling in clinical practice has been initiated. Because the IKZF1 deletion can be detected using DNA, which is less vulnerable to degradation than mRNA, this genomic marker can be more easily implemented in clinical practice. In the recently initiated DCOG ALL-11 protocol, it was therefore decided to first implement the IKZF1 status as new risk factor. In the DCOG ALL-11 trial, medium-risk patients with a deletion in IKZF1 will receive an additional year of maintenance therapy.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE13351).
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Dutch Cancer Society (KWF; grants Erasmus MC Rotterdam 2007-3718 [M.L.D.B., R.P.] and KUN 2009-4298 [R.P.K., P.M.H.]); the Sophia Foundation for Scientific Research (SSWO; grant 2010-138) (A.v.d.V., M.L.D.B.); the Pediatric Oncology Foundation Rotterdam (M.L.D.B., R.P.); the National Institutes of Health National Cancer Institute (research grant R37 CA36401) (W.E.E.); St. Jude Children’s Research Hospital, Memphis, TN (W.E.E.); and the Kay Kendal Leukaemia Fund, United Kingdom (L.J.R, C.J.H.).
These funding sources had no role in the collection, analysis, or interpretation of the results, or in writing the manuscript and the decision to submit this manuscript.
Authorship
Contribution: A.v.d.V., R.P., R.P.K., and M.L.D.B. designed the study; A.v.d.V., M.E.W., W.E.E., and M.L.D.B. performed the gene expression studies to identify BCR-ABL1-like and CRLF2-high expressing patients; A.v.d.V., E.W., M.E.W., S.V.V.R., F.V.L., R.P.K., and M.L.D.B. performed the MLPA analyses, genomic polymerase chain reaction, and array-comparative genomic hybridization to identify and validate deletions in IKZF1; M.E.W., L.J.R., and C.J.H. performed CRLF2 fluorescence in situ hybridization analyses; P.M.H., H.A.D.G.-K., G.E., M.A.H., and E.S. collected and provided patient characteristics and clinical responses; V.H.J.v.d.V. was responsible for the MRD analyses; A.v.d.V., L.M.K., H.A.D.G.-K., and D.R. contributed to the statistical analyses of the data; and all authors read, revised, and approved the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Monique L. Den Boer, Erasmus MC-Sophia Children’s Hospital, Doctor Molewaterplein 60, 3015GJ Rotterdam, The Netherlands; e-mail: m.l.denboer@erasmusmc.nl.
References
Author notes
A.v.d.V., E.W., R.P.K., and M.L.D.B. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal