Key Points
MLL1 does not require interaction with menin to maintain hematopoietic stem cell homeostasis.
Menin and MLL1 are both critical during B-cell differentiation, but largely through distinct pathways.
Abstract
Mixed Lineage Leukemia (MLL1) translocations encode fusion proteins retaining the N terminus of MLL1, which interacts with the tumor suppressor, menin. This interaction is essential for leukemogenesis and thus is a promising drug target. However, wild-type MLL1 plays a critical role in sustaining hematopoietic stem cells (HSCs); therefore, disruption of an essential MLL1 cofactor would be expected to obliterate normal hematopoiesis. Here we show that rather than working together as a complex, menin and MLL1 regulate distinct pathways during normal hematopoiesis, particularly in HSCs and B cells. We demonstrate the lack of genetic interaction between menin and MLL1 in steady-state or regenerative hematopoiesis and in B-cell differentiation despite the fact that MLL1 is critical for these processes. In B cells, menin- or MLL1-regulated genes can be classified into 3 categories: (1) a relatively small group of coregulated genes including previously described targets Hoxa9 and Meis1 but also Mecom and Eya1, and much larger groups of (2) exclusively menin-regulated and (3) exclusively MLL1-regulated genes. Our results highlight the large degree of independence of these 2 proteins and demonstrate that menin is not a requisite cofactor for MLL1 during normal hematopoiesis. Furthermore, our data support the development of menin-MLL1–disrupting drugs as safe and selective leukemia targeting agents.
Introduction
Translocations disrupting the Mixed Lineage Leukemia (MLL1) gene occur frequently in infant and secondary acute leukemia as well as ∼10% of de novo acute myelogenous leukemia.1 Because MLL1 translocations confer very poor prognosis and patients are often young, new strategies that selectively target the actions of oncogenic MLL1 fusion proteins (MLL1-FPs) have been intensively investigated. The most common result of MLL1 translocation is the fusion of the MLL1 N terminus in-frame to a partner protein to produce MLL1-FPs. The native MLL1 protein is a histone methyltransferase at the core of a chromatin-modifying complex.2,3 This complex is widely distributed in most cell types and functions in hematopoietic, neural, and vascular development and homeostasis.4-7 Purification of native MLL1 complexes revealed that menin, a tumor suppressor protein, was a component of this and the related MLL2 chromatin-modifying complex.8,9
Several lines of evidence demonstrate that MLL1-FPs require menin interaction for leukemogenesis. First, mutations in the menin interaction motif of MLL1-ENL abolish its transforming activity in hematopoietic cells.10 Second, menin is required for MLL1-FPs to bind to LEDGF, a PWWP domain–containing protein that participates in targeting of the MLL complex to chromatin.11 Finally, Men1-deficient bone marrow (BM) progenitors cannot sustain leukemogenesis driven specifically by MLL1-FPs.10,12 These data prompted the development of menin-MLL1–disrupting peptides and small molecules as potential targeted therapeutic agents13,14 ; however, it is unclear whether the normal functions of MLL1 similarly depend on menin interaction, because this domain is also present in the native protein.
Menin is a predominantly nuclear protein encoded by the Multiple Endocrine Neoplasia type I (MEN1) gene, responsible for a familial syndrome in which a MEN1 mutation is accompanied by loss of the wild-type MEN1 allele in neuroendocrine tumors.15 Heterozygous Men1 mice also exhibit a similar spectrum of neuroendocrine tumors with loss of the wild-type allele as in the human syndrome.16 Intriguingly, menin is unique in the genome, although the recently solved structure shows the presence of tetratricopeptide repeat motifs and a transglutaminase-like motif.17,18 The tumor-suppressive role of menin is cell type–specific; disruption of Men1 in the liver or hematopoietic system does not result in tumors.19,20 In addition to participation in MLL1/MLL2 complexes, menin interacts with and influences the activity of SMAD proteins, Runx2, JunD, and nuclear factor κB.21-23 Furthermore, Men1-deficient cells lack an S-phase checkpoint, possibly related to interaction with CHES1, a forkhead-related protein.24 Thus, menin likely plays multiple roles even within a particular cell type.
Men1 and Mll1 perform similar functions in several biological settings. Both are essential during embryo development,1,16 and loss of either gene in embryonic stem cells impairs hematopoietic differentiation at a similar stage.25-27 In tissues in which Men1 is a tumor suppressor, the menin-MLL1 complex facilitates expression of several cyclin-dependent kinase inhibitors (CDKIs), including p18Ink4c and p27Kip1, and limits cell proliferation.28,29 In hematopoietic cells, the menin-MLL1 complex maintains Hox gene expression instead.12,20,26
Mll1 is essential for maintaining hematopoietic stem cells (HSCs) and progenitor populations in the BM.5,30,31 Given this critical role in hematopoiesis, disruption of an important chromatin-targeting component of the MLL1 complex would be predicted to result in rapid attrition of HSCs and progenitors. Therefore, we set out to determine the significance of the menin-MLL1 interaction in the normal physiologic functions performed by MLL1 in the hematopoietic system. For these studies, we focused on 3 processes known to be strongly dependent on menin or MLL1 or both: HSC homeostasis, engraftment, and B lymphopoiesis.5,20,31 Our results indicate that MLL1 functions independently from menin for HSC homeostasis, and that both proteins control pathways that function additively in engraftment. Furthermore, we show that menin and MLL1 independently play important roles during B-cell differentiation but control largely independent genetic networks. Importantly, disrupting the menin-MLL1 interaction cannot recapitulate the block in B-cell differentiation observed in individual knockouts. Our data support the concept that selective targeting of aberrant gene expression in vivo can be achieved by disrupting this protein interaction.
Methods
Animals
Men1f/f and Mll1f/f mice5,9 were intercrossed with Mx1-cre transgenic (#003556) or Rag1-cre knock-in32 animals on a B6.SJL background (#002014); RosaYFP (#006148) mice and 8- to 12-week-old C57BL/6J female recipients were from The Jackson Laboratory. For transplantation, Men1f/f mice were backcrossed to B6.SJL at Dartmouse speed-congenic facility (Dartmouth). Polyinosinic:polycytidylic acid (pI:pC) injections were described.5 Excision efficiency was assessed by genomic polymerase chain reaction (PCR) and western blot. Animal protocols were approved by the Institutional Animal Care and Use Committee of Dartmouth College.
Flow cytometry, cell sorting, and transplantation
Cells were stained on ice in Hanks buffered saline solution (Mediatech) plus 2% fetal bovine serum (Invitrogen) and sorted using a FACSAria (BD Biosciences) with purities ranging from 70% to 90%. Analyses were performed on a FACSCalibur or FACSCanto (BD Biosciences) and analyzed using FlowJo software (TreeStar). Competitive transplantation experiments were as described31 with details in the supplemental Methods on the Blood website. Blood was collected from the submandibular vein or cardiac puncture into EDTA-containing microtainer tubes (BD Biosciences). CD45.1+ cells were assessed in recipients up to 25 weeks after engraftment. Reduced lymphocytes in Men1Δ/Δ BM20 necessitated a myeloid pregating based on forward scatter and side scatter parameters (supplemental Figure 1B).
Microarray experiments
Two cohorts (Rag1-cre;Men1f/f vs Men1f/f; Rag1-cre;Mll1f/f vs Mll1f/f, n = 4 each genotype) were used to sort fraction B cells. RNA was purified using Trizol (Invitrogen) followed by RNeasy columns (QIAGEN) and amplified using MessageAmpII aRNA Amplification Kits (Ambion), labeled using BioArray HighYield RNA Transcript Labeling Kits (T7; Enzo Life Sciences), and fragmented and hybridized to mouse 430 2.0 Arrays (Affymetrix) at the Dartmouth Genomics and Microarray Laboratory. Intensities were determined with Microarray Suite version 5.0 (Affymetrix), normalized using GCRMA (BRB-ArrayTools v4.2.1) and differentially expressed probe sets were determined by class comparison using unfiltered expression values with fold-change cutoff of 1.2. A P value cutoff of .001 was used to keep the false discovery rate < 20%. The data reported in this publication have been deposited in NCBI's Gene Expression Omnibus database and are accessible through GEO Series accession number GSE49120.
Recombinant DNA, cell culture, and retroviral infection
The human MLL1 N terminus (wild-type peptide encompassing MLL1 amino acids 1-167 [MBDwt]) was PCR-amplified and cloned into a retrovirus (supplemental Figure 7A). The MBDmut peptide has a 5-alanine substitution of the RWRFP motif. Retroviral supernatant was produced as described in the supplemental Methods. Lineage-negative BM prepared with a lineage-depletion kit (Miltenyi) was spin-infected in growth medium containing recombinant murine cytokines as indicated in the supplemental Methods on retronectin-coated plates (Takara). BM B cells were enriched using anti–B220-microbeads (Miltenyi), then infected in Opti-MEM (Invitrogen)-based growth medium plus 5 µg/mL polybrene (Sigma-Aldrich) and transferred to new plates with fresh growth medium.
Results
Genetic interaction between Men1 and Mll1 in steady-state hematopoiesis
To determine whether MLL1 requires interaction with menin to maintain steady-state hematopoiesis, we assessed genetic interactions during the maintenance of hematopoietic populations. Because MLL1 is critical for maintaining HSCs,5,31,33 we analyzed HSC-enriched populations in double heterozygous BM (from pI:pC-injected Mx1-cre;Men1f/+;Mll1f/+ animals), reasoning that double heterozygotes may exhibit new defects not observed in single heterozygotes. The absolute number of lineage-negative, Sca-1 positive, c-Kit positive (LSK) cells or HSCs (LSK/CD150+/CD48− cells; supplemental Figure 1) in the BM of double heterozygotes was comparable to controls (Figure 1A-B). This observation demonstrates that the removal of a single allele of Men1 is not sufficient to cripple the residual function of the single Mll1. Complete deletion of Mll1 results in rapid attrition of LSK and BM cells5 ; thus, it was not possible to analyze double knockout BM. To determine whether Men1 is required at all for Mll1 function, we removed both alleles of Men1 in the context of Mll1 heterozygosity and analyzed steady-state BM populations. In Men1-deficient BM, HSC numbers were within the normal range (Figure 1D) as previously noted,20 although the LSK population overall was slightly increased (Figure 1C). Surprisingly, Men1 deletion in the context of Mll1 heterozygosity resulted in normal numbers of LSK cells and HSCs in the BM compared with Men1-deficient BM (Figure 1C-D). The complete loss of Men1 was confirmed by western blot (supplemental Figure 2A). These data demonstrate that Mll1 does not require Men1 to sustain HSCs during steady-state hematopoiesis.
Steady-state HSC-enriched populations do not depend on menin-MLL1 collaboration despite a reduction in shared target genes. LSK cells (A) or HSCs (B) (defined as LSK/CD48−/CD150+) are not reduced in double heterozygous BM. Men1 or Mll1 alleles were deleted by pI:pC injection and animals were analyzed 12 to 13 days after the first injection. (A) Controls include both Mx1-cre–negative and Mx1-cre;Men1f/+ animals, whereas controls in panel B are Mx1-cre;Men1f/+ (denoted as Men1Δ/+). Data from 2 experiments were pooled and 8- to 12-week-old mice were analyzed (A, n = 8-12 animals per genotype, P = .1164; B, n = 4-6 animals per genotype, P = .1712). (C) LSK cell quantification in Men1-deficient (Men1Δ/Δ) vs Men1-deficient;Mll1-heterozygous (Men1Δ/Δ;Mll1Δ/+) BM. Cells were produced as described previously, n = 5-6 animals per genotype, P = .1011 for the comparison between Men1Δ/Δ and Men1Δ/Δ;Mll1Δ/+. (D) HSC quantification in Men1Δ/Δ and Men1Δ/Δ;Mll1Δ/+ BM, n = 4-6 animals per genotype, P = .3451 for the comparison between Men1Δ/Δ and Men1Δ/Δ;Mll1Δ/+ BM. Animals were 8 to 9 weeks old when analyzed. (E-F) Gene expression in Men1Δ/Δ and wild-type Men1f/f LSK cells. LSK cells were sorted from 10- to 12-week-old animals 9 days after the first pI:pC injection. Expression levels were normalized to Hprt1, n = 3-4 animals per genotype. Statistical significance was determined using the unpaired Student t test; error bars represent 95% confidence intervals; ***P ≤ .001; **P ≤ .01.
Steady-state HSC-enriched populations do not depend on menin-MLL1 collaboration despite a reduction in shared target genes. LSK cells (A) or HSCs (B) (defined as LSK/CD48−/CD150+) are not reduced in double heterozygous BM. Men1 or Mll1 alleles were deleted by pI:pC injection and animals were analyzed 12 to 13 days after the first injection. (A) Controls include both Mx1-cre–negative and Mx1-cre;Men1f/+ animals, whereas controls in panel B are Mx1-cre;Men1f/+ (denoted as Men1Δ/+). Data from 2 experiments were pooled and 8- to 12-week-old mice were analyzed (A, n = 8-12 animals per genotype, P = .1164; B, n = 4-6 animals per genotype, P = .1712). (C) LSK cell quantification in Men1-deficient (Men1Δ/Δ) vs Men1-deficient;Mll1-heterozygous (Men1Δ/Δ;Mll1Δ/+) BM. Cells were produced as described previously, n = 5-6 animals per genotype, P = .1011 for the comparison between Men1Δ/Δ and Men1Δ/Δ;Mll1Δ/+. (D) HSC quantification in Men1Δ/Δ and Men1Δ/Δ;Mll1Δ/+ BM, n = 4-6 animals per genotype, P = .3451 for the comparison between Men1Δ/Δ and Men1Δ/Δ;Mll1Δ/+ BM. Animals were 8 to 9 weeks old when analyzed. (E-F) Gene expression in Men1Δ/Δ and wild-type Men1f/f LSK cells. LSK cells were sorted from 10- to 12-week-old animals 9 days after the first pI:pC injection. Expression levels were normalized to Hprt1, n = 3-4 animals per genotype. Statistical significance was determined using the unpaired Student t test; error bars represent 95% confidence intervals; ***P ≤ .001; **P ≤ .01.
Hoxa9 and Meis1 are coregulated by Men1 and Mll1,8,12,13 and we have recently defined a set of direct MLL1 target genes important for HSC homeostasis.33 To assess whether these bona fide MLL1 target genes are also regulated by menin, quantitative real-time PCR (qRT-PCR) was performed using purified LSK cells isolated from Mx1-cre;Men1f/f animals injected with pI:pC. Hoxa9 and Meis1 expression levels were both significantly reduced in Men1-deficient LSK cells (Figure 1E), as previously observed.20 Some MLL1-regulated genes were also regulated by menin, including Mecom, Eya1, and most Hoxa cluster genes (Figure 1E; supplemental Figure 3). Other MLL1 targets such as Prdm16 were not affected by Men1 deletion (Figure 1E). We also found examples of genes such as Necdin (Ndn) and Hoxa4, which were selectively regulated by menin and not MLL1 (Figure 1F; supplemental Figure 3).33 These data show that Men1 loss affects only a subset of Mll1 target genes, and more importantly these gene expression changes are not sufficient to impact on HSC homeostasis.
Genetic interaction of Men1 and Mll1 in regenerative hematopoiesis
Hematopoiesis under regenerative conditions such as recovery from myeloablation or competitive transplantation differs from steady-state hematopoiesis in its dependence on distinct genetic pathways.34 Men1-deficient HSCs do not exhibit defects in steady-state hematopoiesis but are impaired in competitive transplantation.20 Thus we evaluated genetic interactions between Men1 and Mll1 in the setting of regenerative hematopoiesis. As noted previously,20 we observed a significant reduction of donor-derived cells in recipients of Men1-deficient BM (Figure 2A,C). However, reduced engraftment was not observed in recipients of double heterozygous BM (Figure 2A,C), indicating that a single copy of each gene provides sufficient activities of both menin and MLL1 to support hematopoietic regeneration. Similarly, myeloid recovery after 5-fluorouracil treatment was not impaired in double heterozygotes either (data not shown). To assess whether a single copy of Mll1 in the complete absence of Men1 was sufficient to support hematopoietic regeneration, we performed competitive transplantation experiments using Men1Δ/Δ;Mll1Δ/+ donor cells. In this case, the impact of Mll1-heterozygosity was a slight but significant reduction in donor-derived cells compared with Men1Δ/Δ BM, when either peripheral blood (Figure 2B) or BM lineage-negative/c-Kit-positive (lin−/c-Kit+) cells (Figure 2D) were assessed at 25 weeks after engraftment. This observation is consistent with a hypothesis that both proteins function additively through independent pathways, which is explored in more detail in the B-cell lineage in the following section. Importantly, these data argue against the view that menin serves as an essential cofactor for MLL1 in regulation of HSC homeostasis.35
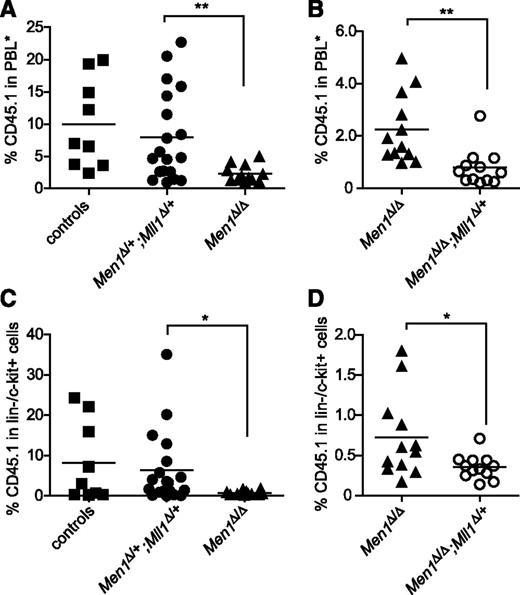
Menin and Mll1 function independently during hematopoietic regeneration. (A-B) Donor-derived (CD45.1) cells in the peripheral blood of recipients 25 weeks posttransplantation. PBL*, myeloid-gated peripheral blood leukocytes used to assess donor contribution independent of the B-cell deficit in Men1-deficient BM (see “Methods”). BM cells of the indicated genotypes were generated by pI:pC injection, harvested 13 days after the first injection, and then mixed with wild-type CD45.2 BM cells and injected into lethally irradiated recipients. Control donor BM was from pI:pC-injected littermates not harboring the Mx1-cre transgene. (C-D) Donor contribution in the lin−/c-Kit+ fraction of BM from the same recipients as in panels A-B. To facilitate pairwise comparisons, data from the Men1Δ/Δ group in A are regraphed in B; n = 9-11 animals per genotype. Statistical significance was determined using the unpaired Student t test; *P ≤ .05; **P ≤ .01. (A) P = .0098. (B) P = .0027. (C) P = .041. (D) P = .0284.
Menin and Mll1 function independently during hematopoietic regeneration. (A-B) Donor-derived (CD45.1) cells in the peripheral blood of recipients 25 weeks posttransplantation. PBL*, myeloid-gated peripheral blood leukocytes used to assess donor contribution independent of the B-cell deficit in Men1-deficient BM (see “Methods”). BM cells of the indicated genotypes were generated by pI:pC injection, harvested 13 days after the first injection, and then mixed with wild-type CD45.2 BM cells and injected into lethally irradiated recipients. Control donor BM was from pI:pC-injected littermates not harboring the Mx1-cre transgene. (C-D) Donor contribution in the lin−/c-Kit+ fraction of BM from the same recipients as in panels A-B. To facilitate pairwise comparisons, data from the Men1Δ/Δ group in A are regraphed in B; n = 9-11 animals per genotype. Statistical significance was determined using the unpaired Student t test; *P ≤ .05; **P ≤ .01. (A) P = .0098. (B) P = .0027. (C) P = .041. (D) P = .0284.
Menin controls B lymphopoiesis at the fraction C-to-C′ transition
Using an estrogen receptor-cre (ER-cre;Men1f/f) model, B lymphopoiesis was the only BM population affected by Men1 loss under steady-state conditions20 ; Mll1 loss also specifically affects B lymphopoiesis.31 Thus we evaluated B-cell differentiation as a process in which menin and MLL1 may function together as a complex. To delete combinations of Men1 and Mll1 alleles specifically in lymphocytes, a Rag1-cre knock-in allele was used.32 Complete loss of menin was confirmed by western blot (supplemental Figure 2B). Rag1-cre;Men1f/f animals exhibited normal T-cell populations (data not shown), but reduced B cells in the BM and spleen. Starting from the small pre-B stage onward, Men1 deletion resulted in a ∼50% reduction in BM B cells and in splenic B cells (Figure 3A-B; supplemental Figure 4). We defined the stage at which B-cell differentiation was affected using the Hardy fractionation scheme36 and found that Men1 deletion results in a partial differentiation block at the fraction C-to-C′ transition (Figure 3C-D; supplemental Figure 4), despite the fact that complete deletion of Men1 is achieved by fraction B (Figure 5A; supplemental Figure 2C). Interestingly, this is the same stage at which Rag1-cre;Mll1f/f animals exhibit a block in B-cell differentiation (Gan et al, in preparation) and both Hoxa9 and Meis1 were reduced in Rag1-cre;Men1f/f pro-B cells (Figure 3E), demonstrating that known target genes of Mll1 are reduced upon menin loss in this cell type as well. In summary, these data demonstrate that menin controls B-cell differentiation at the fraction C-to-C′ transition.
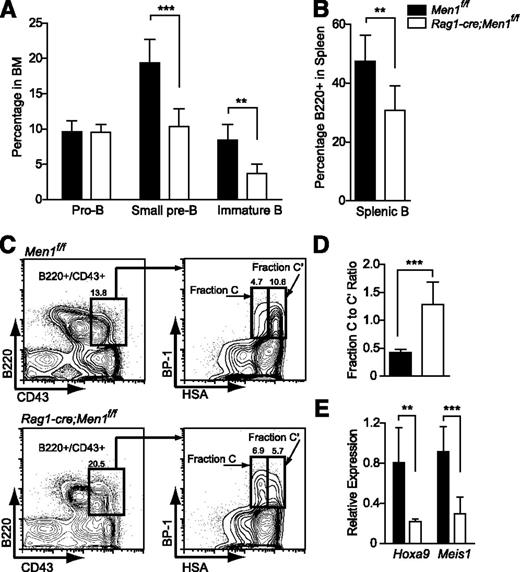
Deletion of Men1 during lymphocyte differentiation results in a partial block in B-cell differentiation. (A) Pro-B (B220+/CD43+), small pre-B (B220+/CD43−/IgM−), immature B (B220+/CD43−/IgM+), and (B) splenic B cells (B220+) were quantified in the BM of Men1f/f or Rag1-cre;Men1f/f animals. (C) Representative fluorescence-activated cell sorter plot illustrating partial fraction C-to-C′ block. (D) Fraction C to fraction C′ ratio comparing Men1f/f to Rag1-cre;Men1f/f mice. Fraction C (B220+/CD43+/BP-1+/HSAlo) to fraction C′ (B220+/CD43+/BP-1+/HSAhi) ratios were determined using absolute numbers of the gated populations in the hind limbs of either genotype. (E) Relative expression of Hoxa9 and Meis1 in pro-B cells (B220+/CD43+) from Men1f/f (black bars) or Rag1-cre;Men1f/f animals (white bars). Relative expression levels were determined using ribosomal RNA as an internal control. Three- to 4-week-old animals were analyzed, n = 3-4 animals per genotype. Statistical significance was determined using the unpaired Student t test; error bars represent 95% confidence intervals; ***P ≤ .001; **P ≤ .01.
Deletion of Men1 during lymphocyte differentiation results in a partial block in B-cell differentiation. (A) Pro-B (B220+/CD43+), small pre-B (B220+/CD43−/IgM−), immature B (B220+/CD43−/IgM+), and (B) splenic B cells (B220+) were quantified in the BM of Men1f/f or Rag1-cre;Men1f/f animals. (C) Representative fluorescence-activated cell sorter plot illustrating partial fraction C-to-C′ block. (D) Fraction C to fraction C′ ratio comparing Men1f/f to Rag1-cre;Men1f/f mice. Fraction C (B220+/CD43+/BP-1+/HSAlo) to fraction C′ (B220+/CD43+/BP-1+/HSAhi) ratios were determined using absolute numbers of the gated populations in the hind limbs of either genotype. (E) Relative expression of Hoxa9 and Meis1 in pro-B cells (B220+/CD43+) from Men1f/f (black bars) or Rag1-cre;Men1f/f animals (white bars). Relative expression levels were determined using ribosomal RNA as an internal control. Three- to 4-week-old animals were analyzed, n = 3-4 animals per genotype. Statistical significance was determined using the unpaired Student t test; error bars represent 95% confidence intervals; ***P ≤ .001; **P ≤ .01.
Genetic interaction between Men1 and Mll1 in B lymphopoiesis
Given that mice can survive without B cells, it was possible to assess the genetic interaction between Men1 and Mll1 in developing lymphocytes using all allelic combinations. Therefore, animals harboring the Rag1-cre allele and combinations of all 4 floxed alleles were generated and B-cell populations were analyzed. Despite the fact that Rag1-cre;Men1f/f and Rag1-cre;Mll1f/f animals phenocopy each other (Figure 4 and data not shown), no defects in B lymphopoiesis were observed in double heterozygotes (Figure 4A-B), demonstrating that combined haploinsufficiency does not significantly impair either Men1 or Mll1 function. In contrast, either Men1 or Mll1 loss individually reduced the BM B220+/CD43− population by approximately 50% (Figure 4A,C). Surprisingly, the analysis of double Men1/Mll1 knockouts revealed a further reduction in B cells compared with the individual knockouts (Figure 4C), a finding that was independent of animal age (supplemental Figure 5A). Furthermore, the block in differentiation occurred at the same stage in all genotypes, as illustrated by the increased ratio of fraction C to fraction C′ cells (Figure 4D). These data suggest that menin and MLL1 both control pathways critical to the fraction C-to-C′ transition, but both proteins do so through independent additive pathways.
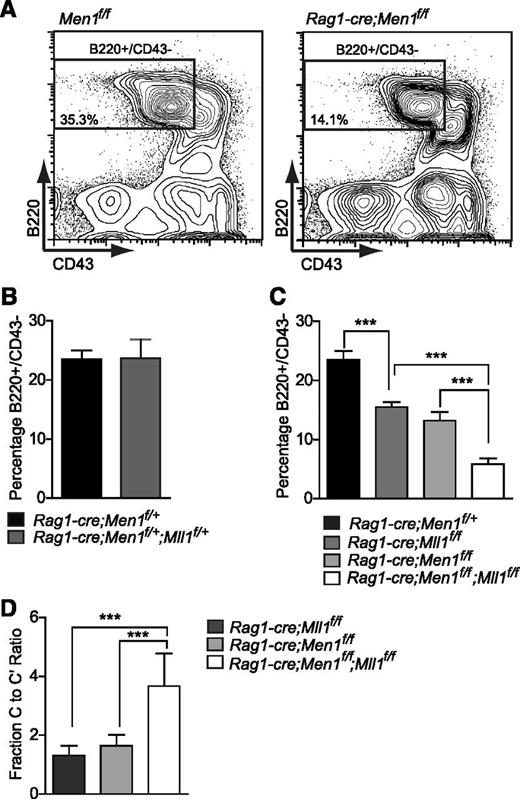
Genetic interactions between Men1 and Mll1 in B lymphopoiesis. (A) Representative example of fluorescence-activated cell sorting gates used in panels B-D. Percentages shown are relative to the total BM. (B) Comparison of control (Rag1-cre;Men1f/+) to double heterozygote (Rag1-cre;Men1f/+;Mll1f/+) BM B220+/CD43− populations, n = 7-9 animals per genotype, P = .5111. (C) Comparison of control (black), single knockout (light and dark gray), and double knockout (white) BM B220+/CD43− populations, n = 4-15 animals per genotype. (D) A more severe fraction C-C′ block in double knockouts compared with either Men1 or Mll1 knockouts, n = 4-12 animals per genotype. Two-week-old animals were analyzed and ratios were determined as in Figure 3. Statistical significance was determined using the unpaired Student t test; error bars represent 95% confidence intervals; ***P ≤ .001; **P ≤ .01.
Genetic interactions between Men1 and Mll1 in B lymphopoiesis. (A) Representative example of fluorescence-activated cell sorting gates used in panels B-D. Percentages shown are relative to the total BM. (B) Comparison of control (Rag1-cre;Men1f/+) to double heterozygote (Rag1-cre;Men1f/+;Mll1f/+) BM B220+/CD43− populations, n = 7-9 animals per genotype, P = .5111. (C) Comparison of control (black), single knockout (light and dark gray), and double knockout (white) BM B220+/CD43− populations, n = 4-15 animals per genotype. (D) A more severe fraction C-C′ block in double knockouts compared with either Men1 or Mll1 knockouts, n = 4-12 animals per genotype. Two-week-old animals were analyzed and ratios were determined as in Figure 3. Statistical significance was determined using the unpaired Student t test; error bars represent 95% confidence intervals; ***P ≤ .001; **P ≤ .01.
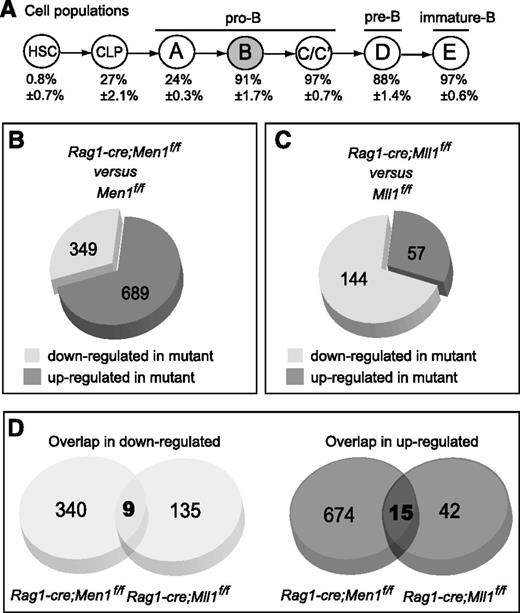
Menin and MLL1 regulate common and distinct genes during B lymphopoiesis. (A) Schematic diagram showing B-cell differentiation using the Hardy fractionation scheme and nomenclature.36 Below each population is shown the YFP+ percentage (thus the penetrance of cre excision) determined using Rag1-cre;RosaYFP reporter animals (Gan et al, in preparation), n = 3-4 animals per population. Fraction B (gray) cells were used for the microarray analyses. (B) Pie chart showing the proportions of differentially expressed probe sets in Men1-deficient vs wild-type fraction B cells. Differentially expressed probe sets were determined as described in the “Methods” section. (C) Pie chart showing the proportions of differentially expressed probe sets in Mll1-deficient vs wild-type fraction B cells. Data are from Gan et al (in preparation) and were analyzed with the same criteria as in panel B. Each group was compared against its own wild-type littermate controls. (D) Pie chart showing the number of probe sets in common between the 2 sets of differentially expressed (down- or upregulated) probe sets in the analyses shown in panels B-C. For details, see supplemental Figure 6.
Menin and MLL1 regulate common and distinct genes during B lymphopoiesis. (A) Schematic diagram showing B-cell differentiation using the Hardy fractionation scheme and nomenclature.36 Below each population is shown the YFP+ percentage (thus the penetrance of cre excision) determined using Rag1-cre;RosaYFP reporter animals (Gan et al, in preparation), n = 3-4 animals per population. Fraction B (gray) cells were used for the microarray analyses. (B) Pie chart showing the proportions of differentially expressed probe sets in Men1-deficient vs wild-type fraction B cells. Differentially expressed probe sets were determined as described in the “Methods” section. (C) Pie chart showing the proportions of differentially expressed probe sets in Mll1-deficient vs wild-type fraction B cells. Data are from Gan et al (in preparation) and were analyzed with the same criteria as in panel B. Each group was compared against its own wild-type littermate controls. (D) Pie chart showing the number of probe sets in common between the 2 sets of differentially expressed (down- or upregulated) probe sets in the analyses shown in panels B-C. For details, see supplemental Figure 6.
Comparison between Men1- and Mll1-dependent genes in B cells
Given the presence of menin in native MLL1 complexes8,9 and detailed analyses of several genes directly regulated or bound by both menin and MLL1,12,28,37 we were surprised by the genetic evidence supporting distinct and additive roles for Men1 and Mll1 in HSCs and B cells. To examine the potential overlap in molecular pathways controlled by either protein in the same cell type, we compared gene expression profiles of Men1- and Mll1-deficient pro-B cells to reveal the degree to which they regulate common or distinct target genes.
The first population in which gene excision is fully penetrant in the Rag1-cre animals is fraction B (Figure 5A and data not shown). Therefore, we performed genome-wide expression analysis using fraction B (B220+/CD43+/HSA+/BP-1−) cells in Rag1-cre;Men1f/f animals. This approach confirmed the downregulation of Hoxa9 and Meis1 in Men1-deficient B cells as observed in other cell types and was validated using independent qRT-PCR (supplemental Figure 5B). Overall, we noted that the majority (66%) of differentially expressed genes were actually upregulated in Men1-deficient B cells (Figure 5B), which is consistent with a documented role of menin as a transcriptional corepressor.21 Mll1 deletion using the same system resulted in only 28% upregulated genes with the majority being downregulated (Figure 5C), consistent with the role of Mll1 as transcriptional activator.38 Comparing downregulated genes, very few were downregulated in common by Men1 or Mll1 deletion (2.6, or 6.3%, respectively) (Figure 5D). Interestingly, this overlap of 9 probe sets, corresponding to 8 genes (supplemental Figure 6), includes Hoxa9, Meis1, and Eya1, recently shown to be directly regulated by MLL1 and MLL1-FPs.33,39 Similarly, only a small proportion was found to be upregulated in common by Men1 or Mll1 deletion (Figure 5D; supplemental Figure 6). Taken together with the genetic data from double heterozygotes and double knockouts (Figure 4B-D), this observation suggests 2 possibilities. First, Men1 and Mll1 may regulate the same stage of B-cell development through distinct mechanisms, using distinct pathways within the nonshared differentially expressed genes. Second, the relevant genes for B lymphopoiesis may be defined by the small overlap between the 2 downregulated gene lists; thus, reduced expression of these few but relevant genes accounts for the block in differentiation in either knockout. To distinguish between these possibilities, we pursued a strategy in which we directly disrupted the menin-MLL1 complex and assessed gene expression changes and B-cell differentiation.
Disruption of the menin-MLL1 interaction affects gene expression but not B-cell differentiation
To determine whether the B-cell differentiation block could be recapitulated by disrupting the menin-MLL1 interaction, we expressed a menin-MLL1 disrupting peptide (supplemental Figure 7A) based on the design used by Caslini and colleagues13 and assessed its effect on gene expression and B-cell differentiation. We first confirmed that the expression of this MLL1 N-terminal peptide (but not an alanine-substitution mutant that lacks interaction with menin) inhibited colony growth of MLL1-AF9 leukemia cells (supplemental Figure 7B). In MLL1-AF9 cells expressing the disrupting peptide, Meis1 expression was reduced as reported13 and we also observed reduced expression of Mecom and Eya1 (supplemental Figure 7C), three of the genes identified as shared targets of menin and MLL1 in LSK cells or B cells (Figure 1D; supplemental Figure 6A). These data demonstrate that genes co-regulated by Men1 and Mll1 can be effectively downregulated by disrupting their interaction between them.
To determine the effect of this disruption strategy on B-cell differentiation, we introduced the peptide into primitive BM cells and assessed B-cell growth and differentiation (Figure 6A). As observed in MLL1-AF9 cells, expression of the wild-type, but not mutant peptide reduced the expression of several menin/MLL1 shared targets including Hoxa9, Meis1, and Eya1 (Figure 6B-C). Expression of the disrupting peptide had no effect on B-cell differentiation or growth (Figure 6D), despite the misregulation of these targets to levels observed in the individual knockouts (Figure 6E-F). Importantly, expression of the disrupting peptide in Men1- or Mll1-single knockout BM B cells had no further effect on the expression of the shared target, Eya1 (supplemental Figure 8), demonstrating the specificity of the disrupting peptide. Collectively, these data support the identification of these target genes as regulated by the menin-MLL1 complex, but also suggest that downregulation of these target genes is not sufficient to result in the block in B-cell differentiation observed in either knockout. Therefore menin and MLL1 more likely function through independent pathways to regulate B lymphopoiesis.
Disrupting the menin-MLL1 interaction affects shared target genes in B cells but not B-cell differentiation. (A) Experimental design for expressing the disrupting peptide in wild-type BM cells and assays to measure its activity. (B-C) The selective effect of the disrupting peptide on the expression levels of several shared target genes in cultured BM B cells. B220-enriched BM cells were infected with peptide-expressing or empty (MIG) retrovirus and GFP+ cells were sorted 6 to 10 days later for qRT-PCR assays. (D) The effect of peptide expression on B-cell growth in vitro. BM was depleted of lineage-positive cells and the resulting lineage-depleted population was cultured overnight and then infected with peptide-expressing or empty retrovirus. The hatched bar shows the B-cell growth using BM from Rag1-cre;Men1f/f infected with empty retrovirus. The B-cell percentages were determined at day 8 by flow cytometry. Error bars represent 95% confidence intervals from 3 replicate infections. (E-F) The effect of the peptides on gene expression in B cells expanded in vitro. GFP+ cells as described in panel D were used for qRT-PCR assays on day 6. Relative expression levels of the genes indicated below each set of bars reflect data normalized to ribosomal RNA expression. All experiments were reproduced 3 times. Statistical significance was determined using the unpaired Student t test; error bars represent 95% confidence intervals; ***P ≤ .001; **P ≤ .01.
Disrupting the menin-MLL1 interaction affects shared target genes in B cells but not B-cell differentiation. (A) Experimental design for expressing the disrupting peptide in wild-type BM cells and assays to measure its activity. (B-C) The selective effect of the disrupting peptide on the expression levels of several shared target genes in cultured BM B cells. B220-enriched BM cells were infected with peptide-expressing or empty (MIG) retrovirus and GFP+ cells were sorted 6 to 10 days later for qRT-PCR assays. (D) The effect of peptide expression on B-cell growth in vitro. BM was depleted of lineage-positive cells and the resulting lineage-depleted population was cultured overnight and then infected with peptide-expressing or empty retrovirus. The hatched bar shows the B-cell growth using BM from Rag1-cre;Men1f/f infected with empty retrovirus. The B-cell percentages were determined at day 8 by flow cytometry. Error bars represent 95% confidence intervals from 3 replicate infections. (E-F) The effect of the peptides on gene expression in B cells expanded in vitro. GFP+ cells as described in panel D were used for qRT-PCR assays on day 6. Relative expression levels of the genes indicated below each set of bars reflect data normalized to ribosomal RNA expression. All experiments were reproduced 3 times. Statistical significance was determined using the unpaired Student t test; error bars represent 95% confidence intervals; ***P ≤ .001; **P ≤ .01.
Discussion
Because disrupting the menin-MLL1 interaction is actively being pursued as a therapeutic strategy for leukemia harboring MLL1 translocations13,14 and loss of endogenous MLL1 in the murine or human hematopoietic system results in cytopenia,5,40 we undertook a genetic analysis of the requirement for Men1-Mll1 interaction during normal hematopoiesis. Because of the widely appreciated role of menin in MLL1-FP–mediated leukemogenesis, the discovery of menin in native MLL1 complexes, and the colocalization of menin, MLL1/MLL2, and H3K4Me3 at many genomic loci, we initially expected that menin would prove to be a critical cofactor for the actions of the MLL1 complex, regardless of tissue or target genes examined.9,10,12,27,29,37 We provide several lines of evidence that MLL1 acts independently from menin in the hematopoietic system. First, heterozygosity for both Men1 and Mll1 had no effect on HSC homeostasis or regeneration in the BM, despite both of these genes displaying haploinsufficiency in a variety of settings.41-43 More significantly, complete Men1 loss in HSCs produces a much milder phenotype than Mll1 loss (Figure 1D; Jude et al5 ) and Mll1-haploinsufficiency contributes additively to the Men1-knockout engraftment defect (Figure 2B,D). Although Rag1-cre–mediated Men1 deletion does phenocopy Mll1 deletion during B lymphopoiesis, the additive severity of the double knockout phenotypes and the limited degree of overlap in the genes deregulated in either mutant strongly suggest that Mll1 acts without Men1 at most target genes. Finally, we show that disruption of the menin-MLL1 complex does not result in a block in B-cell differentiation as do the individual knockouts, despite efficient reduction in shared target gene expression. These data collectively support a model in which a small number of genes depend upon the menin-MLL1 complex, but these genes are not the critical downstream effectors of Mll1 in HSC homeostasis or B lymphopoiesis. Thus, MLL1 can maintain the expression of critical HSC and B-cell genes without menin.
In the case of hematopoietic regeneration after transplantation, Men1-deficient BM cells exhibit a competitive disadvantage compared with wild-type BM, as noted using ER-cre;Men1f/f or Mx1-cre;Men1f/f mice (Figure 2A,C; Maillard et al20 ). Loss of Mll1 in fetal31 or adult HSCs5,30 results in complete loss of competitive transplantation activity and the downregulation of genes encoding transcription factors such as Hoxa9, Meis1, Mecom, and Eya1.33 This common molecular phenotype suggests that downregulation of this series of transcription factors may be sufficient to engender a defect in conditions of replicative stress but not steady-state hematopoiesis, and that additional genes regulated by Mll1 and not Men1 are critical for maintaining HSCs under steady-state conditions. Thus, a more comprehensive analysis between the gene programs affected in the 2 individual mutants may help distinguish pathways required for maintaining HSCs vs those employed under conditions of replicative stress.
The data presented here support a significant role for menin outside of the MLL1 methyltransferase complex, particularly in the control of B-cell differentiation at the fraction C-to-C′ transition. This function is unlikely to involve the MLL2 complex because knockout of Mll2 does not result in B-cell defects.44 Although there have been many menin-interacting proteins reported, the biological significance of such interactions has not been studied in detail, particularly not in B cells. However, several studies demonstrate that Men1-deficient cells are sensitive to double-strand DNA breaks and intrastrand crosslinking agents and fail to arrest in the presence of such damage.24,45 Most of these studies have been performed in fibroblasts or neuroendocrine cells, but patients’ lymphocytes also exhibit chromosome instability and sensitivity to DNA-damaging agents.46,47 Therefore, it is possible that both the defect in HSCs under replicative stress and the defect in B-cell differentiation relate to an inappropriate response to spontaneous or Rag-mediated double-strand DNA breaks, although further studies will be required to test this hypothesis.
Curiously, the absolute requirement for MLL1-FP to interact with menin stands in contrast to the minimal role for menin in the function of endogenous MLL1 in normal hematopoiesis described here. This suggests that MLL1-FPs regulate a very unique subset of MLL1 target genes that depend strictly on menin for targeting to particular chromatin landscapes. We provide evidence for this concept by showing that Hoxa9, Eya1, and Mecom genes, all implicated in MLL1-FP leukemogenesis, are affected by menin loss. In contrast, Prdm16, which is not upregulated in MLL1-translocation leukemia, is not affected by Men1 loss (Figure 1F) but is critical for HSC maintenance.48,49 These observations are consistent with a model in which MLL1 complex recruitment to some target genes requires menin, presumably to facilitate interaction with LEDGF and thus a particular chromatin landscape. Other genes as dependent on MLL1 but unaffected by Men1 deletion may rely on distinct recruitment mechanisms through other domains of MLL1. MLL1-response elements in mammals have been very difficult to define and predict based on DNA sequence features alone, partially because of an incomplete understanding of the range and diversity of bona fide MLL1 target genes in specific cell types. The work presented here as well as a recent study dissecting Mll2-dependent genes in macrophages50 suggest that the analysis of tissue-specific MLL-family target genes may facilitate the identification of regulatory elements with common sequence features rendering them responsive to MLL family members. In other words, target specificity of MLL family members may be defined by combinatorial use of distinct cofactors (such as menin) at different biologically relevant classes of target genes.
Overall, our data support the selectivity of menin-MLL1–disrupting drugs against the actions of leukemogenic fusion proteins and predict minimal effects on normal hematopoietic functions. However, the menin-MLL1 complex may play an important tumor suppressive role in neuroendocrine cells. Disruption of this complex in pancreatic β cells may downregulate p18Ink4c and p27Kip1 expression, thus disrupting tumor suppression activity.28,29 These CDKIs are not regulated by Mll1 in hematopoietic cells34 ; thus, we were unable to predict whether inhibition of the menin-MLL1 interaction would affect expression of these CDKIs in neuroendocrine cells. Further studies such as targeting inhibitory molecules to endocrine cells will be important to determine whether this strategy presents a risk for inhibiting the tumor suppression activity enforced by menin.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the members of our group for helpful discussions; Xianxin Hua, Ivan Maillard, and Rachel Gerstein for advice and critical review; and Kristin Zaffuto for excellent animal husbandry.
This work was supported by grants from the American Cancer Society (RSG-10-242-LIB), Gabrielle’s Angel Foundation for Cancer Research, Lauri Strauss Leukemia Foundation, the National Institutes of Health, National Heart, Lung, and Blood Institute (HL090036), and program grants from Leukaemia and Lymphoma Research and the Medical Research Council (T.H.R.). The Dartmouth Genomics and Microarray Laboratory received support from a Cancer Center Core Grant (CA23108), and Dartmouse was established by funding from the National Center for Research Resources (P20-RR16437).
Authorship
Contribution: B.E.L. performed most of the experiments, analyzed the data, and wrote the manuscript; T.G. performed experiments and provided important advice; M.M. and T.H.R. provided critical strains; and P.E. conceived of and directed the project and wrote the manuscript.
Conflict-of-interest disclosure: P.E. owns Amgen stock. The remaining authors declare no competing financial interests.
Correspondence: Patricia Ernst, Geisel School of Medicine at Dartmouth, Department of Genetics, HB 7400, Remsen 725, Hanover, NH 03755; e-mail: patricia.ernst@dartmouth.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal