Key Points
FOXP3 functions as a negative regulator of T-cell proliferation and cytokine production in human conventional T cells.
Expression of FOXP3 in human Th17 cells functions to suppress IFN-γ production.
Abstract
The role of forkhead box P3 (FOXP3) is well-established in T-regulatory cells, but the function of transient FOXP3 expression in activated human conventional T (Tconv) cells is unknown. In the present study, we used 2 approaches to determine the role of FOXP3 in human Tconv cells. First, we obtained Tconv clones from a female subject who is hemizygous for a null mutation in FOXP3, allowing the comparison of autologous T-cell clones that do or do not express FOXP3. Second, we knocked down activation-induced FOXP3 in Tconv cells from healthy donors with small interfering RNA against FOXP3. We found that FOXP3-deficient Tconv cells proliferate more and produce more cytokines than wild-type Tconv cells and have differential expression of 274 genes. We also investigated the role of FOXP3 in Th1 and Th17 cells and found that the expression of activation-induced FOXP3 was higher and more sustained in Th17 cells compared with Th1 cells. Knocking down FOXP3 expression in Th17 cells significantly increased the production of IFN-γ and decreased the expression of CCR4, but had no effect on IL-17 expression. These data reveal a novel function of FOXP3 in Tconv cells and suggest that expression of this protein is important in the function of multiple CD4+ T-cell lineages.
Introduction
Expression of the transcription factor forkhead box P3 (FOXP3) in T-regulatory cells (Tregs) is necessary and sufficient for Tregs to suppress the effector function of conventional T (Tconv) cells. In cooperation with other transcription factors, including NFAT and Runx1, and the Th17-associated transcription factors ROR-γt and RORα, FOXP3 establishes the Treg program by repressing or trans-activating defined genes.1,2 The molecular mechanisms of FOXP3-mediated regulation of gene transcription are not clearly defined, but repression involves interactions with the histone acetyl-transferase TIP60, the histone deacetylase HDAC7, and linker histone H1.5.3,4
After its discovery in Tregs, it was soon demonstrated that FOXP3 is also expressed transiently in human Tconv cells after TCR activation.5-11 Similarly, another Treg-associated transcription factor, Helios, can also be expressed on activation.12 The major differences between FOXP3 in Tregs and Tconv cells are in stability and expression levels. In Tregs, the Treg-specific demethylated region (TSDR) region of the FOXP3 promoter is demethylated, permitting high and stable expression.13 Conversely, in Tconv cells, the TSDR is methylated, resulting in transient expression of FOXP3 that never reaches the intensity of that in similarly activated Tregs.6,7,9,13 Transient FOXP3 expression in Tconv cells does not prevent cytokine production and/or confer suppressive capacity, although this has been a point of controversy.5-11
Another role for FOXP3 is to antagonize Th17 cell development. Interaction of FOXP3 with ROR-γt or Runx1 inhibits IL-17 production,1 whereas ROR-γt together with hypoxia-inducible factor 1α inhibits FOXP3.14 Therefore, in a tolerogenic environment that includes TGFβ, FOXP3 suppresses Th17 cell development and Treg differentiation prevails. In contrast, in inflammatory environments hypoxia inducible factor1α promotes ROR-γt expression, causing degradation of FOXP3, promotion of Th17 cell development, and blockade of Treg differentiation.14 Beyond this “tug-of-war” during differentiation, Tregs and Th17 cells may have the ability to interconvert. FOXP3+ IL-17–secreting cells exist in vivo,15-17 but it is not clear whether these cells are Tregs that have begun to secrete IL-17 or if they are Th17 cells that have begun to express FOXP3.
Despite clear evidence that FOXP3 is expressed in Tconv cells, its function remained unknown. In the present study, we investigated the role of FOXP3 in human Tconv cells and found that FOXP3-deficient Tconv cells proliferated to a greater extent and produced greater amounts of cytokines than wild-type (WT) Tconv cells. Furthermore, FOXP3 was highly expressed in activated Th17 cells and contributed to their phenotype by suppressing IFN-γ production and up-regulating the expression of CCR4. These data reveal a novel function for FOXP3 and indicate that this transcription factor intrinsically regulates multiple T-cell subsets.
Methods
Isolation and cloning of human T cells
Peripheral blood was obtained from healthy volunteers and a female carrier of a T → C mutation at the second coding nucleotide in FOXP3 (carrier number 3 in Di Nunzio et al18 ), who gave written informed consent per the Declaration of Helsinki in accordance with protocols approved by the University of British Columbia Clinical Research Board and the HSR Internal Ethical Committee (TIGET02). APCs, CD4+ T cells, and CD4+CD25−CD45RO− (naive) T cells were isolated and expanded as described previously.19,20 CD4+CD25− T cells were sorted from the PBMCs of the carrier of a 2 T → C mutation in FOXP318 on a FACSAria cell sorter (BD Biosciences) and cloned as described previously.21 X-chromosome inactivation analysis was performed18 to determine which allele of FOXP3 was active.
Microarray analysis and validation
RNA was isolated from FOXP3 WT or null T-cell clones, which were unstimulated (0 hours) or stimulated (72 hours) with immobilized anti-CD3 (10 μg/mL) and soluble anti-CD28 mAbs (1 μg/mL; BD Biosciences) at 106 cells/mL. Total RNA was hybridized to a GeneChip Human Gene 1.0 ST array (Affymetrix) according to the manufacturer's instructions. Hybridized arrays were washed and stained on a gene Chip Fluidics Station 450 and scanned on a Gene Chip Scanner 3000 7G (Affymetrix), generating an intensity file (*.CEL). CEL files were analyzed by RMA quantile normalization using the R package oligo.22 Differential gene expression was assessed using linear models through the R package limma.23 A cluster of HLA genes was removed because of their sequence similarity and thus difficulty in evaluating the significance of differential expression. Genes showing P < .05 (Benjamini and Hochberg correction for multiple comparisons) were considered significant. Bioinformatics analysis showed that the most statistically significant differences were attributable to FOXP3 genotype rather than activation state. Therefore, data from unstimulated and stimulated cells were combined and differential expression based on genotype was determined. The dataset is in the National Center for Biotechnology Information Gene Expression Omnibus as number GSE41087.
To validate gene expression, expression of mRNAs was quantitatively assessed by real-time PCR using KAPA SYBR green FAST qPCR Kit (Kapa Biosystems) in a ViiA7 real-time system (Applied Biosystem). Each sample was assayed in duplicate for at least 3 runs and results were normalized to β2 microglobulin and hypoanthine-guanine phosphoribosyl transferase.
Pathway analysis
Gene set enrichment analysis (GSEA)24 was performed using RefSeq annotated genes that were differentially expressed between FOXP3 WT and null clones. The resulting pathways (C2) were filtered using an adjusted P < .05 (Benjamini and Hochberg correction, R function: p.adjust25 ) and cross-validated using randomized lists of background genes. Briefly, randomized lists of the same length as the differentially expressed list were obtained from the list of nonsignificantly changed genes between FOXP3 WT and null clones. The lists were used as input for GSEA canonical pathways analysis and the pathways arising from those analyses were removed from the list of differentially expressed genes. Only those pathways with more than 4 genes in overlap are shown.
Isolation and expansion of Th1 and Th17 cells
CD4+ T cells were depleted of CD25+ cells with CD25 microbeads (Miltenyi Biotec). Th1 (CD4+CD25−CXCR3+CCR4−CCR6−) and Th17 (CD4+CD25−CXCR3−CCR4+CCR6+) cells were sorted from CD4+CD25− T cells on a FACSAria cell sorter (BD Biosciences) to > 95% purity based on cell-surface markers.26 Supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article) contains detailed information on all of the Abs used for flow cytometry. Immediately after sorting, cells were stained for FOXP3 and Helios to determine if there were Tregs present in the various populations. Although the Th1- and Th17-enriched cells had been depleted of CD25+ cells, they both contained a proportion of FOXP3+Helios+ cells that was similar to that in unsorted CD4+ T cells (supplemental Figure 1). However, unlike FOXP3+Helios+ cells in CD4+ T cells or CD25+-enriched T cells, FOXP3+Helios+ cells in Th1- and Th17-enriched populations were not CD25 high, suggesting that they are not classic Tregs.
Alternatively, IL-17A+ and IL-17A− T cells were captured after stimulating CD4+CD25− T cells with phorbol 12-myristate 13-acetate (PMA, 10 ng/mL; Sigma-Aldrich), ionomycin (500 ng/mL), and anti-CD28 (500 ng/mL, BD Biosciences). IL-17A–secreting cells were labeled with the IL-17 Secretion Assay Detection Kit (Miltenyi Biotec) according to the manufacturer's instructions and then sorted on the FACSAria.
Chemokine receptor–sorted or cytokine-captured T cells were expanded with autologous APCs, soluble anti-CD3 (OKT3, 1 μg/mL), and IL-2 (100 U/mL) for 2 weeks. Th1 or Th17 cell lines were rested before restimulation with anti-CD3/anti-CD28–coated beads (Invitrogen) at a ratio of 1 bead to 32 cells or with APC plus anti-CD3 with IL-2 (100 U/mL).
Lentiviral vectors and transduction
Lentiviral vectors encoding siRNA against FOXP3 (siFOXP3) or luciferase (siLuc), and the truncated nerve growth factor receptor (ΔLNGFR), were provided by Luigi Naldini.27 Naive CD4+CD25−CD45RO− T cells or Th1 and Th17-enriched cells were transduced as described previously.19,20 ΔLNGFR+ cells were purified (≥ 80% ΔLNGFR+) and analysis was performed on ΔLNGFR+ gated cells. The level of activation-induced FOXP3 expression increased with each round of stimulation, with the optimal difference between control (siLuc) and siFOXP3-expressing T-cell lines observed after 3 cycles of stimulation (supplemental Figure 2). Therefore, all experiments were performed at or after this time point.
Proliferation assays and expansion curves
T-cell clones or lines were labeled with CFSE and stimulated with anti-CD3/anti-CD28–coated beads. Four days later, T cells were stained with fixable viability dye (eBiosciences), CD4, and, in some cases, ΔLNGFR mAbs. Data were acquired on a FACSCanto flow cytometer (BD Biosciences) and analyzed with the FlowJo Proliferation Platform Version 7.6 software (TreeStar) to determine the division index (ie, the average number of divisions undergone by a cell in the starting population). Expansion curves were determined by counting the number of live (fixable viability dye negative) ΔLNGFR+ cells with counting beads (surfactant-free white sulfate latex beads, 0.5μM; Interfacial Dynamics). Fold expansion was determined by dividing the number of cells at each time point by the number of live, unstimulated cells.
Determination of cytokine production
For CD4+CD25− T-cell clones and lines, IFN-γ and IL-2 production were measured 20 hours after activation and IL-17A after 48 hours by ELISA from supernatants of CFSE-proliferation assay cultures. For Th1 and Th17 cells, IFN-γ, IL-17A, and IL-2 production were measured on days 0, 8, and 11 after restimulation for 5.5 hours with PMA (10 ng/mL) and ionomycin (100 ng/mL), with brefeldin A (10 μg/mL) added for the last 3.5 hours. Cells were stained with anti-ΔLNGFR and then with anti–IFN-γ, anti–IL-17A, anti–IL-2, and anti-FOXP3. Th1 and Th17 cells were washed and replated in fresh medium with IL-2 (100 U/mL, 1 × 106 cells/mL) 8 days after activation. After 48 hours, the amounts of cytokines in supernatants were determined by the Th1/Th2/Th17 cytometric bead array (BD Biosciences).
Suppression assay
FOXP3-null Tconv clone responders were labeled with CFSE or Cell Proliferation Dye eFluor 670 (CPD; eBiosciences) and cocultured with WT or FOXP3-null Tconv clones labeled with CPD or CFSE. Cocultures were activated with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells; Invitrogen) and after 4 days were restimulated with PMA and ionomycin in the presence of 1 μg/mL of brefeldin A. Cells were then stained with anti–IFN-γ and anti-FOXP3 (236/AE7).
Statistical analysis
All statistical analyses were performed with the 2-tailed Student paired (where applicable) or unpaired t test. Data were first transformed with log (base 10) to account for variability between donors before the t test was performed.
Results
FOXP3-null human Tconv clones proliferate more and produce more IFN-γ and IL-2 than WT Tconv clones
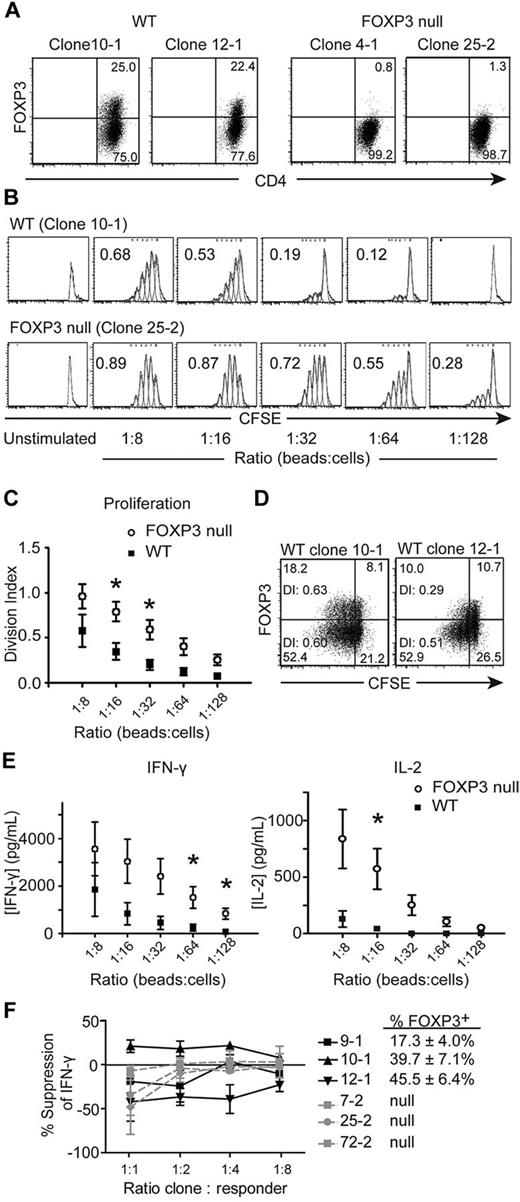
FOXP3 is located on the X-chromosome and, in females, is subject to X-chromosome inactivation.18 Tregs from carriers of FOXP3 mutations have a skewed pattern of X-inactivation with demethylation of the WT allele, whereas Tconv cells have a random X-inactivation profile.18 We sorted CD4+CD25− Tconv cells from a female subject who is hemizygous for a mutation in FOXP3 (2 T → C), which disrupts the initiating methionine and results in loss of protein expression,28,29 and isolated Tconv cell clones. Seven WT and 11 FOXP3-null Tconv clones were identified and analyzed by flow cytometry and molecular X-chromosome inactivation analysis (data not shown) to determine whether the WT or null allele of FOXP3 was active. Figure 1A shows representative examples of FOXP3 expression 4 days after activation for 2 WT and 2 FOXP3-null Tconv clones. As expected,5-11 WT Tconv clones did not express FOXP3 in the resting state, but up-regulated FOXP3 on activation; approximately 25% of cells with an active WT FOXP3 allele expressed FOXP3 4 days after activation. Clones with an active mutant allele had no detectable FOXP3 protein at any time point.
FOXP3-null CD4+CD25− Tconv cell clones proliferate to a greater extent and produce more IFN-γ and IL-2 than WT Tconv cell clones. Tconv cell clones from a subject who is hemizygous for a null mutation in FOXP3 were labeled with CFSE and stimulated with different ratios of anti-CD3/anti-CD28–coated beads. After 4 days, Tconv clones were stained with anti-CD4 and anti-FOXP3 Abs (236A/E7) and read on a FACSCanto. (A) Activation-induced FOXP3 expression in WT but not FOXP3-null Tconv cell clones. Two representative WT and 2 FOXP3-null Tconv clones are shown for clones that were stimulated with 1 anti-CD3/anti-CD28–coated bead per 32 cells for 4 days. (B) One representative experiment; numbers in each plot represent the division index. (C) Average division index of multiple WT (n = 7) and FOXP3-null (n = 11) Tconv clones. (D) Division indices (DI) of the FOXP3+ and FOXP3− populations are given within each plot of 2 representative WT Tconv clones stimulated with 1 anti-CD3/anti-CD28–coated bead per 32 cells for 4 days. (E) Average IFN-γ and IL-2 production by WT (n = 7) and FOXP3-null (n = 11) Tconv clones (5 × 105 cells/mL). Supernatants from cultures were collected 20 hours after T-cell activation and analyzed by ELISA. (F) Percent suppression of IFN-γ production in Tconv clone cocultures. FOXP3-null clones were labeled with CFSE and cocultured at the indicated ratios with WT (black) or FOXP3-null (gray) Tconv clones labeled with CPD. Cocultures were activated with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells). After 4 days, cocultures were restimulated with PMA and ionomycin and stained for IFN-γ. Shown is the average percent suppression against 3 different FOXP3-null responders by each WT or null clone. The average FOXP3 expression on day 4 for the WT clones is also shown (n = 3). Error bars represent SEM. *P < .05.
FOXP3-null CD4+CD25− Tconv cell clones proliferate to a greater extent and produce more IFN-γ and IL-2 than WT Tconv cell clones. Tconv cell clones from a subject who is hemizygous for a null mutation in FOXP3 were labeled with CFSE and stimulated with different ratios of anti-CD3/anti-CD28–coated beads. After 4 days, Tconv clones were stained with anti-CD4 and anti-FOXP3 Abs (236A/E7) and read on a FACSCanto. (A) Activation-induced FOXP3 expression in WT but not FOXP3-null Tconv cell clones. Two representative WT and 2 FOXP3-null Tconv clones are shown for clones that were stimulated with 1 anti-CD3/anti-CD28–coated bead per 32 cells for 4 days. (B) One representative experiment; numbers in each plot represent the division index. (C) Average division index of multiple WT (n = 7) and FOXP3-null (n = 11) Tconv clones. (D) Division indices (DI) of the FOXP3+ and FOXP3− populations are given within each plot of 2 representative WT Tconv clones stimulated with 1 anti-CD3/anti-CD28–coated bead per 32 cells for 4 days. (E) Average IFN-γ and IL-2 production by WT (n = 7) and FOXP3-null (n = 11) Tconv clones (5 × 105 cells/mL). Supernatants from cultures were collected 20 hours after T-cell activation and analyzed by ELISA. (F) Percent suppression of IFN-γ production in Tconv clone cocultures. FOXP3-null clones were labeled with CFSE and cocultured at the indicated ratios with WT (black) or FOXP3-null (gray) Tconv clones labeled with CPD. Cocultures were activated with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells). After 4 days, cocultures were restimulated with PMA and ionomycin and stained for IFN-γ. Shown is the average percent suppression against 3 different FOXP3-null responders by each WT or null clone. The average FOXP3 expression on day 4 for the WT clones is also shown (n = 3). Error bars represent SEM. *P < .05.
Because anergy is associated with FOXP3 expression in Tregs, we investigated how FOXP3 affects the proliferation of Tconv cells. WT and FOXP3-null Tconv clones were labeled with CFSE, activated, and after 4 days the division index was calculated. As shown in Figure 1B-C and supplemental Figure 3, FOXP3-null Tconv clones proliferated significantly more than WT Tconv clones. Within each WT clone culture, the population of WT clones that did not express FOXP3 4 days after TCR-stimulation (approximately 75%) had a similar or higher division index than the population of WT clones expressing FOXP3 (approximately 25%), suggesting that the lower division index of WT clones was not because of suppression of the FOXP3− population by the FOXP3+ population within the culture (Figure 1D).
FOXP3 negatively regulates a variety of cytokines, including IFN-γ and IL-2, so we investigated whether cytokine production differed between FOXP3-null and WT Tconv clones. T-cell clones were stimulated and IFN-γ and IL-2 production were determined. The FOXP3-null Tconv clones produced significantly more IFN-γ and IL-2 than the WT Tconv clones (Figure 1E and supplemental Figure 4). For IL-2, at ratios of 1:32 beads to cells and lower, WT Tconv clones failed to produce detectable levels of IL-2, whereas the FOXP3-null Tconv clones continued to produce IL-2 even at ratios as low as 1:128 beads to cells. None of the Tconv cell clones produced detectable IL-17 after 48 hours (data not shown).
We carried out suppression assays to determine whether the increased activation of FOXP3-null Tconv clones was because of loss of intrinsic regulation by FOXP3 versus loss of acquired suppressive function by the FOXP3-expressing cells within the culture. FOXP3-null Tconv clones were responders, labeled with CFSE, and cocultured with WT or null Tconv clones that were labeled with CPD. Four days after activation, cocultures were stained for IFN-γ. As shown in Figure 1F and supplemental Figure 5, there was no consistent ability of the WT Tconv clones to suppress IFN-γ production by the FOXP3-null clones. Similarly, the FOXP3-null Tconv clones did not suppress IFN-γ production by FOXP3-null Tconv clones.
Differential gene expression in FOXP3-null versus WT Tconv clones
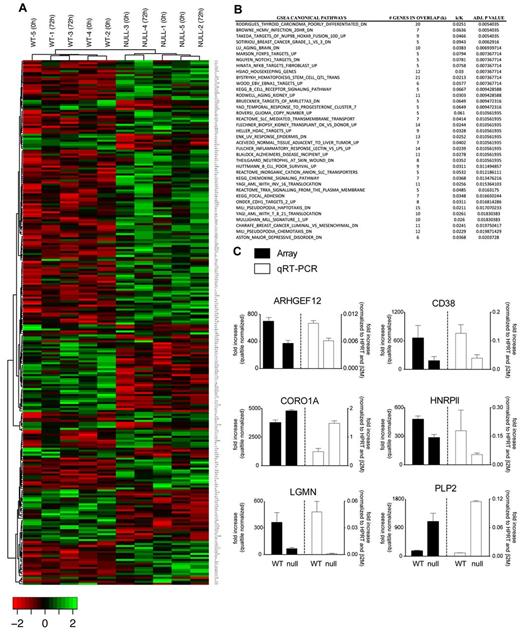
To identify genes regulated by FOXP3 in Tconv cells, we performed microarray gene expression analysis in FOXP3-null and WT clones and found significant differences (Benjamini-Hochberg adjusted P < .05) in the expression of 274 genes (182 down-regulated and 92 up-regulated), which permitted hierarchical clustering on the basis of the FOXP3 genotype (Figure 2A). GSEA was applied and the results highlighted FOXP3-regulated expression of processes linked to immune cell biology, signaling, and inflammatory response (Figure 2B). Eleven genes were selected to validate based on their fold change and biologic relevance, and differential expression of 6 of these was confirmed (Figure 2C). CORO1A and HNRPll belong to a previously reported list of FOXP3 targets.30
Gene-expression analysis of FOXP3 WT and null Tconv clones. Total RNA from T-cell clones was hybridized to a GeneChip Human Gene 1.0 ST array. (A) Hierarchical clustering revealed differential expression of 274 RefSeq annotated genes with a P < .05 (Benjamini correction). (B) Differentially expressed genes were subject to GSEA after cross-validation using randomization from background genes. (C) Differential expression of 6 genes was confirmed by quantitative RT-PCR. Mean values ± SEM determined in triplicate are shown.
Gene-expression analysis of FOXP3 WT and null Tconv clones. Total RNA from T-cell clones was hybridized to a GeneChip Human Gene 1.0 ST array. (A) Hierarchical clustering revealed differential expression of 274 RefSeq annotated genes with a P < .05 (Benjamini correction). (B) Differentially expressed genes were subject to GSEA after cross-validation using randomization from background genes. (C) Differential expression of 6 genes was confirmed by quantitative RT-PCR. Mean values ± SEM determined in triplicate are shown.
Knockdown of activation-induced FOXP3 expression in human Tconv cells with siRNA increases their proliferation
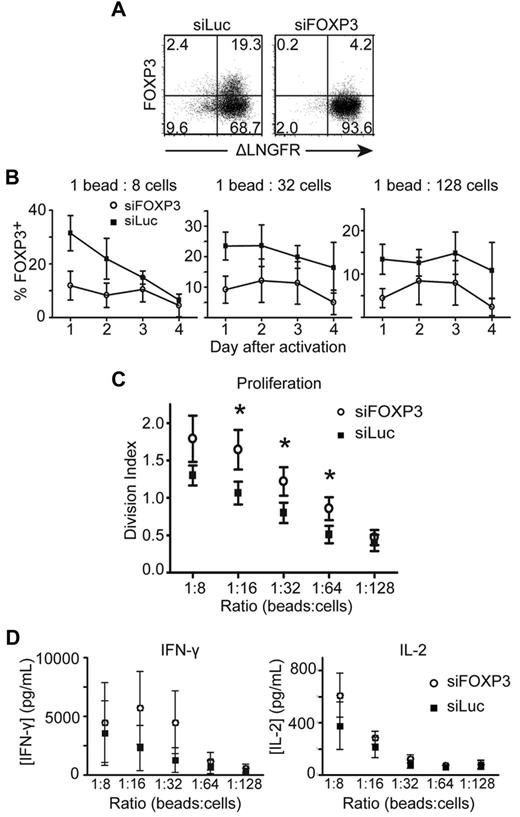
To exclude the possibility that the effects of FOXP3 deficiency were restricted to carriers of FOXP3 mutations, we knocked down the expression of activation-induced FOXP3 by transducing naive Tconv cells from healthy donors with a lentiviral vector encoding siFOXP3. In Tregs, this vector decreases expression of FOXP3 by an average of 64%.27 Purified transduced T-cell lines were activated, and expression of siFOXP3 resulted in an average reduction in FOXP3 expression of 54.9% ± 3.29% (Figure 3A-B).
CD4+CD25− T cells transduced with siRNA against FOXP3 proliferate to a greater extent than CD4+CD25− T cells transduced with control siRNA against luciferase. CD4+CD25− T cells transduced with siFOXP3 or siLuc were purified as ΔLNGFR+ cells, then labeled with CFSE and stimulated with different ratios of anti-CD3/anti-CD28–coated beads. Transduced T cells were stained with anti-CD4, anti-ΔLNGFR, and anti-FOXP3 Abs. (A) Representative FACS plots of FOXP3 expression in purified siFOXP3 and siLuc-transduced T cells 3 days after activation at a 1:32 bead to cell ratio. (B) FOXP3 expression in ΔLNGFR+ siFOXP3-transduced T cells and control siLuc-transduced T cells after anti-CD3/anti-CD28 bead stimulation (n = 4). (C) Average division index of siFOXP3-transduced T cells and control siLuc-transduced T cells (n = 4). (D) CD4+CD25− T cells transduced with siFOXP3 or siLuc were stimulated with different ratios of anti-CD3/anti-CD28–coated beads at 5 × 105 cells/mL. Supernatants were collected 20 hours later and analyzed for IFN-γ and IL-2 by ELISA (n = 4). Error bars represent SEM. *P < .05.
CD4+CD25− T cells transduced with siRNA against FOXP3 proliferate to a greater extent than CD4+CD25− T cells transduced with control siRNA against luciferase. CD4+CD25− T cells transduced with siFOXP3 or siLuc were purified as ΔLNGFR+ cells, then labeled with CFSE and stimulated with different ratios of anti-CD3/anti-CD28–coated beads. Transduced T cells were stained with anti-CD4, anti-ΔLNGFR, and anti-FOXP3 Abs. (A) Representative FACS plots of FOXP3 expression in purified siFOXP3 and siLuc-transduced T cells 3 days after activation at a 1:32 bead to cell ratio. (B) FOXP3 expression in ΔLNGFR+ siFOXP3-transduced T cells and control siLuc-transduced T cells after anti-CD3/anti-CD28 bead stimulation (n = 4). (C) Average division index of siFOXP3-transduced T cells and control siLuc-transduced T cells (n = 4). (D) CD4+CD25− T cells transduced with siFOXP3 or siLuc were stimulated with different ratios of anti-CD3/anti-CD28–coated beads at 5 × 105 cells/mL. Supernatants were collected 20 hours later and analyzed for IFN-γ and IL-2 by ELISA (n = 4). Error bars represent SEM. *P < .05.
To determine whether siFOXP3-expressing T cells had changes in proliferation and/or cytokine production, siRNA-transduced Tconv cells were labeled with CFSE and proliferation was measured 4 days after stimulation. For FOXP3-null T-cell clones, siFOXP3-transduced Tconv cells proliferated significantly more than control-transduced cells (Figure 3C). There was also a trend toward greater IFN-γ and IL-2 production by siFOXP3-expressing T cells (Figure 3D), but this result did not reach statistical significance.
In addition to repressing cytokines, FOXP3 positively regulates cell-surface markers including CD25 and CTLA-4.19,31 These markers were analyzed after activation with various strengths of stimulus, but no difference was found in expression of CTLA-4 or CD25 between control- or siFOXP3-transduced Tconv cells (data not shown).
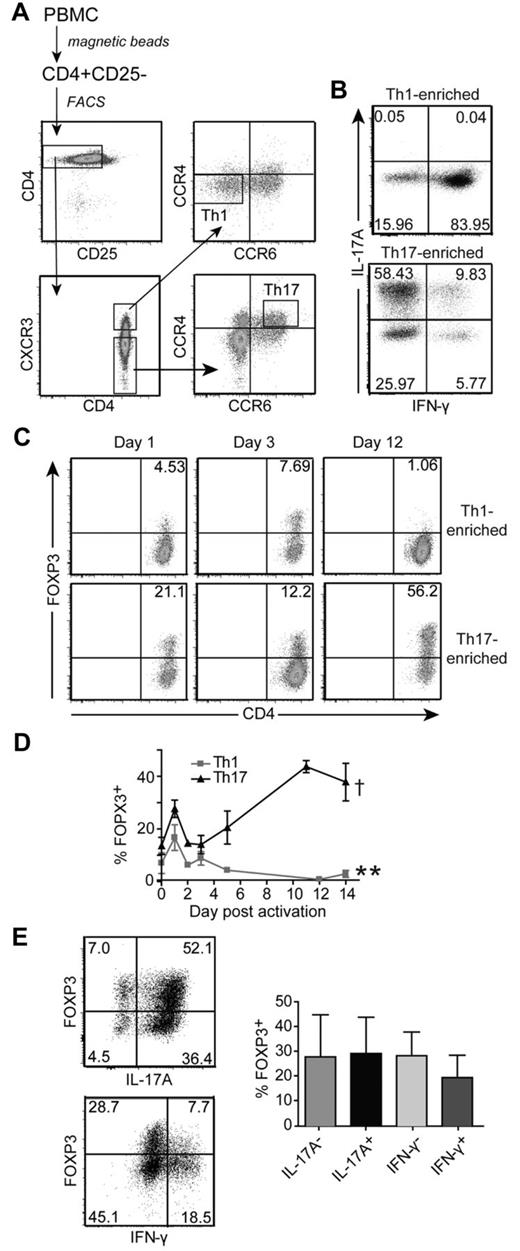
In vivo–differentiated Th17 cells express significantly more activation-induced FOXP3 than Th1 cells
FOXP3 controls the differentiation of induced Tregs versus Th17 cells, but little is known about its function in fully differentiated Th17 cells. We sorted Th1 and Th17 cells on the basis of chemokine receptor expression.26 Th1 cells were CD4+CD25−CXCR3+CCR4−CCR6− and Th17 cells were CD4+CD25−CXCR3−CCR6+CCR4+ (Figure 4A). These Th1-enriched and Th17-enriched cells were expanded and cytokine expression was determined. As expected, Th17 cells produced IL-17A but little IFN-γ, whereas the Th1 cells produced IFN-γ but no detectable IL-17A (Figure 4B). To determine FOXP3 expression, Th1- and Th17-enriched cells were restimulated. As was the case for previous studies on undifferentiated CD4+ T cells,5-11 FOXP3 expression was induced in Th1-enriched cells but returned to baseline levels after 4-7 days (Figure 4C-D). In contrast, Th17-enriched cells expressed significantly more activation-induced FOXP3 compared with Th1 cells. Moreover, the expression of FOXP3 did not wane and remained high throughout the 14-day activation cycle. Similar results were obtained when T cells were reactivated with APC and soluble anti-CD3 (supplemental Figure 6). In the resting state, the FOXP3-expressing cells did not coexpress Helios (data not shown).
Enriched Th17 cells express high levels of FOXP3 on activation. (A) Sorting procedure for human Th1 (CD4+CD25−CXCR3+CCR4−CCR6−) and Th17 (CD4+CD25−CXCR3−CCR4+CCR6+) cells. CD4+ T cells were isolated from human PBMCs and depleted of CD25+ cells before sorting. (B) Phenotype of Th1-enriched and Th17-enriched cells after 2 weeks of expansion with APC and anti-CD3 Abs. T cells were restimulated and IL-17A and IFN-γ production were determined by intracellular cytokine staining. The cytokine profiles from 1 representative donor of 6 are shown. (C) Representative plots of FOXP3 expression in Th1-enriched and Th17-enriched cells from one donor of 4 on days 1, 3, and 12 after reactivation with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells) are shown. (D) Average FOXP3 expression in Th1-enriched and Th17-enriched cells after reactivation with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells) over 14 days. Each time point represents the average FOXP3 expression from 2-5 different donors. **Significant difference in FOPX3 expression between Th1-enriched and Th17-enriched cells 14 days after reactivation (P = .0028). There is no difference at day 0 (P = .1080). †Significant difference in FOXP3 expression between day 0 and 14 in Th17-enriched cells (P = .0126). (E) Representative (left panel) and average (right panel; n = 3 for IL-17, n = 2 for IFN-γ) FOXP3 expression in IL-17A+, IL-17A−, IFN-γ+, and IFN-γ− subsets within Th17-enriched cultures 12 days after restimulation with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells).
Enriched Th17 cells express high levels of FOXP3 on activation. (A) Sorting procedure for human Th1 (CD4+CD25−CXCR3+CCR4−CCR6−) and Th17 (CD4+CD25−CXCR3−CCR4+CCR6+) cells. CD4+ T cells were isolated from human PBMCs and depleted of CD25+ cells before sorting. (B) Phenotype of Th1-enriched and Th17-enriched cells after 2 weeks of expansion with APC and anti-CD3 Abs. T cells were restimulated and IL-17A and IFN-γ production were determined by intracellular cytokine staining. The cytokine profiles from 1 representative donor of 6 are shown. (C) Representative plots of FOXP3 expression in Th1-enriched and Th17-enriched cells from one donor of 4 on days 1, 3, and 12 after reactivation with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells) are shown. (D) Average FOXP3 expression in Th1-enriched and Th17-enriched cells after reactivation with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells) over 14 days. Each time point represents the average FOXP3 expression from 2-5 different donors. **Significant difference in FOPX3 expression between Th1-enriched and Th17-enriched cells 14 days after reactivation (P = .0028). There is no difference at day 0 (P = .1080). †Significant difference in FOXP3 expression between day 0 and 14 in Th17-enriched cells (P = .0126). (E) Representative (left panel) and average (right panel; n = 3 for IL-17, n = 2 for IFN-γ) FOXP3 expression in IL-17A+, IL-17A−, IFN-γ+, and IFN-γ− subsets within Th17-enriched cultures 12 days after restimulation with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells).
FOXP3 expression in Th17-enriched cells was not because of loss of a Th17 cell phenotype because the percentage of IL-17–expressing cells did not decrease as the expression of FOXP3 increased (supplemental Figure 7). There was coexpression of FOXP3 and IL-17 (Figure 4E), but a trend to lower FOXP3 expression within the IFN-γ+ cells (Figure 4E).
Enrichment on the basis of chemokine receptor expression does not result in a 100% pure population of Th17 cells, and non-Th17 cells in the cultures could influence the results. We therefore attempted to use an alternate method to isolate Th17 cells: cytokine capture of IL-17A+ cells. IL-17A+ cells were sorted and expanded, and expression of cytokines and FOXP3 were measured after activation. In contrast to the chemokine receptor-sorted cells,39 which always contained less than 20% of IFN-γ–producing cells (supplemental Figure 7), a significant (always > 50%) proportion of the IL-17A–captured cells produced IFN-γ (supplemental Figure 8). This finding is consistent with Nistala et al,32 and meant this approach could not be further pursued because of the impurity of the cells.
Activation-induced FOXP3 limits the expansion of Th17 cells
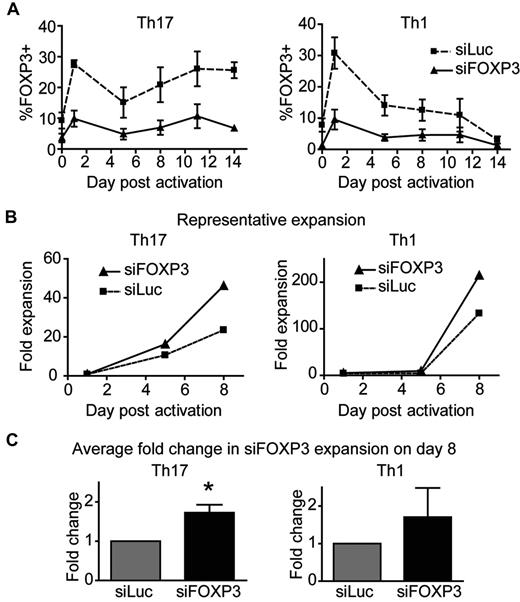
To investigate the function of FOXP3 in Th1 and Th17 cells, chemokine receptor–sorted Th1 and Th17 cells were transduced with control or siFOXP3 lentivirus and expanded for 2 weeks. Similar to data with undifferentiated CD4+ T-cell lines, activation-induced FOXP3 expression was reduced by 65.54% ± 2.11% in siFOXP3-Th1 cells and by 67.30% ± 4.15% in siFOXP3-Th17 cells (Figure 5A). The fold expansion of the T-cell lines was determined, and siFOXP3-Th17 cells had a greater expansion potential than control Th17 cells (Figure 5B-C), with FOXP3-deficient Th17 cells expanding 1.73 ± 0.20-fold times more than control Th17 cells (P = .0229). In contrast, the expansion potential of FOXP3-deficient Th1 cells was variable, with no significant difference in proliferation. The difference in expansion potential between Th1 and Th17 cells confirms our previous finding and recent reports.33,34
FOXP3-deficient human Th17 cells have a greater expansion potential than control Th17 cells. Th1 and Th17 cells transduced with siFOXP3 or control siLuc were stimulated with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells) in the presence of IL-2. (A) FOXP3 expression in siFOXP3 and control siLuc-transduced Th1 and Th17 cells over 2 weeks (n = 4). (B-C) At the indicated time points, cells were collected and stained with viability dye and anti-ΔLNGFR. Live, ΔLNGFR+ cells were counted by flow cytometry with counting beads. Fold expansion was determined by dividing the number of cells at each time point by the number of live, unstimulated cells counted by the same method on day 1. One representative experiment of 4 is shown in panel B and the average fold expansion of siFOXP3-transduced T cells over control siLuc-transduced T cells on day 8 after activation is shown in panel C. Error bars represent SEM. *P < .05.
FOXP3-deficient human Th17 cells have a greater expansion potential than control Th17 cells. Th1 and Th17 cells transduced with siFOXP3 or control siLuc were stimulated with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells) in the presence of IL-2. (A) FOXP3 expression in siFOXP3 and control siLuc-transduced Th1 and Th17 cells over 2 weeks (n = 4). (B-C) At the indicated time points, cells were collected and stained with viability dye and anti-ΔLNGFR. Live, ΔLNGFR+ cells were counted by flow cytometry with counting beads. Fold expansion was determined by dividing the number of cells at each time point by the number of live, unstimulated cells counted by the same method on day 1. One representative experiment of 4 is shown in panel B and the average fold expansion of siFOXP3-transduced T cells over control siLuc-transduced T cells on day 8 after activation is shown in panel C. Error bars represent SEM. *P < .05.
Activation-induced FOXP3 limits IFN-γ production by Th17 cells
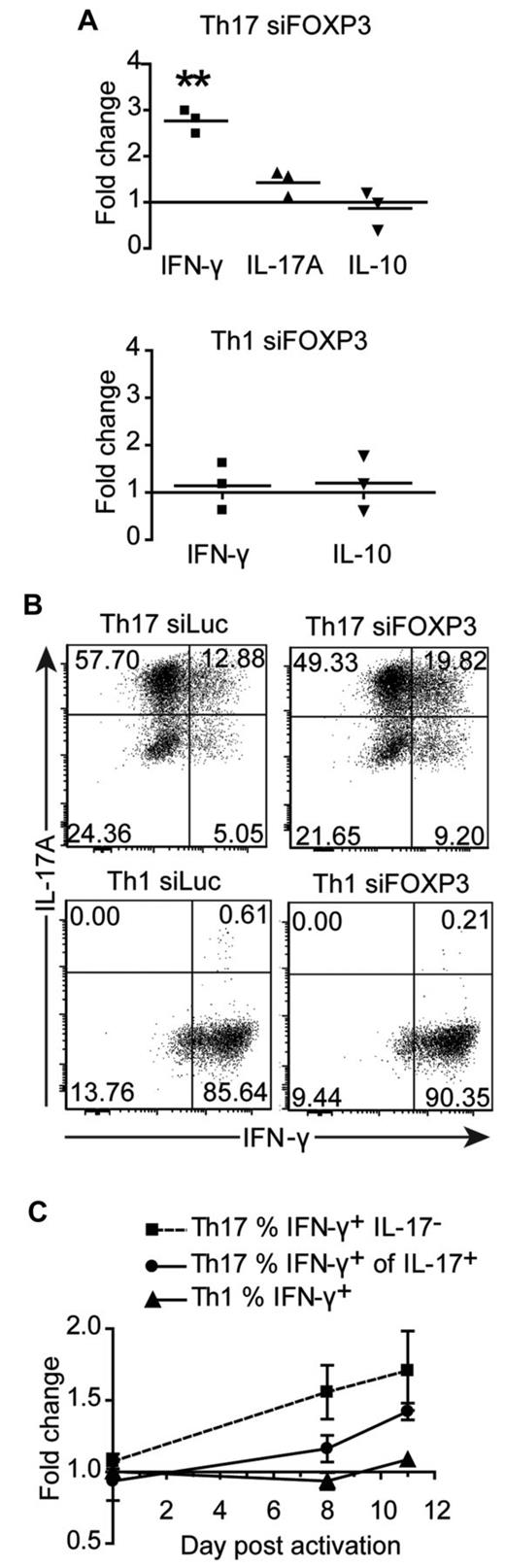
We next investigated whether cytokine production was altered in siFOXP3-tranduced Th1 and Th17 cells. Because FOXP3 expression remained high in the second week after TCR activation of Th17 cells, we examined cytokine production at this time point. Eight days after activation, cells were washed and replated and supernatants were collected after 48 hours. There was an average of 2.77 ± 0.15 times more IFN-γ in the supernatants of siFOXP3-Th17 cells compared with control Th17 cells (Figure 6A, P = .0028), but no difference in IL-17 or IL-10. Similarly, siFOXP3-Th17 cells had higher proportions of IFN-γ–producing IL-17+ cells and IFN-γ+IL-17− cells (Figure 6B-C). Conversely, expression of siFOXP3 had no effect on the proportion of cells expressing IL-17 (Figure 6B) or IL-2 (data not shown). Cytokine production from siFOXP3-Th1 cells was analyzed, but there were no differences in IFN-γ, IL-10 (Figure 6A-C), or IL-2 (data not shown).
Cytokine production by FOXP3-deficient Th17 and Th1 cell lines. Th1 and Th17 cells transduced with siFOXP3 or control siLuc and purified based on ΔLNGFR expression were restimulated with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells) in the presence of IL-2. (A) On day 8 after stimulation, cells were washed and replated at 1 × 106 cells/mL, and supernatants were collected 48 hours later for analysis by cytometric bead array. Each dot represents the fold change in cytokine production by siFOXP3-transduced T cells relative to control siLuc-transduced T cells for one donor. (B-C) On days 0, 8, and 11 of the expansion, Th1 and Th17 cells were restimulated with PMA and ionomycin and stained intracellularly for IFN-γ and IL-17A. Analysis was conducted on ΔLNGFR+ cells. (B) Representative plots of IL-17A and IFN-γ expression 11 days after activation. (C) The average fold changes in the percentage of IFN-γ+IL-17− Th17 cells (n = 4), IFN-γ+ of IL-17+ Th17 cells (n = 4), and IFN-γ+ Th1 cells (n = 3) in siFOXP3 relative to control siLuc over the course of the experiment. At day 8 after activation, the fold change in the percentage of IFN-γ+IL-17− Th17 cells is significant (P = .0436) and at day 11 after activation, the fold change in the percentage of IFN-γ+ of IL-17+ is significant (P = .0033).
Cytokine production by FOXP3-deficient Th17 and Th1 cell lines. Th1 and Th17 cells transduced with siFOXP3 or control siLuc and purified based on ΔLNGFR expression were restimulated with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells) in the presence of IL-2. (A) On day 8 after stimulation, cells were washed and replated at 1 × 106 cells/mL, and supernatants were collected 48 hours later for analysis by cytometric bead array. Each dot represents the fold change in cytokine production by siFOXP3-transduced T cells relative to control siLuc-transduced T cells for one donor. (B-C) On days 0, 8, and 11 of the expansion, Th1 and Th17 cells were restimulated with PMA and ionomycin and stained intracellularly for IFN-γ and IL-17A. Analysis was conducted on ΔLNGFR+ cells. (B) Representative plots of IL-17A and IFN-γ expression 11 days after activation. (C) The average fold changes in the percentage of IFN-γ+IL-17− Th17 cells (n = 4), IFN-γ+ of IL-17+ Th17 cells (n = 4), and IFN-γ+ Th1 cells (n = 3) in siFOXP3 relative to control siLuc over the course of the experiment. At day 8 after activation, the fold change in the percentage of IFN-γ+IL-17− Th17 cells is significant (P = .0436) and at day 11 after activation, the fold change in the percentage of IFN-γ+ of IL-17+ is significant (P = .0033).
Activation-induced FOXP3 up-regulates CCR4 in Th17 cells
We next examined whether activation-induced FOXP3 positively regulates cell-surface markers in Th1 and Th17 cells. CCR4 expression was significantly decreased in siFOXP3-Th17 cells compared with control Th17 cells (Figure 7A and supplemental Figure 9). Similarly, expression of CCR6 expression was reduced in siFOXP3-Th17, but this difference was not statistically significant (Figure 7B and supplemental Figure 9). In contrast, there was no difference in CTLA-4 expression between siFOXP3-Th17 and siLuc-Th17 or Th1 cells (Figure 7C-D). There was also no difference in CCR4 or CCR6 expression in siFOXP3-transduced Th1 cells because they do not express these proteins highly (data not shown).
Cell-surface marker expression in FOXP3-deficient Th1 and Th17 cells. Th1 and Th17 cells transduced with siFOXP3 or control siLuc and purified based on ΔLNGFR expression were restimulated with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells) in the presence of IL-2. At the indicated days after activation, cell-surface marker expression was determined by flow cytometry. Analysis was conducted on ΔLNGFR+ cells. The top panels of A-D show mean fluorescence intensities (MFIs) of one representative experiment. The bottom panels of A-D show the average fold change in MFI of siFOXP3 relative to siLuc (MFI siFOXP3/ MFI siLuc). (A) Th17 CCR4 (n = 3-4). (B) Th17 CCR6 (n = 2-3). (C) Th17 CTLA-4 (n = 2-3). (D) Th1 CTLA-4 (n = 2-3). Error bars represent SEM. *P < .05; **P < .01.
Cell-surface marker expression in FOXP3-deficient Th1 and Th17 cells. Th1 and Th17 cells transduced with siFOXP3 or control siLuc and purified based on ΔLNGFR expression were restimulated with anti-CD3/anti-CD28–coated beads (1 bead: 32 cells) in the presence of IL-2. At the indicated days after activation, cell-surface marker expression was determined by flow cytometry. Analysis was conducted on ΔLNGFR+ cells. The top panels of A-D show mean fluorescence intensities (MFIs) of one representative experiment. The bottom panels of A-D show the average fold change in MFI of siFOXP3 relative to siLuc (MFI siFOXP3/ MFI siLuc). (A) Th17 CCR4 (n = 3-4). (B) Th17 CCR6 (n = 2-3). (C) Th17 CTLA-4 (n = 2-3). (D) Th1 CTLA-4 (n = 2-3). Error bars represent SEM. *P < .05; **P < .01.
Discussion
In the present study, we investigated the role of activation-induced FOXP3 in human CD4+ T cells and found that this protein acts as an intrinsic negative regulator of T-cell proliferation and cytokine production. Tconv cells with ectopically reduced or genetically absent FOXP3 expression proliferated to a greater extent and produced more IFN-γ and IL-2 compared with WT T cells. We also found that compared with Th1 cells, Th17 cells expressed significantly more FOXP3. Functionally, expression of FOXP3 in Th17 cells limited their expansion and prevented IFN-γ expression while up-regulating CCR4 expression. In contrast, although Th1 cells also expressed FOXP3 on activation, its expression did not appear to contribute significantly to their ability to expand or produce IFN-γ or IL-2.
FOXP3-deficient Tconv cells had a greater proliferative capacity than WT Tconv cells. This FOXP3-mediated inhibition of proliferation in WT Tconv cells is likely an intrinsic effect of FOXP3 rather than a transient acquisition of suppressive function by FOXP3+ Tconv clones, because if FOXP3+ Tconv cells suppressed FOXP3− Tconv cells within the same culture, the FOXP3− Tconv cells would have a lower division index than the FOXP3+ T cells. In fact, the FOXP3− Tconv cells had a similar or higher division index than the FOXP3+ Tconv cells. The fact that WT FOXP3 clones did not suppress IFN-γ production by FOXP3-null clones further supports this conclusion.
To identify FOXP3 target genes in Tconv cells, we compared the gene-expression profiles of WT versus FOXP3-null T-cell clones. A total of 274 genes were differentially expressed regardless of the state of activation. Among these genes, several were previously identified as targets of mouse FOXP3,30 highlighting the conservation of FOXP3-regulated gene expression across species. Differential gene expression was validated by quantitative RT-PCR for 6 genes and further studies are required to understand the biologic function of these FOXP3-regulated genes in Tconv cells.
We also examined how FOXP3 affects cytokine production by Tconv cells and found that FOXP3-null T-cell clones had significantly increased cytokine production. This phenotype, however, was not as robust in siFOXP3-transduced Tconv cells; although there was a trend of increased cytokine production, it was not statistically significant. This difference is likely because the FOXP3-null T cells clones do not express any FOXP3, whereas the knockdown only reduced FOXP3 expression by approximately 50%. In contrast, in Th17 cells, there is a strong intrinsic role for FOXP3 in regulating activation: a significant increase in IFN-γ production was evident even with only approximately 65% reduction in FOXP3 expression. These data suggest that FOXP3 may restrain Th17 cells from becoming the IL-17+IFN-γ+ T cells that are implicated in many inflammatory diseases.32,35-39 FOXP3 is known to suppress the Th1 developmental program during Th17 differentiation,40 and our data suggest that this function continues in fully differentiated Th17 cells.
Whether FOXP3-expressing Tconv cells acquire suppressive capacity has been debated.5,7-11,41 In contrast to unpolarized CD4+ T cells or Th1 cells, our present findings suggest that with repeated activation, Th17 cells may gain regulatory function. In support of this possibility, after repetitive stimulation, Th17 cell clones acquire an increasingly demethylated TSDR and regulatory capacity42 and there is evidence for suppressive IL-17+FOXP3+ T cells in the blood of both healthy subjects16,17 and colitic patients.15 The origin of IL-17+FOXP3+ cells was assumed to be the result of FOXP3+ Tregs becoming unstable and acquiring IL-17 production, but our data suggest that the opposite may also be happening: Th17 cells may acquire FOXP3. A caveat is that the Th17-enriched cells contained a small proportion of cells that expressed FOXP3 and Helios, but were not CD25 high, ex vivo. Because both FOXP3 and Helios can be expressed on TCR activation,12 it is possible that these are ex vivo–activated Th17 cells rather than classic thymus-derived Tregs. Furthermore, contaminating Tregs would not be expected to preferentially expand and, after expansion, no FOXP3+ cells were Helios+. Therefore, although we cannot exclude the possibility that contaminating Tregs contribute to the reported phenotype of the Th17-enriched cells, the data argue against this possibility.
FOXP3 may be induced to different degrees depending on the environment. New evidence shows that FOXP3 can be transiently expressed in mouse Tconv cells if they are activated without memory T cells or APCs.43 In human Tconv cells, FOXP3 is expressed in the presence of APCs, but it is notable that induction of FOXP3 expression in Th17 cells was not as consistent when cells were activated with APCs (supplemental Figure 6) versus anti-CD3/anti-CD28–coated beads. Therefore, different types of APCs may affect activation-induced FOXP3 expression and activation-induced FOXP3-expression was indeed lower in cells stimulated with mature versus immature DCs.6
In conclusion, FOXP3 is an important regulator of T-cell activation and proliferation that extrinsically confers Tregs with the ability to suppress Tconv cells, but also intrinsically limits Tconv cells. CTLA-4 also intrinsically negatively regulates T-cell proliferation,44 but FOXP3 likely acts in parallel to CTLA-4 instead of upstream, because knocking down FOXP3 had no effect on CTLA-4 expression. Although FOXP3 is expressed continuously at high levels in Tregs, it is only induced in Tconv cells after TCR activation. Therefore, on TCR activation, Tconv cells are first able to respond, but then expression of FOXP3 limits their effector function. This role of FOXP3 as a regulator of activation appears to be especially relevant in Th17 cells, and the function of FOXP3 in these cells in the context of disease warrants further investigation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the families affected by immunodysregulation polyendocrinopathy enteropathy X-linked syndrome for their willingness to participate in this research; Anne Junker for facilitating clinical research; Mario Amendola and Luigi Naldini at HSR-TIGET, Milan, for generously providing the FOXP3 and control luciferase siRNA vectors; Massimiliano Cecconi from the Genetics Laboratory at Galliera Hospital in Genoa, Italy, for X-inactivation analysis on carrier T-cell clones; Claudia Sartirana at HSR-TIGET for excellent help in T-cell cloning; Lisa Xu for excellent flow cytometry support; and the Cell Separator Unit at Vancouver General Hospital, NetCAD at Canadian Blood Services, and the Transfusion Safety Unit at BC Children's & Women's Hospital for blood collection.
This work was supported by grants from the Canadian Institutes for Health Research (CIHR; MOP-93793), and StemCell Technologies. A.N.M holds a CIHR Canada Vanier Scholarship, a Michael Smith Foundation for Health Research Junior Graduate Studentship, and a CIHR Transplantation Training Program award. R.B. is supported by the Italian Telethon Foundation, Rome. M.K.L. holds a Canada Research Chair in Transplantation. A.N.M. is a doctoral candidate at the University of British Columbia and this work is submitted in partial fulfillment of the requirement for the degree.
Authorship
Contribution: A.N.M. performed the research, analyzed the data, and wrote the manuscript; J.G. and S.D.N. performed the research; M.C.G. performed cell culture, RNA extraction, and quantitative RT-PCR for validation experiments and edited the manuscript; M.R. and J.M.G.-M. performed the microarray and quantitative RT-PCR data analysis and edited the manuscript; D.C. and E.S. supervised the data analysis and edited the manuscript; D.L. supervised the quantitative RT-PCR experiments; I.S.P. and A.B. performed the arrays; M.G.R. provided scientific guidance; R.B. designed and coordinated the research and edited the manuscript; and M.K.L. designed the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Megan Levings, Department of Surgery, University of British Columbia, Room A4-186, 950 West 28th Ave, Vancouver, BC V5Z 4H4 Canada; e-mail: megan.levings@ubc.ca.
References
Author notes
R.B. and M.K.L. are co–senior authors.