Key Points
Crystal structure of human USB1 identifies it as a member of the LigT-like superfamily of 2H phosphoesterases.
USB1 protects spliceosomal U6 small nuclear RNA from aberrant 3′ oligoadenylation.
Abstract
The recessive disorder poikiloderma with neutropenia (PN) is caused by mutations in the C16orf57 gene that encodes the highly conserved USB1 protein. Here, we present the 1.1 Å resolution crystal structure of human USB1, defining it as a member of the LigT-like superfamily of 2H phosphoesterases. We show that human USB1 is a distributive 3′-5′ exoribonuclease that posttranscriptionally removes uridine and adenosine nucleosides from the 3′ end of spliceosomal U6 small nuclear RNA (snRNA), directly catalyzing terminal 2′, 3′ cyclic phosphate formation. USB1 measures the appropriate length of the U6 oligo(U) tail by reading the position of a key adenine nucleotide (A102) and pausing 5 uridine residues downstream. We show that the 3′ ends of U6 snRNA in PN patient lymphoblasts are elongated and unexpectedly carry nontemplated 3′ oligo(A) tails that are characteristic of nuclear RNA surveillancetargets. Thus, our study reveals a novel quality control pathway in which posttranscriptional 3′-end processing by USB1 protects U6 snRNA from targeting and destruction by the nuclear exosome. Our data implicate aberrant oligoadenylation of U6 snRNA in the pathogenesis of the leukemia predisposition disorder PN.
Introduction
Poikiloderma with neutropenia (PN; OMIM 604173) is an autosomal recessive cancer predisposition syndrome, characterized by poikiloderma, nail dystrophy (pachonychia), short stature, pulmonary disease, and neutropenia with significant predisposition to myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML).1,2 To date, 38 PN patients have been reported, harboring 19 different mutations in the C16orf57 gene1-3 that encodes a 265 amino acid protein, herein called USB1 (U6 biogenesis 1). Biallelic mutations in the C16orf57 gene also produce phenotypes that overlap with dyskeratosis congenita and Rothmund-Thomson syndrome.2 The C16orf57 mutations are all predicted to generate truncated, nonfunctional proteins.3 However, the function of the USB1 protein remains poorly characterized and the pathogenesis of PN remains obscure.
The removal of introns from pre-mRNA by the process of splicing is essential for gene expression and is catalyzed by the spliceosome, which contains approximately 200 proteins and 5 small ribonucleoprotein particles (the U1, U2, U4, U5, and U6 snRNPs).4 Indicating its critical importance, the U6 small nuclear RNA (snRNA) is the most highly conserved across species of all 5 of the snRNAs involved in the spliceosome, with at least 49 copies of the U6 gene in the human genome (hg18). The snRNPs assemble onto the pre-mRNA in an ordered and stepwise manner to generate the active spliceosome (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). First, the U1 snRNP interacts with the 5′ splice site and the U2 snRNP binds to the branch point sequence. Next, the association of the U4/U6.U5 tri-snRNP with the pre-mRNA promotes dissociation of the U1 and U4 snRNPs from the spliceosome. The extensive base pairing interaction between the U4 and U6 snRNPs in the U4/U6.U5 tri-snRNP is unwound by the RNA helicase Brr2/U5-200K to enable U6 snRNA to base pair with the U2 snRNA and the pre-mRNA at the catalytic core of the spliceosome during the first step of splicing. After completion of both steps of the splicing reaction, the spliceosome disassembles and the snRNPs are recycled for another round of splicing.5
The biogenesis of U6 snRNA differs in several respects from the other spliceosomal snRNAs. U1, U2, U4, and U5 snRNAs are transcribed by RNA polymerase II, have a trimethylated guanosine cap and a cytoplasmic maturation phase. In contrast, U6 snRNA is transcribed by RNA polymerase III, has a γ-monomethyl triphosphate cap6 and probably does not leave the nucleus.7 Newly transcribed U6 snRNA contains a 3′ stretch of 4 templated uridine residues that are immediately bound by the La protein.8-10 In mammalian cells, the majority of mature U6 snRNA contains 4 templated and 1 nontemplated uridine with a 2′, 3′ cyclic phosphate (> p) modification (UUUU(U) > p) at the 3′ end (denoted 3′ > p)11 and is associated with the set of 7 LSm proteins (LSm2-8) instead of the La protein.12-17 The presence of the 3′ > p modification appears to enhance the affinity of U6 snRNA for the heptameric LSm2-8 complex and inversely reduce binding to the La chaperone,13 thereby promoting U6 retention in the nucleus.18 In human cells, there is a dynamic balance between 2 opposing enzyme activities that elongate and trim the 3′ end of U6 snRNA.19-21 U6 is posttranscriptionally 3′ oligouridylated by a terminal uridyl transferase (TUTase).21,22 The identification of a series of 3′ end > p modified U6 snRNAs that differ in length by 1 nucleotide and derive from the α-phosphate of UMP, suggested the activity of an opposing exoribonuclease that deletes the 3′ terminal nucleosides of U6 snRNA by cleaving a phosphodiester bond to form a 3′ > p.20 However, the identity of this putative 3′-5′ exoribonuclease is unknown, the molecular mechanisms underlying the generation of the terminal > p and the functional significance of the U6 snRNA 3′ end modifications remains obscure.
In this report, we identify human USB1 protein as a 3′-5′ exoribonuclease that removes both uridine and adenosine residues from the 3′ end of U6 snRNA, generating a 3′ terminal > p modification. We reveal that the 3′ ends of U6 snRNA in PN patient lymphoblasts not only have elongated nontemplated oligo(U) tracts but also unexpectedly, carry nontemplated 3′ oligo(A) tails characteristic of targets of nuclear RNA surveillance. Thus, our study reveals a novel quality control pathway in which posttranscriptional 3′-end processing by USB1 protects U6 snRNA from targeting and destruction by the exosome and implicates aberrant oligoadenylation of U6 snRNA in the pathogenesis of the leukemia predisposition disorder PN.
Methods
Yeast strains and plasmids
S cerevisiae strains used in this study are listed in supplemental Table 1, primers in supplemental Table 2, and plasmids in supplemental Table 3.
Protein expression and purification
Recombinant wild-type and mutant human USB1 proteins were expressed and purified from Escherichia coli C41 (DE3) cells. The protein was purified using Ni-NTA (GE Healthcare) affinity chromatography, Resource S (Amersham) ion-exchange, and Hiload 26/60 Superdex 75 (Amersham Pharmacia) gel filtration and stored in 20mM HEPES, pH 7.0, 100mM NaCl, and 1mM DTT.
Crystallization and structure determination
The detailed crystallographic methods are provided in supplemental Methods. Coordinates for the human USB1 structure have been deposited in the Protein Data Bank (ID 4H7W).
3′-5′ exoribonuclease assay
Wild-type or mutant USB1 (30 μg/mL) was incubated at 25°C in 60 μL of reaction buffer (20mM Tris-HCl pH 8.0, 1mM DTT) containing 10 to 60 pmol of oligoribonucleotide substrate. An equal volume of 1M Tris-HCl pH 7.2, 8M urea was added to terminate the reaction and the products visualized by gel electrophoresis in 15% to 20% polyacrylamide containing 8M urea. Calf intestinal alkaline phosphatase (1 U/mL, Promega) was incubated at 37°C for 16 hours, and exonuclease T (5000 U/mL, New England Biolabs) was incubated at 25°C.
Genetic complementation assays
Diploid USB1 usb1Δ::NatMX4 cells were transformed with empty vectors (pRS316 or pRS424), with plasmids expressing wild-type or mutant USB1 alleles, or with a plasmid (pRS424-snR6) overexpressing U6 snRNA (gift from D. Brow, University of Wisconsin). Cells were sporulated and cultured on solid medium that selects for the germination of MATa meiotic progeny carrying the indicated plasmid and containing 100 mg/mL nourseothricin, which selects for usb1Δ::NatMX4 cells. Plates were incubated for 3 days at 30°C.
Lymphoblast cell lines
Human lymphoblast cell lines were maintained in RPMI 1640 + 10% FCS (Invitrogen) at 37°C in 5% CO2.
qPCR analysis
RNA was purified with a miRNeasy kit (QIAGEN) and reverse transcribed with random hexamers to generate cDNA. qPCRs were performed on the cDNAs on an Illumina Eco thermal cycler using SYBR Green PCR master mix (Applied Biosystems) and the GAPDH and U6 snRNA primer pairs (supplemental Table 2).
U6 snRNA sequence analysis
RNA was prepared using miRNeasy (QIAGEN) and RNeasy MinElute (QIAGEN) kits. Additional detail is provided in supplemental Methods.
Synthesis of oligoribonucleotides
RNA oligomers were purchased from Integrated DNA Technologies and Sigma-Aldrich. Oligoribonucleotides are listed in supplemental Table 4 and detailed oligonucleotide synthesis protocols are provided in supplemental Methods.
N-terminal Edman sequencing
Samples were prepared by electroblotting from gels and immobilized on polyvinylidene difluoride membrane. Samples were subjected to automated N-terminal Edman degradation on an Applied Biosystems Procise 494HT protein sequencer using protocols and reagents recommended by the manufacturer.
Results
Crystal structure of human USB1
To elucidate the specific function of the protein encoded by the C16orf57 open reading frame (herein called USB1), we determined its structure using x-ray crystallography. The data collection, phasing, and refinement statistics are summarized in supplemental Tables 5 and 6. The current model includes residues N80-K265 of human USB1 and has been refined to a resolution of 1.1 Å, yielding R factor and Rfree values of 0.136 and 0.154, respectively.
USB1 has a globular α-β–type architecture consisting of eleven β-strands, 4 α-helices, and two 310 helices with approximate dimensions of 50 Å × 40 Å × 30 Å (Figure 1A-B). USB1 comprises 2 lobes containing 2 antiparallel β-sheets at the core related by pseudo 2-fold rotational symmetry with 2 peripheral α-helices lying on the outside of each lobe. Following previous nomenclature, the terminal lobe contains the N and C-termini, whereas the second lobe is termed the transit lobe. The terminal lobe consists of a 6-stranded antiparallel β-sheet (strands β1, 3, 4, 9, 10, and 11) and 2 α-helices (α2 and α3). The transit lobe contains a 5-stranded antiparallel β-sheet (strands β2, 5, 6, 7, and 8) and 2 α-helices (α1 and α4). A groove containing a layer of ordered water molecules lies between the 2 lobes, penetrating approximately 17 Å into 1 face of the protein. At the base of this positively charged groove contributed by both the transit and terminal lobes (Figure 1C-D), the antiparallel strands β3 and β8 each harbor an H-x-S motif that defines the 2H phosphoesterase family. Residues H208, S210, H120, and S122 within the H-x-S motifs are well defined in the electron density map, which indicates the presence of 2 conformations for the side-chains of H120 and S122 (Figure 1E). The main chain atoms of residues Q203 and D204 within the loop connecting α3 and β8 are relatively mobile (<B> main-chain of 26.5 Å2 for residues Q202-D204, compared with overall <B> main-chain of 11.8 Å2; supplemental Figure 2). This region lies at one entrance to the active site groove adjacent to another flexible loop region (residues Q164-E166, connecting β6 and β7) that also possesses relatively high B factors (<B> main-chain of 24.96 Å2 for residues Q164-E166).
Crystal structure of human USB1. (A-B) Ribbon representation of human USB1. The active site residues H208, S210, H120, and S122 are shown. The 2 conformers for H120 and S122 observed in the electron density map are indicated. The terminal (magenta) and transit (cyan) lobes are labeled. Figures were prepared using the program PyMOL (http://www.pymol.org). (C-D) Representation of the electrostatic surface potential of human USB1, calculated by the program APBS,42 and colored using a linear color ramp from − 7.0 kT (red) to + 7 kT (blue). The orientation of USB1 in panels C and D is identical to panels A and B, respectively. White arrow indicates the active site. (E) Section of the σA-weighted, solvent flattened 2Fo-Fc electron density map contoured at 0.75 σ for residues F119 (left) to L123 (right), indicating the presence of 2 conformations for the H120 and S122 side-chains. The figure was generated using CCP4mg.43 (F) Schematic diagram of USB1 secondary structure topology. Secondary structure elements are shown as arrows (β-strands) and cylinders (α-helices). Transit lobe is colored cyan and the terminal lobe, magenta.
Crystal structure of human USB1. (A-B) Ribbon representation of human USB1. The active site residues H208, S210, H120, and S122 are shown. The 2 conformers for H120 and S122 observed in the electron density map are indicated. The terminal (magenta) and transit (cyan) lobes are labeled. Figures were prepared using the program PyMOL (http://www.pymol.org). (C-D) Representation of the electrostatic surface potential of human USB1, calculated by the program APBS,42 and colored using a linear color ramp from − 7.0 kT (red) to + 7 kT (blue). The orientation of USB1 in panels C and D is identical to panels A and B, respectively. White arrow indicates the active site. (E) Section of the σA-weighted, solvent flattened 2Fo-Fc electron density map contoured at 0.75 σ for residues F119 (left) to L123 (right), indicating the presence of 2 conformations for the H120 and S122 side-chains. The figure was generated using CCP4mg.43 (F) Schematic diagram of USB1 secondary structure topology. Secondary structure elements are shown as arrows (β-strands) and cylinders (α-helices). Transit lobe is colored cyan and the terminal lobe, magenta.
The USB1 protein belongs to the LigT-like superfamily, defined in the SCOP database23 as a β barrel domain with a duplicated beta/alpha/beta/alpha/beta topology (Figure 1F). The invariance of the H-x-S motif within the USB1 protein family, despite low overall amino acid sequence identity (Figure 2A), supports a critical role in catalysis. The closest structural homologs (DALI server)24 are the 2′, 5′ RNA ligases from Pyrococcus horikoshii (PDB 1VDX),25 Pyrococcus furiosus (PDB 2FYH),26 and Thermus thermophilus (PDB 1IUH),27 the central domain of the mammalian A-kinase anchoring protein AKAP18δ (PDB 2VFY),28 and a 1′-2′ cyclic nucleotide 2′ phosphodiesterase (CNPase) from Arabidopsis thaliana involved in tRNA splicing (PDB 1FSI).29 The signature motif residues of rat AKAP18δ and human USB1 lie in the same position (Figure 2B).
USB1 sequence alignment and structural homology. (A) Structure-based sequence alignment of human USB1 homologues from various species. Shading intensity indicates the degree of amino acid identity. Secondary structure elements for H sapiens USB1 are shown below the alignment with the terminal lobe colored magenta, the transit lobe cyan. The invariant H-x-S motifs in the active site cleft are indicated in red. GenBank accession numbers: H sapiens (NP_078874.2), D rerio (NP_001003460.1), S pombe (NP_593641.2), S cerevisiae (NP_013233.1), D melanogaster (NP_649911.1), and C glabrata (CAG62512.1). The secondary structure elements of USB1 were calculated from the crystal structure using DSSP44 and the alignment generated using T-coffee and Jalview.45,46 (B) Superposition of the active site of human USB1 (PDB 4H7W, Cα chain, light blue; side-chains, blue) and rat AKAP18δ (PDB 2VFY, Cα chain, light red; side-chains, red). The catalytic residues are shown as ball-and-stick models. Human USB1 is numbered in blue, AKAP18δ in red.
USB1 sequence alignment and structural homology. (A) Structure-based sequence alignment of human USB1 homologues from various species. Shading intensity indicates the degree of amino acid identity. Secondary structure elements for H sapiens USB1 are shown below the alignment with the terminal lobe colored magenta, the transit lobe cyan. The invariant H-x-S motifs in the active site cleft are indicated in red. GenBank accession numbers: H sapiens (NP_078874.2), D rerio (NP_001003460.1), S pombe (NP_593641.2), S cerevisiae (NP_013233.1), D melanogaster (NP_649911.1), and C glabrata (CAG62512.1). The secondary structure elements of USB1 were calculated from the crystal structure using DSSP44 and the alignment generated using T-coffee and Jalview.45,46 (B) Superposition of the active site of human USB1 (PDB 4H7W, Cα chain, light blue; side-chains, blue) and rat AKAP18δ (PDB 2VFY, Cα chain, light red; side-chains, red). The catalytic residues are shown as ball-and-stick models. Human USB1 is numbered in blue, AKAP18δ in red.
USB1 is a 3′-5′ exoribonuclease
We set out to test the hypothesis that USB1 is a cyclic nucleotide phosphodiesterase (CPDase) using a modified CPDase assay.26 Full-length recombinant USB1 was incubated with a 5′-FAM (6-carboxyfluorescein)–labeled substrate (CAUG) carrying a 2′, 3′ cyclic phosphate (> p), a 2′ monophosphate (2′p) or a 3′ monophosphate (3′p) modification at the 3′ end (supplemental Figure 3A-E). CAUG > p (but not CAUG-2′p or 3′p) was converted over time to a product that migrated faster than CAUG-2′p or CAUG-3′p (supplemental Figure 3C-D). In addition, CAUG-2′p and CAUG-3′p, but not the USB1 reaction product or a CAUG > p control, gel shifted when incubated with calf intestinal phosphatase (CIP; supplemental Figure 3D). Taken together, these experiments suggested that USB1 had cleaved the terminal > p nucleoside from CAUG > p to generate a CIP-resistant product containing a > p modification (CAU > p). We confirmed this hypothesis by MALDI-TOF mass spectrometry (supplemental Figure 3D). Mutation of the catalytic histidines (H120A and H208A) inactivated a functional N-terminally truncated fragment (USB1Δ, residues G70-K265), excluding cross-contamination with a nonspecific exoribonuclease (supplemental Figure 3B-E).
Overexpression of U6 snRNA rescues USB1 deletion in yeast
The exoribonuclease activity of USB1 CAUG > p was evident only on prolonged incubation, indicating that the substrate was not optimal. In mammalian cells, identification of a range of cyclic phosphate-modified U6 snRNAs in vivo that differ in length by 1 nucleotide suggested the activity of a 3′-5′ exoribonuclease that deletes the terminal nucleoside of U6 snRNA by cleaving the phosphodiester bond and generating a cyclic phosphate.20 We therefore hypothesized that U6 snRNA might be a potential physiologic target for the 3′-5′ exoribonuclease activity of USB1 in vivo. To test this, we initially turned to S cerevisiae and identified USB1 (YLR132C) as the homologue of the human C16orf57 gene (Figure 2A). We disrupted the endogenous USB1 allele in wild-type diploid cells by replacing USB1 with the NatMX4 selection cassette and showed that USB1 is essential for yeast viability in random sporulation assays (Figure 3A). We confirmed that haploid usb1Δ cells were fully complemented for growth by expressing wild-type USB1 on a centromeric plasmid (supplemental Table 7).
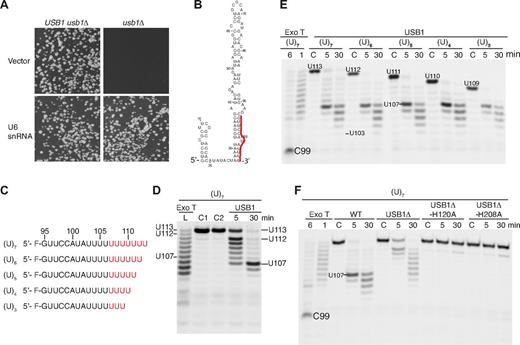
USB1 is a U6 snRNA-specific 3′-5′ exoribonuclease. (A) Overexpression of U6 snRNA rescues the lethality of USB1 deletion in yeast. USB1 usb1Δ diploid (left) or usb1Δ haploid cells (right) transformed with empty vector (pRS426) or plasmid expressing U6 snRNA (pSNR6). (B) Secondary structure model of human U6 snRNA.13 The 3′ end sequence (G95-U106) is marked in red. (C) Fluorescently labeled oligoribonucleotides used in the figure corresponding in sequence to the 3′ end of U6 snRNA (nucleotides G95-U106) carrying 3 to 7 nontemplated uridine residues (indicated in red). (D) USB1 has distributive 3′-5′ exoribonuclease activity. Oligo(U)7 RNA was incubated with exonuclease T (Exo T) for 1 minute (L, RNA ladder) or with USB1 for the indicated times. C1: oligo(U)7 dissolved in water; C2: oligo(U)7 incubated in buffer alone for 30 minutes. The position on the gel of full-length oligo(U)7 RNA (U113), the initial oligo(U)7 degradation product (U112), and the strong USB1 pause site (U107) are indicated. (E) USB1 trims back oligo(U) tracts to nucleotide U107 independent of their length. The USB1 pause site at nucleotide U107 and the processing termination site at U103 after 30 minutes are indicated. C: RNA incubated with buffer alone for 30 minutes. The major product of Exo T degradation (C99) is indicated. (F) The conserved H-x-S motifs are required for USB1 function in vitro. The oligo(U)7 RNA substrate was incubated with Exo T, wild-type USB1, N-terminally truncated USB1 (USB1Δ, residues G70-K265), or USB1Δ containing active site histidine mutations (USB1Δ-H120A and USB1Δ-H208A).
USB1 is a U6 snRNA-specific 3′-5′ exoribonuclease. (A) Overexpression of U6 snRNA rescues the lethality of USB1 deletion in yeast. USB1 usb1Δ diploid (left) or usb1Δ haploid cells (right) transformed with empty vector (pRS426) or plasmid expressing U6 snRNA (pSNR6). (B) Secondary structure model of human U6 snRNA.13 The 3′ end sequence (G95-U106) is marked in red. (C) Fluorescently labeled oligoribonucleotides used in the figure corresponding in sequence to the 3′ end of U6 snRNA (nucleotides G95-U106) carrying 3 to 7 nontemplated uridine residues (indicated in red). (D) USB1 has distributive 3′-5′ exoribonuclease activity. Oligo(U)7 RNA was incubated with exonuclease T (Exo T) for 1 minute (L, RNA ladder) or with USB1 for the indicated times. C1: oligo(U)7 dissolved in water; C2: oligo(U)7 incubated in buffer alone for 30 minutes. The position on the gel of full-length oligo(U)7 RNA (U113), the initial oligo(U)7 degradation product (U112), and the strong USB1 pause site (U107) are indicated. (E) USB1 trims back oligo(U) tracts to nucleotide U107 independent of their length. The USB1 pause site at nucleotide U107 and the processing termination site at U103 after 30 minutes are indicated. C: RNA incubated with buffer alone for 30 minutes. The major product of Exo T degradation (C99) is indicated. (F) The conserved H-x-S motifs are required for USB1 function in vitro. The oligo(U)7 RNA substrate was incubated with Exo T, wild-type USB1, N-terminally truncated USB1 (USB1Δ, residues G70-K265), or USB1Δ containing active site histidine mutations (USB1Δ-H120A and USB1Δ-H208A).
We next sought evidence for a genetic interaction between USB1 and U6 snRNA. We reasoned that if USB1 is required for U6 snRNA biogenesis or stability, then overexpression of wild-type U6 snRNA might rescue the growth defect of usb1Δ haploid cells. Remarkably, the fitness defect of haploid usb1Δ cells was fully complemented by U6 snRNA expressed on a multicopy plasmid (Figure 3A), indicating that U6 snRNA is the essential target of USB1 activity in vivo and that USB1 is required, directly or indirectly, for U6 snRNA biogenesis or stability.
USB1 N-terminal domain and the conserved H-x-S motifs are essential in vivo
To determine whether the signature H-x-S motifs are critical for USB1 function in vivo, we used genetic complementation (supplemental Table 7). In contrast to wild-type USB1, mutant usb1 alleles carrying mutations in the catalytic histidine residues (usb1-H133A or usb1-H231A) failed to complement the fitness defect of USB1 deleted (usb1Δ) cells in vivo, despite appropriate expression. Mutant usb1-S135A or usb1-S233A alleles both conferred severe growth defects, whereas a usb1 allele carrying an N-terminal deletion (usb1Δ1-76) failed to complement. We conclude that the N-terminal domain and the catalytic histidines are essential for the function of the USB1 family in vivo.
USB1 removes uridine nucleosides from the 3′ end of U6 snRNA
To investigate whether human USB1 might be a 3′-5′ exoribonuclease involved in U6 snRNA processing, we set up in vitro degradation assays using defined synthetic 5′ fluorescently labeled RNA substrates corresponding to nucleotides G95-U106 from the 3′ end of U6 snRNA containing a variable number of nontemplated uridine residues (Figure 3B-F). Incubation of exonuclease T (Exo T/RNase T, a control 3′-5′ exoribonuclease) or USB1 with an oligo containing 7 nontemplated uridines (oligo(U)7) generated a series of fragments that are progressively shortened from the 3′ end, characteristic of a distributive mode of hydrolysis (Figure 3D). Regardless of the number of nontemplated uridines added, USB1 paused at nucleotide U107 after 5 minutes and trimmed back to nucleotide U103 after 30 minutes. In contrast, Exo T degraded oligo(U)7 back to residue C99 within 6 minutes (Figure 3E). Compared with wild-type USB1, USB1Δ processed the oligo(U)7 tail inefficiently and 2 active site mutants (USB1Δ-H120A and USB1Δ-H208A) showed no significant activity (Figure 3F). Furthermore, oligo(U)7 terminating in a 3′OH or a 3′ > p, but not a 3′p group, was efficiently degraded by USB1 (supplemental Figure 4). We conclude that USB1 has distributive 3′-5′ exoribonuclease activity on U6 snRNA and shows a strong preference for a 3′OH or 3′ > p group for its activity. Furthermore, USB1 pauses at a specific position, rather than removing a fixed number of residues from the 3′ end.
USB1 directly catalyzes formation of a terminal 2′, 3′ cyclic phosphate
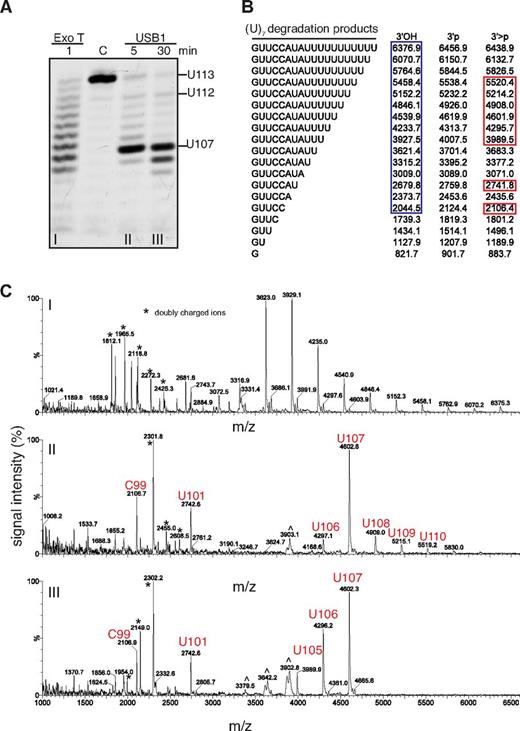
Incubation of USB1 with oligo(U)7 generated a series of degradation fragments with increased electrophoretic gel mobility compared with the Exo T products (Figures 3D and 4A). MALDI-TOF mass spectrometry analysis revealed observed mass/charge ratios for the Exo T RNA degradation products that were consistent with a terminal 3′OH group as expected (Figure 4A-C).30 In contrast, the observed mass/charge ratios for the USB1 RNA degradation products were consistent with the presence of a 3′ > p modification (Figure 4A-C). Thus, human USB1 is a 3′-5′ exoribonuclease that catalyzes the sequential liberation of 3′ terminal uridine nucleosides from U6 snRNA, directly generating a 3′ > p modification.
USB1 directly catalyzes 3′ terminal 2′, 3′ cyclic phosphate formation. (A) Degradation of oligo(U)7 RNA by Exo T (i) or USB1 (ii, iii). C: oligo(U)7 RNA incubated in buffer for 30 minutes. (B) Expected masses of potential oligo(U)7 RNA degradation products containing 3′OH, 3′p or 3′ > p groups. Blue box: observed masses after incubation with Exo T; red boxes: observed masses after incubation with USB1. (C) MALDI-TOF mass spectrometric analysis of oligo(U)7 RNA degradation products. m/z, mass-charge ratio. ∧ indicates artifacts. USB1 degradation products with 3′ end > p modifications are marked in red.
USB1 directly catalyzes 3′ terminal 2′, 3′ cyclic phosphate formation. (A) Degradation of oligo(U)7 RNA by Exo T (i) or USB1 (ii, iii). C: oligo(U)7 RNA incubated in buffer for 30 minutes. (B) Expected masses of potential oligo(U)7 RNA degradation products containing 3′OH, 3′p or 3′ > p groups. Blue box: observed masses after incubation with Exo T; red boxes: observed masses after incubation with USB1. (C) MALDI-TOF mass spectrometric analysis of oligo(U)7 RNA degradation products. m/z, mass-charge ratio. ∧ indicates artifacts. USB1 degradation products with 3′ end > p modifications are marked in red.
USB1 degrades oligo(U) and oligo(A) tracts in vitro
Given the observed activity of USB1 on oligo(U) tails, we next asked whether USB1 could degrade other oligonucleotide tracts in vitro. USB1 was able to process RNA substrates containing 3′ oligo(U)7 and oligo(A)7 tracts, but not oligo(G)7 or oligo(C)7 (supplemental Figure 4A-C). Interestingly, in contrast to the oligo(U)7 substrate where USB1 paused at nucleotide U107, the initial pause position on oligo(A)7 was residue U106. USB1 was equally active on oligo(A) tails containing a 3′OH or 3′ > p, but not a 3′p (supplemental Figure 5B). We conclude that USB1 can process both oligo(U) and oligo(A) tails in vitro, with a strong preference for a 3′OH or 3′ > p modification, but a terminal 3′p inhibits USB1 processing activity.
USB1 reads U6 snRNA nucleotide A102 as a pause signal
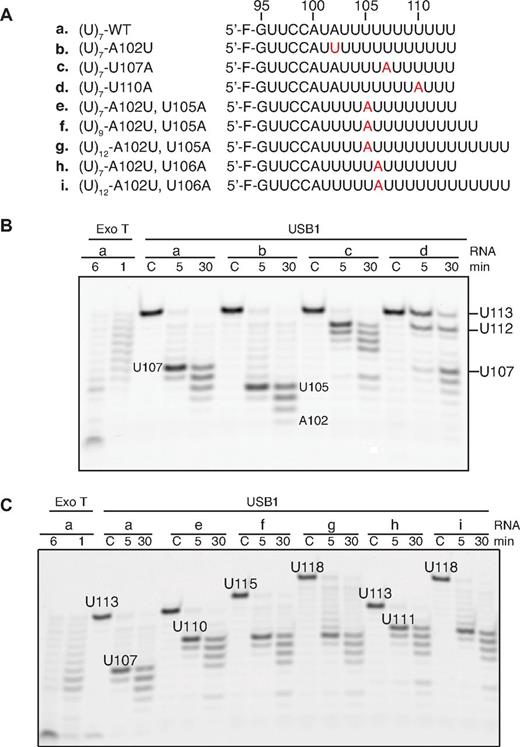
We hypothesized that USB1 might recognize adenosine 102 as a pause signal that restricts U6 snRNA degradation. USB1 paused at residue U107 when incubated with wild-type oligo(U)7, but paused at residue U105 when incubated with an A102U mutant (Figure 5A-B). Introduction of an adenine nucleotide 3′ to A102 (U107A) induced pausing of USB1 a further 5 nucleotides downstream (U112), whereas a U110A mutation severely perturbed USB1 3′-end processing. The adenosine-dependent pausing activity of USB1 was independent of the number of nontemplated uridines present at the 3′ end (Figure 5C). We conclude that USB1 pauses 5 uridine residues downstream of an adenosine residue (A102), providing a rationale for the observation that the major 3′ end sequence of U6 snRNA in human cells is AUUUU(U)> p, where (U) represents a nontemplated uridine nucleotide.11
USB1 pauses 5 nucleotides 3′ to an adenosine. (A) Wild-type and mutant 5′ fluorescently labeled oligoribonucleotides used in the assay. Mutations are highlighted in red. (B) USB1 reads the position of an upstream adenine nucleotide. The initial length of the oligos and the positions of the strong pause sites after incubation with USB1 are indicated. (C) USB1 pauses 5 nucleotides downstream of an adenine nucleotide independent of the total number of 3′ uridines.
USB1 pauses 5 nucleotides 3′ to an adenosine. (A) Wild-type and mutant 5′ fluorescently labeled oligoribonucleotides used in the assay. Mutations are highlighted in red. (B) USB1 reads the position of an upstream adenine nucleotide. The initial length of the oligos and the positions of the strong pause sites after incubation with USB1 are indicated. (C) USB1 pauses 5 nucleotides downstream of an adenine nucleotide independent of the total number of 3′ uridines.
U6 snRNA is aberrantly 3′ oligoadenylated in PN patient lymphoblasts
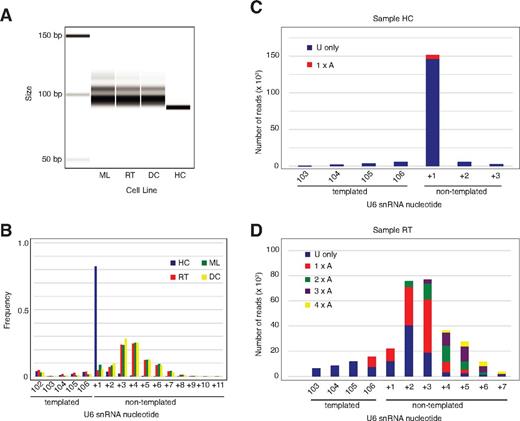
Our data demonstrate that U6 snRNA is the essential target of Usb1 in yeast and that human USB1 is a 3′-5′ exoribonuclease that degrades oligo(U) and oligo(A) tails at the 3′ end of U6 snRNA in vitro. We therefore hypothesized that U6 snRNA from PN patient-derived B-lymphoblastoid cell lines harboring mutations in the C16orf57 gene (supplemental Table 8) would exhibit 3′-end processing defects in vivo. Amplification and separation of cDNA fragments encompassing the 3′ end of U6 snRNA on an Agilent Bioanalyzer revealed a discrete band in the heterozygous carrier control, but elongation and peak broadening in 3 independent PN patient-derived lymphoblastoid cell lines (Figure 6A). Sequence analysis of the 3′ termini from the control sample revealed that 93% of the U6 snRNAs terminated with a single nontemplated residue, although a further 4% terminated in 2 (Figure 6B), consistent with a previous study.31 In contrast, the U6 snRNA 3′ termini in the PN patient samples were elongated and heterogeneous. Deep sequencing analysis revealed that 90% of U6 snRNA 3′ termini from control lymphoblasts contained a uridine at the first (+1) nontemplated position and approximately 2% contained an adenosine, consistent with previous reports (Figure 6C).32,33 In contrast, in the PN patient samples, a high proportion of reads unexpectedly contained ≥ 2 nontemplated adenosines at the 3′ end, particularly as the overall length of the 3′ extension increased (Figure 6D, supplemental Figure 6A-B). However, we found no significant difference in steady state U6 snRNA levels in patient samples compared with control (supplemental Figure 7). Furthermore, deep sequencing of the entire transcriptome of the lymphoblast cell lines failed to identify evidence for generalized splicing defects. We conclude that, as in yeast, U6 snRNA is a major target for human USB1 activity and that USB1 functions as a 3′-5′ exoribonuclease to remove 3′ terminal nontemplated adenosine and uridine residues in vivo. As nontemplated, 3′ terminal oligo(A) tails are a hallmark of nuclear RNA surveillance targets, our data suggest that USB1-mediated processing of the U6 oligo(U) tails protects U6 snRNA from oligoadenylation and subsequent degradation by the nuclear exosome.
U6 snRNA in PN patient lymphoblasts is aberrantly 3′ oligoadenylated. (A) PCR analysis of U6 snRNA 3′ tails from control (HC) and 3 independent PN patient (DC, ML, RT) lymphoblasts separated on an Agilent bioanalyzer. (B) Frequency distribution of the number of nontemplated nucleotides at the U6 3′ terminus in the same 4 lymphoblast cell lines as indicated determined by MiSeq analysis. (C) Sequences of nontemplated nucleotides at the 3′ end of U6 snRNA in control lymphoblasts (sample HC). Reads containing uridine residues alone are shown in blue, reads with a single 3′ terminal adenine nucleotide are in red. Sequences represented by less than 1000 reads are excluded. (D) Sequences of nontemplated nucleotides at the 3′ end of U6 snRNA from PN patient lymphoblasts (sample RT). Sequences are grouped according to the number of consecutive adenine nucleotides at the U6 3′ end. Sequences represented by less than 1000 reads are not included.
U6 snRNA in PN patient lymphoblasts is aberrantly 3′ oligoadenylated. (A) PCR analysis of U6 snRNA 3′ tails from control (HC) and 3 independent PN patient (DC, ML, RT) lymphoblasts separated on an Agilent bioanalyzer. (B) Frequency distribution of the number of nontemplated nucleotides at the U6 3′ terminus in the same 4 lymphoblast cell lines as indicated determined by MiSeq analysis. (C) Sequences of nontemplated nucleotides at the 3′ end of U6 snRNA in control lymphoblasts (sample HC). Reads containing uridine residues alone are shown in blue, reads with a single 3′ terminal adenine nucleotide are in red. Sequences represented by less than 1000 reads are excluded. (D) Sequences of nontemplated nucleotides at the 3′ end of U6 snRNA from PN patient lymphoblasts (sample RT). Sequences are grouped according to the number of consecutive adenine nucleotides at the U6 3′ end. Sequences represented by less than 1000 reads are not included.
Discussion
Using a crystallographic, biochemical, and genetic approach, we conclude that (1) USB1 is a member of the LigT-like superfamily of 2H phosphoesterases containing 2 conserved H-x-S motifs that are essential for function in vitro and in vivo; (2) USB1 has distributive 3′-5′ exoribonuclease activity that trims U6 snRNA 3′ oligo(U) and oligo(A) tracts; (3) USB1 directly catalyzes formation of the > p modification found at the 3′ end of ∼ 90% of cellular U6 snRNA; (4) USB1 reads the position of nucleotide A102, pausing 5 uridine nucleotides downstream, suggesting a mechanism to maintain the appropriate length of the U6 oligo(U) tract; and (5) USB1 protects U6 snRNA from aberrant 3′ oligoadenylation in vivo. Thus, our data independently support, but significantly extend the recent conclusions reached for S cerevisiae Usb134 and its S pombe ortholog Mpn1,35 in particular through elucidation of a high resolution crystal structure of human USB1 and by providing a potential mechanism to explain the reduced stability of U6 snRNA in PN patient cells.35
Catalytic mechanism of USB1
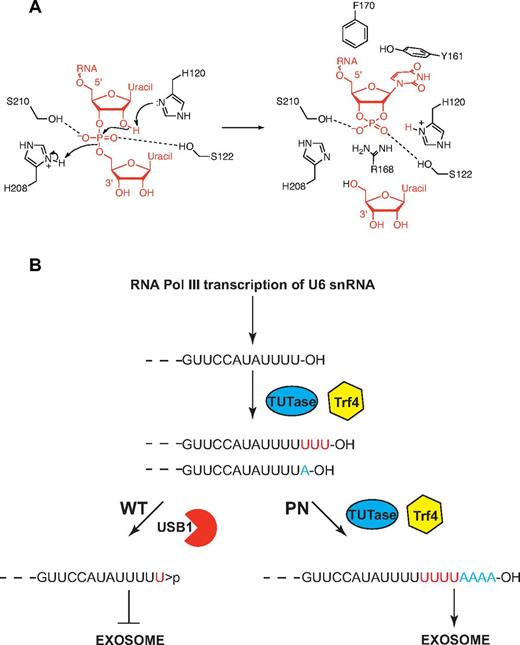
We propose a catalytic mechanism for the USB1 family (Figure 7A). The mechanism is based on (1) the role of the 2 catalytic histidines of RNAse A that cleave the RNA phosphodiester bond to generate a terminal 2′, 3′ cyclic phosphate36 ; and (2) the putative catalytic mechanism of rat AKAP18δ28 and Thermus thermophilus 2′-5′ ligase.27 We suggest that H120 in its unprotonated form removes a proton from the 2′ oxygen of the phospho-ribosyl moiety to facilitate an attack on the phosphorus atom. H208 acts as a general acid, protonating the 5′ oxygen to facilitate displacement of a uridine nucleoside. The 2 nonessential (supplemental Table 7), but highly conserved serine residues (S122 and S210) probably coordinate the phosphate oxygen atoms. Conserved residues lining the USB1 catalytic groove are probably functionally equivalent to similarly positioned residues in other 2H phosphoesterases. In particular, USB1 residues Y161 and F170 (equivalent to AKAP18δ F179 and V183) probably interact with the base and ribose moieties of U6 snRNA. Furthermore, USB1 R168 (AKAP18δ K229) occupies the same spatial position as R135 in the T thermophilus 2′-5′ RNA ligase structure, potentially stabilizing the phosphate oxygen atoms. In human USB1, the electron density indicates that in one conformation, the H120 side-chain is directed toward the center of the cavity where there is an ordered water molecule (TP165) at a distance of 2.7 Å from H120 (atom Nϵ). We therefore favor H120 as the initiating residue of catalysis, but the reverse mechanism cannot be excluded in the absence of additional experimental data.
Model for USB1 function in catalysis and in U6 snRNA quality control. (A) Model for USB1 3′-5′ exonuclease activity. The side-chain of H120 acts as a general base, abstracting a proton from the 2′ oxygen of the RNA substrate to facilitate nucleophilic attack on the phosphorus atom. The resulting in-line displacement of the leaving 5′ oxygen is favored by proton transfer from the side-chain of H208, which serves as a general acid. The RNA substrate is depicted in red. The figure was prepared using ChemBioDraw Ultra. (B) Model for USB1 function in U6 snRNA quality control. The length and 3′ end modification of U6 snRNA is dynamically regulated through the competing addition of UMP by terminal uridyl transferase (TUTase; 3′OH groups) and trimming by USB1 (3′ cyclic phosphate groups). In the absence of functional USB1 in poikiloderma with neutropenia, the polyA polymerase Trf4 3′ adenylates free 3′OH groups on U6 snRNA, targeting it for destruction by the exosome.
Model for USB1 function in catalysis and in U6 snRNA quality control. (A) Model for USB1 3′-5′ exonuclease activity. The side-chain of H120 acts as a general base, abstracting a proton from the 2′ oxygen of the RNA substrate to facilitate nucleophilic attack on the phosphorus atom. The resulting in-line displacement of the leaving 5′ oxygen is favored by proton transfer from the side-chain of H208, which serves as a general acid. The RNA substrate is depicted in red. The figure was prepared using ChemBioDraw Ultra. (B) Model for USB1 function in U6 snRNA quality control. The length and 3′ end modification of U6 snRNA is dynamically regulated through the competing addition of UMP by terminal uridyl transferase (TUTase; 3′OH groups) and trimming by USB1 (3′ cyclic phosphate groups). In the absence of functional USB1 in poikiloderma with neutropenia, the polyA polymerase Trf4 3′ adenylates free 3′OH groups on U6 snRNA, targeting it for destruction by the exosome.
U6 snRNA 3′ end modification and nuclear RNA surveillance
The exosome plays major roles in RNA processing and surveillance, but is dependent on cofactors including the Trf4-Air-Mtr4 polyadenylation (TRAMP) complex.37 In Drosophila and budding yeast, a fraction of U6 snRNA is 3′ adenylated by the TRAMP-associated polyA polymerase Trf4, targeting it for degradation by the nuclear exosome. Indeed, higher levels of mature U6 snRNA and 3′ extended (∼ 3 nucleotides) forms were observed in yeast cells carrying mutations in the RNase II homologue Rrp44, a core component of both the nuclear and cytoplasmic exosome, the key player in cellular RNA metabolism.38 As Trf4 requires a free 3′OH terminal group, it is probable that a terminal > p or monophosphate group would inhibit Trf4-mediated oligoadenylation. Targeting of oligoadenylated U6 snRNA for degradation by the exosome in the absence of USB1 is consistent with our data showing that overexpression of U6 snRNA suppresses the lethality of usb1Δ yeast cells and provides a potential mechanism to explain how USB1 might regulate the reduced U6 snRNA stability shown both in S cerevisiae,34 in S pombe, and in human cells.35 Thus, our data support the existence of a novel quality control pathway in which USB1 function alters the 3′ end modification of U6 snRNA to target (3′OH) or protect (3′ > p or 3′p) it from Trf4-dependent 3′ oligoadenylation and destruction by the exosome (Figure 7B).
Dynamic regulation of the U6 snRNA 3′ end length
Although the major 3′ end sequence of mammalian U6 RNA is reported to be (5′) GUUCCAUAUUUU(U) > p (3′),11 there is considerable length heterogeneity in vivo.11,19 Biochemical studies in HeLa cell extracts suggest that the removal and addition of uridylic acid residues occurs continuously in dynamic equilibrium.11,20 The addition of nucleotides to U6 snRNA is mediated by a specific poly(U) polymerase (TUTase) that adds UMP to the free 3′OH group of primary U6 snRNA RNA polymerase III transcripts.21,22 Here, we identified USB1 as the 3′-5′ exoribonuclease that trims U6 snRNA by catalyzing cleavage of the 3′ phosphodiester bond at the 3′ end of the oligo(U) tract, thereby generating a 3′ > p modification. Support for the dynamic interplay between TUTase and USB1 in vivo in human cells is provided by our data from PN patient lymphoblasts showing that in the absence of functional USB1, the 3′ oligo(U) tract of U6 snRNA is extended and heterogeneous because of the unopposed poly(U) polymerase activity of TUTase.
How does USB1 know where to terminate U6 3′-end processing? Our data indicate that USB1 reads the position of nucleotide A102 to measure the appropriate length of the 3′ oligouridine tract (Figure 7B), providing a rationale for the observation that the predominant in vivo form of U6 snRNA in human cells carries 4 templated and a single nontemplated uridine residue. We suggest that critical residues within the N-terminal domain of USB1 may mediate the interaction with the U6 snRNA nucleotide A102.
Pathogenesis of PN
Our analysis of PN patient-derived lymphoblasts indicates that perturbation of USB1 protein function does not dramatically impair global splicing efficiency or the steady state levels of U6 snRNA in transformed lymphoblast cell lines. However, the ratio of U6 snRNA with specific 3′-end modifications is known to vary during development in Drosophila and X laevis.11 Furthermore, U6 snRNA expression levels vary during development in different tissues and cell types.39 Thus, the PN phenotype may reflect variation in U6 expression or different ratios of 3′ end modified U6 snRNA at key stages during development. Alternatively, USB1 may target small RNAs in addition to U6 snRNA. For example, > p is found at the 3′ end of SRP and MRP RNA.31
PN is an inherited bone-marrow failure disorder associated with neutropenia and increased propensity to MDS and AML.2 By identifying the specific function of USB1 in spliceosomal U6 snRNA 3′-end processing and protection from nuclear RNA surveillance, our data link the pathogenesis of PN to acquired forms of MDS that are characterized by mutations in multiple components of the pre-mRNA splicing machinery.40,41 To date, the factors mutated in MDS have largely been components of the 3′ splice-site recognition complex. By identifying the specific function of USB1 in U6 snRNA quality control, our study strongly underscores the relevance of mutations in components of the splicing apparatus to the pathogenesis of human bone-marrow failure and cancer predisposition disorders.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr David Brow, University of Wisconsin, for plasmid pRS426; Dr Lidia Larizza (University of Milan), Dr Alessandra Renieri (University of Siena), and the Telethon Network of Genetic Biobanks for lymphoblast cell lines; Phil Evans for expert assistance with crystallography; David Neuhaus for assistance with APBS; John Sutherland for helpful discussion; Christopher Chan and Colm Duffy for assistance with ChemBioDraw; and Mike Weldon (University of Cambridge) for N-terminal sequencing analysis. The authors thank EASIH, Addenbrooke's Hospital, for sequencing and bioinformatics analysis. Data were collected on beam line I-02 at the Diamond Light Source in Oxford, United Kingdom.
This work was supported by Leukemia and Lymphoma Research, the Medical Research Council (U105161083), the MDS Foundation, the Tesni Parry Memorial Fund, and the Cambridge NIHR Biomedical Research Center.
Authorship
Contribution: C.H., P.J.S., A.J.F., F.R.B., M.J.C., L.J., L.C.P., A.S., P.C., and A.J.W. designed and performed research, analyzed data, and wrote the paper; and M.K. and I.D. contributed vital new reagents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alan Warren, University of Cambridge, MRC Laboratory of Molecular Biology, Hills Road, Cambridge, CB2 0QH, United Kingdom; e-mail: ajw@mrc-lmb.cam.ac.uk.