Key Points
This study has uncovered an oncogenic role of EZH2 independent of its methyltransferase activity in NKTL.
This study suggests that targeting EZH2 may have therapeutic usefulness in NKTL.
Abstract
The role of enhancer of zeste homolog 2 (EZH2) in cancer is complex and may vary depending on the cellular context. We found that EZH2 is aberrantly overexpressed in the majority of natural killer/T-cell lymphoma (NKTL), an aggressive lymphoid malignancy with very poor prognosis. We show that EZH2 upregulation is mediated by MYC-induced repression of its regulatory micro RNAs and EZH2 exerts oncogenic properties in NKTL. Ectopic expression of EZH2 in both primary NK cells and NKTL cell lines leads to a significant growth advantage. Conversely, knock-down of EZH2 in NKTL cell lines results in cell growth inhibition. Intriguingly, ectopic EZH2 mutant deficient for histone methyltransferase activity is also able to confer growth advantage and rescue growth inhibition on endogenous EZH2 depletion in NKTL cells, indicating an oncogenic role of EZH2 independent of its gene-silencing activity. Mechanistically, we show that EZH2 directly promotes the transcription of cyclin D1 and this effect is independent of its enzymatic activity. Furthermore, depletion of EZH2 using a PRC2 inhibitor 3-deazaneplanocin A significantly inhibits growth of NK tumor cells. Therefore, our study uncovers an oncogenic role of EZH2 independent of its methyltransferase activity in NKTL and suggests that targeting EZH2 may have therapeutic usefulness in this lymphoma.
Introduction
Nasal-type natural killer/T-cell lymphoma (NKTL) is an aggressive lymphoid malignancy associated with very poor survival outcomes.1 A better understanding of the molecular abnormalities underlying this disease will provide important insights into the biology of this tumor; however, studies on NKTL are often limited by the lack of adequate tissue in small nasal biopsies and the presence of necrosis in biopsy specimens. Although more effective therapy is now available, treatment is still completely reliant on radiotherapy and combinations of chemotherapy.2 ,3
We and others have recently performed whole-genome gene expression studies and identify a number of genes that are differentially expressed in NKTL as well as pathways that are activated in NKTL. Enhancer of zeste homolog 2 (EZH2), one of the genes identified in our study to be aberrantly overexpressed in NKTL,4 is a H3K27-specific histone methyltransferase and a component of the polycomb repressive complex 2 (PRC2), which plays a key role in the epigenetic maintenance of repressive chromatin mark. EZH2 protein contains a catalytic domain (SET domain) at the COOH-terminus that provides the methyltransferase activity. The catalytic domain must partner with other noncatalytic proteins, such as EED and SUZ12, to form the PRC2 in order to attain robust histone methyltransferase activity. Genome-wide approaches have demonstrated the importance of the PRC2 complex in the transcriptional regulation through H3K27 methylation and gene repression.5
Published literature reveals a number of possible mechanisms of EZH2 upregulation in different types of human cancers.6 It has been shown that EZH2 expression can be transcriptionally activated by a fusion oncoprotein EWS-FLI1 in Ewing sarcoma.7 EZH2 expression in the breast tumor–initiating cell population is particularly enhanced by hypoxia through HIF1a-mediated transactivation.8 In addition to transcriptional regulation, the EZH2 transcript is known to be regulated by tumor suppressor micro RNAs (miRNAs). For example, miR-26a binds to and inhibits EZH2 transcript expression in B-cell lymphoma.9 miR-101 is frequently lost in metastatic prostate cancer, thus releasing EZH2 from miR-101–mediated repression.10 EZH2 can also be modulated by post-translational modifications through phosphorylation by AKT and cyclin-dependent kinase.11,12 To the best of our knowledge, the mechanism of EZH2 overexpression in NKTL has not yet been described.
A high level of EZH2 expression is associated with aggressiveness and poor outcome in solid tumors such as prostate, breast, and endometrial cancers. The oncogenic role of EZH2 overexpression in these tumor types has been studied extensively. In human B-cell malignancies, mutations of Y641 and A677 have been documented to be associated with profoundly increased activity for methylated H3K27, which may promote the development of lymphoma.13-15 On the other hand, recent discoveries of recurrent somatic EZH2 mutations in myelodysplastic syndromes and myeloproliferative neoplasms indicate that inactivation of EZH2 may contribute to the pathogenesis of myeloid malignancies.16,17 Genetic inactivation of EZH2 has also been identified in T-cell acute lymphoblastic leukemia, and the study by Ntziachristos and colleagues suggests a tumor suppressor role for EZH2 in human leukemia by a hitherto unrecognized dynamic interplay between oncogenic NOTCH1 and EZH2.18 Taken together, the role of EZH2 and the underlying mechanisms of gene regulation by EZH2 in cancer are complex, and further studies need to be performed in a cell context–dependent manner.
In our study, we demonstrated the overexpression of EZH2 in NKTL, deciphered the molecular mechanisms underlying the overabundance of EZH2, and investigated its functional role as an oncogene in this disease. Contrary to our expectations, we found that EZH2 overexpression is not associated with H3K27 trimethylation in NKTL, and its oncogenic activity does not require its histone methyltransferase activity. Instead, EZH2 directly promotes cyclin D1 expression. Thus, this study demonstrates a noncanonic role of EZH2 in NKTL.
Methods
Immunohistochemistry
A total of 38 clinical cases of NKTL that fulfill the World Health Organization diagnostic criteria were used for immunohistochemistry (IHC) studies. The clinicopathologic data of the cases are included in supplemental Table 1. There were 27 cases that came from the tissue microarray used in our previous study (GEO accession no. GSE31377).4 IHC analysis was performed for EZH2, Ki67, and H3K27me3 on 4-µm sections of NKTL using the conditions listed in supplemental Table 2. IHC study was also performed on cell blocks of normal NK cells for comparison. Appropriate positive tissue controls were used. Details of scoring and imaging are appended in the supplemental Methods.
Primary NK cell isolation and retroviral transduction
Highly purified (90%-99%) normal human NK cells were isolated and cultured as described previously.19 Retroviruses were generated by transfection of empty plasmid vector polymorphonuclear neutrophil (pMN)-enhanced green fluorescence protein (EGFP) or vectors containing EZH2 using Fugene HD6 into Phoenix-amphotropic packaging cells. At 48 hours after transfection, the supernatants were collected and filtered. A total of 200 000 cells were mixed with 1.6 mL of retroviral supernatant in 12-well plates with 10 μg/mL of Polybrene added. The infection was repeated at 72 hours after transfection.
Luciferase reporter assay
The cyclin D1 (CCND1) promoter construct pGL4-CCND1-Luc has been described previously.20 Cells were harvested 24 hours after transfection and were analyzed with the Dual Luciferase system (Promega). See the supplemental Methods and Materials for details.
ChIP assay
Chromatin immunoprecipitation (ChIP) assays were performed as described previously.21 See the supplemental Methods and Materials for details of the antibodies used and primer sequences.
Western blotting
Cells were lysed in radioimmunoprecipitation assay buffer and were subjected to sonication. The primary antibodies used included cell-signaling antibody EZH2 (4905), H3K27me3 (07-449), H3K27me2 (9755), Total H3 (9715) and SantaCruz antibody CCND1 (DCS-6 and A12), and cleaved poly(ADP-ribose)polymerase (PARP; F2, sc-8007).
Full methods are provided in the supplemental information.
Results
EZH2 is overexpressed in NKTL
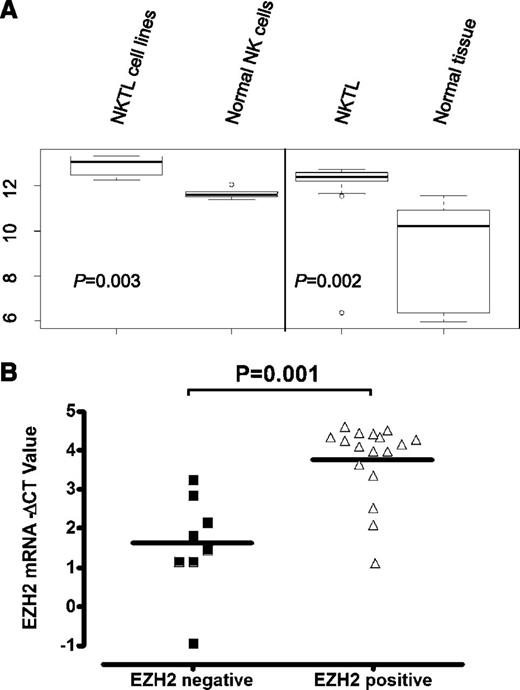
In our previously published genome-wide gene expression profiling (GEP) of extranodal nasal-type NKTL,4 the EZH2 transcript level was significantly higher in NKTL compared with normal NK cells (Figure 1A). In corroboration with the GEP findings, we observed a significant percentage of cases (61%) showing positive expression of EZH2 protein in the tumor cells in our 38 cases of NKTL (tissue microarrays or whole-tissue sections) by IHC studies (supplemental Figure 1; supplemental Table 3), whereas the normal NK cells only showed a minimal level of EZH2 (staining in ≤5% cells). Indeed, EZH2-positive samples have significantly higher EZH2 messenger RNA (mRNA) levels (Figure 1B). These data confirm that EZH2 is overexpressed in NKTL at both the mRNA and protein levels.
EZH2 mRNA levels are elevated in NKTL and cell lines. (A) Expression score for EZH2 mRNAs in NKTL GEP dataset. EZH2 gene expressions in NKTL FFPE samples were compared with that in respective normal FFPE tissue controls, as well as the NK cell lines and normal NK cells using significance analysis of microarray. (B) Correlation between EZH2 transcript levels determined by qRT-PCR and EZH2 protein levels measured by IHC for NKTL samples.
EZH2 mRNA levels are elevated in NKTL and cell lines. (A) Expression score for EZH2 mRNAs in NKTL GEP dataset. EZH2 gene expressions in NKTL FFPE samples were compared with that in respective normal FFPE tissue controls, as well as the NK cell lines and normal NK cells using significance analysis of microarray. (B) Correlation between EZH2 transcript levels determined by qRT-PCR and EZH2 protein levels measured by IHC for NKTL samples.
Loss of miR-26 and miR-101 contributes to the EZH2 upregulation in NKTL
Next, we sought to identify the mechanisms leading to EZH2 upregulation in NKTL. The genomic locus containing EZH2 is not commonly amplified in NKTL.22 In our previous miRNA expression–profiling study, we found several miRNAs that were predicted to target EZH2 by various computational algorithms (supplemental Table 4) to be downregulated in NKTL.19 Among these, the expression of miR-26a, miR-26b, and miR-101 has a significant inverse correlation with EZH2 when our previous GEP and miRNA profiling data were analyzed. miRNAs negatively regulate protein translation by predominantly destabilizing and, hence, decreasing their target mRNA levels.23 In prostate, muscle, and B-cell lymphoma, miR-101, miR-26a, and miR-26b can negatively regulate EZH2 expression by binding to the highly conserved predicted binding sites within the 3′UTR of EZH2.9,10 Using lentiviral transduction to express miR-101, miR-26a, and miR-26b in NKYS (supplemental Figure 2A, left), we observed that EZH2 expression was effectively attenuated (supplemental Figure 2A, right). These data suggest that EZH2 overexpression may be attributed to the deregulation of miR-101, miR-26a, and miR-26b in NK tumor cells.
MYC activation suppresses the expression of miR-26 and miR-101 in NKTL
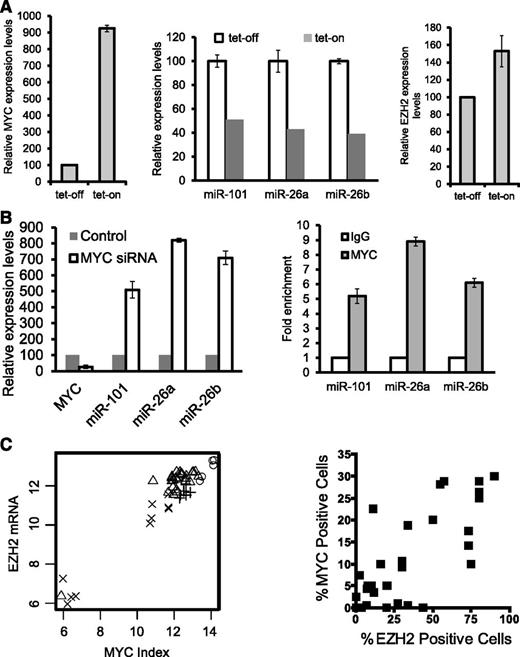
As the genomic loci containing miR-101, miR-26a, and miR-26b are not recurrently deleted in the NKTL (data not shown), we looked for other mechanisms for their repression. On the basis of our previous study of gene expression in NKTL, which showed MYC activation,4 and a recent paper showing that MYC activation can lead to repression of a many miRNAs in tumorigenesis,24 we investigated whether MYC is involved in the suppression of miR-101 and miR-26 in malignant NK cells. When MYC expression was induced in NKYS cells using a tet-on system (Figure 2A, left), miR-101, miR-26a, and miR-26b was downregulated (Figure 2A, middle) with a corresponding (1.53-fold) increase in EZH2 mRNA (Figure 2A, right). Conversely, these 3 miRNAs were up-regulated by depletion of MYC using small interfering RNA (siRNA)–mediated knockdown (Figure 2B, left), and MYC depletion reduced EZH2 3′UTR luciferase reporter activity (supplemental Figure 2B). However, this effect was still seen when the miR-101 and miR-26 binding sites in the EZH2 3′UTR were mutated, suggesting that other miRNAs or factors regulated by MYC may be involved. Furthermore, ChIP-quantitative polymerase chain reaction (qPCR) results revealed that MYC binds to the genomic locus of miR-26a and miR-26b (Figure 2B, right), consistent with previous reports.24 Interestingly, it also revealed an association of MYC specifically with a conserved region upstream of miR-101 on chromosome 1 (Figure 2B, right). In summary, MYC activation suppresses the expression of miR-26a, miR-26b, and miR-101 in NKTL cell lines by direct binding to their genomic locus, and that MYC may stimulate EZH2 overexpression by repression of its negative regulatory miRNAs. Consistent with these in vitro findings, we observed a strong correlation between EZH2 transcript levels and MYC activation as measured by the gene expression–based MYC activation index4 in our clinical GEP dataset (Figure 2C, left). In addition, there is also a positive correlation between the percentage of cells expressing EZH2 and nuclear MYC, which is a marker for MYC activation, detected by IHC analysis in NKTL tissue microarrays (Figure 2C, right). In addition, a significantly greater number of cells with nuclear MYC are detected in EZH2-positive samples (supplemental Figure 3A), suggesting that the data from NKTL cell lines are applicable to clinical samples.
Inhibition of EZH2 expression by miRNA-101 and miRNA-26, which are suppressed by MYC in NKTL cells. (A, left) MYC induction by tet-on. MYC overexpression was induced by treating cells with doxycycline. MYC expression was quantified by qRT-PCR analysis. (A, middle) Decreased levels of miR-101, miR-26a, and miR-26b in NKYS upon MYC induction by tet-on. (A, right) MYC induction by tet-on increases EZH2 mRNA levels. (B, left) Depletion of MYC by siRNAs results in induction of miR-101, mir-26a, and miR-26b transcription in NKYS cells. Cells were transfected with MYC siRNA or nontargeting siRNA as a control. Cells were harvested 48 hours after transfection for mRNA analysis of miR-101 and miR-26 gene levels by real-time PCR. (B, right) ChIP-qPCR for endogenous MYC binding to miR-101, miR-26a, and miR-26b genes. Fold enrichment in the ChIP experiment represents the signal obtained after MYC immunoprecipitation followed by qPCR amplified by primer pairs that spanned gene promoters. (C, left) Correlation between MYC activation index and EZH2 mRNA levels. Cross: Normal tissue; Plus sign: Normal NK; Triangle: NKTL; Circle: Cell Lines. R>0.95, P < 2.2*10–16. (C, right) Scatterplot showing the correlation between IHC MYC staining and EZH2 expression. Spearman correlation coefficient r for MYC v EZH2 = 0.76; P < .0001.
Inhibition of EZH2 expression by miRNA-101 and miRNA-26, which are suppressed by MYC in NKTL cells. (A, left) MYC induction by tet-on. MYC overexpression was induced by treating cells with doxycycline. MYC expression was quantified by qRT-PCR analysis. (A, middle) Decreased levels of miR-101, miR-26a, and miR-26b in NKYS upon MYC induction by tet-on. (A, right) MYC induction by tet-on increases EZH2 mRNA levels. (B, left) Depletion of MYC by siRNAs results in induction of miR-101, mir-26a, and miR-26b transcription in NKYS cells. Cells were transfected with MYC siRNA or nontargeting siRNA as a control. Cells were harvested 48 hours after transfection for mRNA analysis of miR-101 and miR-26 gene levels by real-time PCR. (B, right) ChIP-qPCR for endogenous MYC binding to miR-101, miR-26a, and miR-26b genes. Fold enrichment in the ChIP experiment represents the signal obtained after MYC immunoprecipitation followed by qPCR amplified by primer pairs that spanned gene promoters. (C, left) Correlation between MYC activation index and EZH2 mRNA levels. Cross: Normal tissue; Plus sign: Normal NK; Triangle: NKTL; Circle: Cell Lines. R>0.95, P < 2.2*10–16. (C, right) Scatterplot showing the correlation between IHC MYC staining and EZH2 expression. Spearman correlation coefficient r for MYC v EZH2 = 0.76; P < .0001.
EZH2 overexpression in NKTL confers a growth advantage independently of histone methyltransferase activity
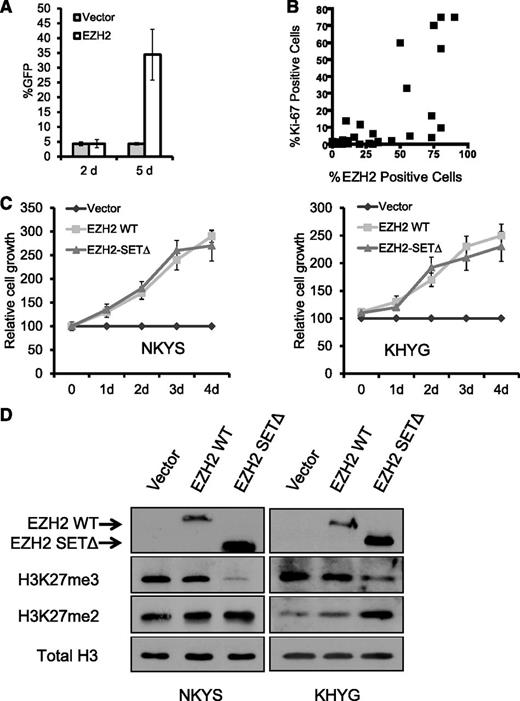
Next, we investigated whether EZH2 is functionally important in NKTL. When EZH2 was introduced into primary NK cells purified from normal human peripheral blood, we observed an eightfold increase in the percentage of GFP(+) cells from day 2 to day 5 compared with vector control (Figure 3A), demonstrating that the overexpression of EZH2 is able to provide a growth advantage in normal primary human NK cells. We next analyzed the effect of increasing EZH2 on cell proliferation in NKTL cell lines. Ectopic expression of EZH2 through transient cotransfection with the GFP expression construct in NKYS cells for 3 days resulted in significantly higher percentage of viable GFP(+) cells in EZH2-transfected cells than in empty vector-transfected cells (supplemental Figure 4). This result suggests that EZH2 overexpression by transfection leads to a competitive cell growth advantage. Consistent with these observations in the cell lines, a positive correlation between the expression of EZH2 and Ki-67, a marker of cell proliferation, was also observed in NKTL tumor samples (Figure 3B). Furthermore, EZH2 positivity is associated with a greater percentage of tumor cells expressing Ki-67 (supplemental Figure 3B).
EZH2 overexpression in NKTL promotes cell growth independent of histone methyltransferase activity. (A) Primary NK cells transduced by EZH2 exhibit a growth advantage. Primary NK cells expressing ectopic EZH2 were monitored by a coexpressed GFP marker. Using our established viral infection protocol, we routinely obtained a transduction efficiency of 4.3% for normal NK cells. If EZH2 infection does not alter the cell growth, the EZH2-infected cell will not gain a growth advantage; thus, the percentage of GFP+ cells should remain at 4.3%. However, the percentage of GFP+ cells increased from approximately 4.3% at day 2 to ∼34.4% at day 5, indicating an acquired growth advantage in these EZH2-infected cells. Antibiotic selection of positively infected cells was not done. (B) Scatterplot representation of the correlation between the percentage of Ki-67–positive cells and the percentage of EZH2-positive cells. Spearman correlation coefficient r for EZH2 v Ki67 = 0.73; P < .0001. (C) MTS proliferation assay showing that ectopic expression of EZH2 promotes cell growth of NKTL cell lines without requiring SET domain activity. Cells were cotransfected with pMAX-GFP and the control empty vector pcDNA4.1 or EZH2 expression plasmids. Cells transfected were subjected to proliferation assays for up to 96 hours. The cell growth (expressed as a percentage of the empty vector control) was determined by MTS assay as described in Materials and Methods. The mean values of triplicate samples are shown, and error bars indicate standard deviations. (D) Western blot analysis of EZH2, H3K27m3, and H3K27m2 in indicated samples. Expression of EZH2 WT and the SET-domain mutant was detected by the MYC-tag antibody. H3 was used as a loading control.
EZH2 overexpression in NKTL promotes cell growth independent of histone methyltransferase activity. (A) Primary NK cells transduced by EZH2 exhibit a growth advantage. Primary NK cells expressing ectopic EZH2 were monitored by a coexpressed GFP marker. Using our established viral infection protocol, we routinely obtained a transduction efficiency of 4.3% for normal NK cells. If EZH2 infection does not alter the cell growth, the EZH2-infected cell will not gain a growth advantage; thus, the percentage of GFP+ cells should remain at 4.3%. However, the percentage of GFP+ cells increased from approximately 4.3% at day 2 to ∼34.4% at day 5, indicating an acquired growth advantage in these EZH2-infected cells. Antibiotic selection of positively infected cells was not done. (B) Scatterplot representation of the correlation between the percentage of Ki-67–positive cells and the percentage of EZH2-positive cells. Spearman correlation coefficient r for EZH2 v Ki67 = 0.73; P < .0001. (C) MTS proliferation assay showing that ectopic expression of EZH2 promotes cell growth of NKTL cell lines without requiring SET domain activity. Cells were cotransfected with pMAX-GFP and the control empty vector pcDNA4.1 or EZH2 expression plasmids. Cells transfected were subjected to proliferation assays for up to 96 hours. The cell growth (expressed as a percentage of the empty vector control) was determined by MTS assay as described in Materials and Methods. The mean values of triplicate samples are shown, and error bars indicate standard deviations. (D) Western blot analysis of EZH2, H3K27m3, and H3K27m2 in indicated samples. Expression of EZH2 WT and the SET-domain mutant was detected by the MYC-tag antibody. H3 was used as a loading control.
Although EZH2 commonly exerts its oncogenic properties through H3K27 trimethylation (H3K27me3) and gene repression, we did not observe an association between EZH2 expression and the abundance of H3K27me3 by IHC studies in our clinical NKTL samples (supplemental Figure 3C). Interestingly, a lack of association between EZH2 and H3K27me3 has also been described in breast tumor subtypes25,26 and in ovarian and pancreatic cancers.26 This finding raises the possibility that EZH2 may have functions other than its activity on H3K27me3 in cancers, including NKTL. To investigate this possibility, we compared the ability of EZH2 wild-type (WT) and an EZH2 SET domain deletion mutant (EZH2 SETΔ) to increase the cell growth of NKTL cells. Interestingly, EZH2 SETΔ, which lacked the methyltransferase activity for H3K27me3, was still able to strongly promote cell growth of NKYS cells. This effect was as potent as EZH2 WT, as indicated by a similar increase in the percentage of GFP(+) cells with time (supplemental Figure 4), and increase in cell growth as measured by the MTS assay (Figure 3C). Both western blot (Figure 3D) and quantitative reverse-transcription PCR (qRT-PCR) analysis using a pair of primers that specifically amplifies the SET domain (supplemental Figure 5) confirmed the ectopic expression of EZH2 SETΔ in transfected cells. The ability of EZH2 SETΔ to deplete the H3K27me3 was also validated (Figure 3D; supplemental Figure 6C). These results indicate that the proproliferative property of EZH2 in NKTL is not mediated by its histone methyltransferase activity.
EZH2 directly activates CCND1 transcription by binding to its promoter independent of its methyltransferase activity in NKTL
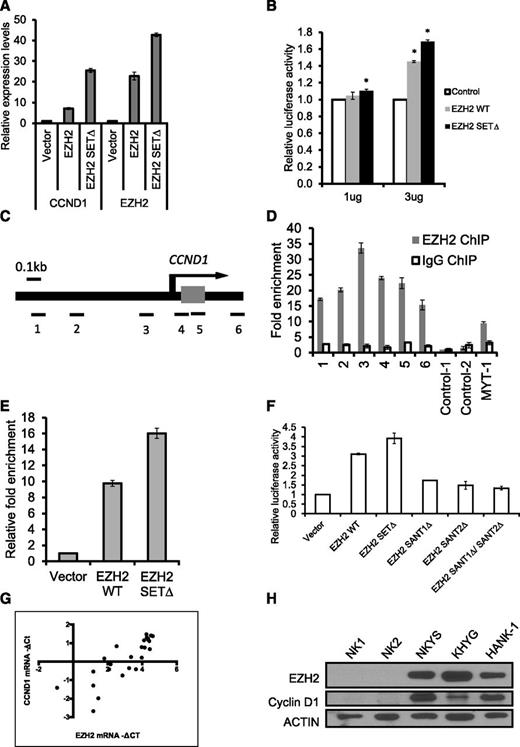
To better understand how EZH2 promotes proliferation in NK cell lines, we explored the mechanism by which EZH2 regulates cell cycle genes. The study by Bracken and colleagues showed that suppression of EZH2 by RNA interference (RNAi) significantly decreased positive regulators of cell proliferation such as G1/S-cyclins,27 so it is tempting to speculate that EZH2 directly regulates the transcription of these genes by binding to their genomic locus. The CCND1 transcript is reported to be upregulated in NKTL tissues compared with normal NK cells,28 and high expression of CCND1 correlates with poor prognosis and decreased survival duration in NKTL.29 Therefore, we examined whether the transcription of CCND1 is affected by EZH2 overexpression. Consistent with our findings on cell growth, qRT-PCR indicated that CCND1 mRNA levels increased substantially after EZH2 overexpression in NKYS (Figure 4A). Higher induction of CCND1 by EZH2-SETΔ could be explained by a previous study that ectopic EZH2 SETΔ depletes endogenous EZH2 and consistently displays higher levels of protein accumulation compared with the ectopic EZH2 WT.30
EZH2 positively regulates CCND1 transcription by binding to its promoter in NKTL cells. (A) Overexpression of EZH2 induces the expression of CCND1. Vectors expressing EZH2 (pcDNA-EZH2 or pcDNA-EZH2 SETΔ) were transiently transfected into NKYS cells. The RNA harvested from the cells at 24 hours after transfection was isolated, reverse-transcribed, subjected to qPCR by using primers specific for CCND1 mRNA, and normalized with GAPDH. (B) Luciferase promoter assay showing that EZH2 activates CCND1 transcription. NKYS cells were transfected with the luciferase reporter construct pGL4 containing the CCND1 promoter and various amounts of EZH2 WT/SETΔ plasmid or a control vector. Luciferase activities were measured after 24 hours. Luciferase readings were further normalized to the internal control pRL null. Results are presented as averages of triplicate experiments. Error bars represent standard deviation. * denotes P < .01 with respect to cells transfected with the same amount of control vector. (C) Genomic structure of human CCND1. The locations of the 6 pairs of primer sets used to detect the ChIP-enriched DNA fragments are indicated. (D) ChIP-qPCR for endogenous EZH2 binding to the CCND1 gene. ChIP assays were performed by using NKYS. Real-time PCR was performed with immunoprecipitated chromatin fragments obtained by using an anti-EZH2 antibody or an irrelevant antibody (IgG) as a control. A known EZH2 binding site in the promoter region of the MYT-1 gene was amplified as a positive control for the ChIP assays. (E) ChIP-qPCR for ectopically expressed EZH2 WT and EZH2 SETΔ binding to the CCND1 gene. ChIP assays were performed by using NKYS transfected by a pcDNA4.1/Myc-His vector or a plasmid expressing EZH2 WT and EZH2-SETΔ. Real-time PCR was performed with immunoprecipitated chromatin fragments obtained by using an anti-His antibody. Primer set 3, which amplifies a region representing EZH2 binding, was used to detect the ChIP-enriched DNA fragments. (F) Luciferase promoter assay showing the inability of EZH2 SANT domain deletion mutants to activate CCND1 transcription. NKYS were transfected with 3µg of EZH2 WT/SETΔ/SANTΔ plasmids or a control vector. Luciferase activities were measured after 24 hours. (G) Correlation between mRNA levels for EZH2 and CCND1 determined by qRT-PCR in NKTL clinical samples. Spearman correlation coefficient = 0.8608; P < .0001. (H) Western blot analysis of protein levels of CCND1 and EZH2 in NK cell lines and normal NK cells.
EZH2 positively regulates CCND1 transcription by binding to its promoter in NKTL cells. (A) Overexpression of EZH2 induces the expression of CCND1. Vectors expressing EZH2 (pcDNA-EZH2 or pcDNA-EZH2 SETΔ) were transiently transfected into NKYS cells. The RNA harvested from the cells at 24 hours after transfection was isolated, reverse-transcribed, subjected to qPCR by using primers specific for CCND1 mRNA, and normalized with GAPDH. (B) Luciferase promoter assay showing that EZH2 activates CCND1 transcription. NKYS cells were transfected with the luciferase reporter construct pGL4 containing the CCND1 promoter and various amounts of EZH2 WT/SETΔ plasmid or a control vector. Luciferase activities were measured after 24 hours. Luciferase readings were further normalized to the internal control pRL null. Results are presented as averages of triplicate experiments. Error bars represent standard deviation. * denotes P < .01 with respect to cells transfected with the same amount of control vector. (C) Genomic structure of human CCND1. The locations of the 6 pairs of primer sets used to detect the ChIP-enriched DNA fragments are indicated. (D) ChIP-qPCR for endogenous EZH2 binding to the CCND1 gene. ChIP assays were performed by using NKYS. Real-time PCR was performed with immunoprecipitated chromatin fragments obtained by using an anti-EZH2 antibody or an irrelevant antibody (IgG) as a control. A known EZH2 binding site in the promoter region of the MYT-1 gene was amplified as a positive control for the ChIP assays. (E) ChIP-qPCR for ectopically expressed EZH2 WT and EZH2 SETΔ binding to the CCND1 gene. ChIP assays were performed by using NKYS transfected by a pcDNA4.1/Myc-His vector or a plasmid expressing EZH2 WT and EZH2-SETΔ. Real-time PCR was performed with immunoprecipitated chromatin fragments obtained by using an anti-His antibody. Primer set 3, which amplifies a region representing EZH2 binding, was used to detect the ChIP-enriched DNA fragments. (F) Luciferase promoter assay showing the inability of EZH2 SANT domain deletion mutants to activate CCND1 transcription. NKYS were transfected with 3µg of EZH2 WT/SETΔ/SANTΔ plasmids or a control vector. Luciferase activities were measured after 24 hours. (G) Correlation between mRNA levels for EZH2 and CCND1 determined by qRT-PCR in NKTL clinical samples. Spearman correlation coefficient = 0.8608; P < .0001. (H) Western blot analysis of protein levels of CCND1 and EZH2 in NK cell lines and normal NK cells.
Next, we evaluated whether EZH2 can activate the activity of the CCND1 promoter in NKYS cells. Cotransfection of the EZH2 WT expression vector with the pGL4-CCND1-Luc reporter resulted in considerable activation of CCND1 promoter activity (Figure 4B). Consistent with prior results, activation of luciferase activity was also observed in cells transfected with EZH2 SETΔ (Figure 4B). Collectively, these results indicate that EZH2 positively regulates CCND1 transcription independent of its histone methyltransferase activity.
We then addressed whether EZH2 binds to the CCND1 promoter using ChIP-qPCR assays. Six pairs of primers, located sequentially along the proximal promoter, first exon, and intron 1 of CCND1 were used to quantify the ChIP-enriched DNA by real-time PCR (Figure 4C). A peak representing EZH2 binding was observed (∼33-fold above background) at a region very close to the transcriptional start site of CCND1 (Figure 4D). ChIP using a control IgG showed no significant enrichment over the entire surveyed region (Figure 4D). By performing ChIP assays using a His-Tag antibody in NKYS transfected with His-Tagged EZH2 SETΔ, we showed that ectopically expressing EZH2 SETΔ resulted in significant enrichment of EZH2-DNA complexes, which was even higher than EZH2 WT (Figure 4E). This effect is specific to the SET domain, as EZH2 SANT domain deletion mutants abolished EZH2 transcriptional activity on the CCND1 promoter (Figure 4F). Furthermore, data from tumor samples that CCND1 mRNA levels correlate well with EZH2 mRNA levels are consistent with this finding (Figure 4G). Consistently, western blot analyses showed that CCND1 protein is expressed together with EZH2 in NK cell lines but not in normal NK cells (Figure 4H). Taken together, our findings identify CCND1 as a bona fide direct target of EZH2 in NKTL. Importantly, EZH2 acts as a transcriptional activator for the CCND1 gene without requiring histone methylation catalytic activity, which provides a mechanistic explanation for the proproliferative role of EZH2 SETΔ in NKTL.
Growth inhibition on depletion of endogenous EZH2 can be rescued by exogenous EZH2 SET mutants
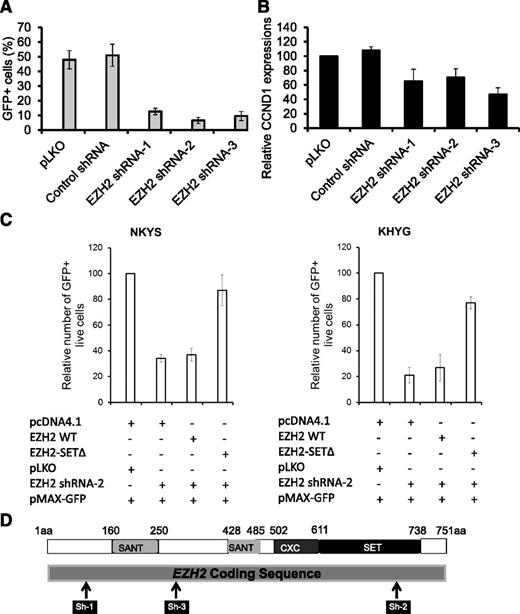
Given that EZH2 is of functional importance in NKTL raises the possibility that targeting EZH2 may be a feasible strategy in NKTL. We first investigated the effects of depleting EZH2 on cell growth. Using 3 different short hairpin RNAs (shRNAs) targeting the EZH2 gene (Figure 5D; supplemental Figure 7A) in NKYS, we showed that depletion of EZH2 resulted in a substantial decrease in cell numbers, as revealed by the percentage of GFP+ cells (Figure 5A), and a specific concomitant decrease of CCND1 expression but not in other gene transcripts such as PRDM1 and IGF1 (Figure 5B; supplemental Figure 8). This result strengthens the postulation that EZH2 is required for expression of the proliferative gene CCND1 and suggests that downregulation of CCND1 is responsible for the reduction in cell growth in NKTL cells after EZH2 depletion.
EZH2 depletion inhibits cell growth of NKTL tumor cells. (A) Effects of EZH2 knockdown on cell growth of NKYS cells. EZH2 shRNAs or control shRNA plasmids were cotransfected with a GFP-expressing plasmid pMAX-GFP in cells by electroporation. The percentage of GFP+ cells was determined by a Tali image-based cytometer at 18 hours after transfection. Cells transfected with EZH2 shRNA, but not those transfected with an empty vector or unrelated shRNA, exhibited a severe competitive growth disadvantage and cell death, as indicated by a significant depletion of GFP+ cells with time. (B) qRT-PCR analysis of CCND1 transcription in NKYS cells with EZH2 depleted by RNAi. (C) Rescue of EZH2 shRNA induced cell viability loss by forced expression of EZH2 SETΔ. Endogenous EZH2 was knocked down by an EZH2 shRNA-2, or was restored to physiological levels by ectopically expressing EZH2 shRNA2-resistant EZH2 SETΔ in NKYS and KHYG1 cells. Cells were cotransfected with EZH2 shRNA, together with either EZH2 WT or EZH2 SETΔ. Control transfections included a pLKO shRNA and pcDNA4.1, respectively. pMAX-GFP was cotransfected with other plasmids in each transfection to mark successfully transfected cells. The percentage of GFP+ live cells was determined by a Tali image-based cytometer at 18 hours after transfection. (D) Schematic description of EZH2 protein as well as the relative positions of regions targeted by EZH2 shRNAs. shRNAs expressed from a pLKO.1 vector targeting 3 regions of EZH2 are shown as black bars in relationship to the protein-coding regions.
EZH2 depletion inhibits cell growth of NKTL tumor cells. (A) Effects of EZH2 knockdown on cell growth of NKYS cells. EZH2 shRNAs or control shRNA plasmids were cotransfected with a GFP-expressing plasmid pMAX-GFP in cells by electroporation. The percentage of GFP+ cells was determined by a Tali image-based cytometer at 18 hours after transfection. Cells transfected with EZH2 shRNA, but not those transfected with an empty vector or unrelated shRNA, exhibited a severe competitive growth disadvantage and cell death, as indicated by a significant depletion of GFP+ cells with time. (B) qRT-PCR analysis of CCND1 transcription in NKYS cells with EZH2 depleted by RNAi. (C) Rescue of EZH2 shRNA induced cell viability loss by forced expression of EZH2 SETΔ. Endogenous EZH2 was knocked down by an EZH2 shRNA-2, or was restored to physiological levels by ectopically expressing EZH2 shRNA2-resistant EZH2 SETΔ in NKYS and KHYG1 cells. Cells were cotransfected with EZH2 shRNA, together with either EZH2 WT or EZH2 SETΔ. Control transfections included a pLKO shRNA and pcDNA4.1, respectively. pMAX-GFP was cotransfected with other plasmids in each transfection to mark successfully transfected cells. The percentage of GFP+ live cells was determined by a Tali image-based cytometer at 18 hours after transfection. (D) Schematic description of EZH2 protein as well as the relative positions of regions targeted by EZH2 shRNAs. shRNAs expressed from a pLKO.1 vector targeting 3 regions of EZH2 are shown as black bars in relationship to the protein-coding regions.
Next, we sought to clarify if the reduction in cell growth is dependent on the methyltransferase activity of EZH2. Because EZH2 SETΔ is immune to EZH2 shRNA-2, which targets the SET domain, EZH2 shRNA-2 only depleted endogenous WT EZH2 but not the EZH2 SETΔ after being introduced into the cells, as expected (supplemental Figure 7B). Indeed, EZH2 SETΔ was able to prevent the reduction in cell numbers mediated by EZH2 shRNA-2, whereas reintroduction of EZH2 WT could not (Figure 5C, left). These rescue experiments excluded the possibility that the cell death phenotype occurred because of an off-target effect of EZH2 shRNA and, at the same time, confirming that the prosurvival effect of endogenous EZH2 does not require its enzymatic activity. Similar results were obtained in KHYG1 (Figure 5C, right), another NKTL cell line, indicating a consistent requirement for expression of EZH2 in the growth and survival of NK tumor cells.
Inhibition of EZH2 by 3-deazaneplanocin A (DZNep) induced growth inhibition and apoptosis of malignant NK cell lines
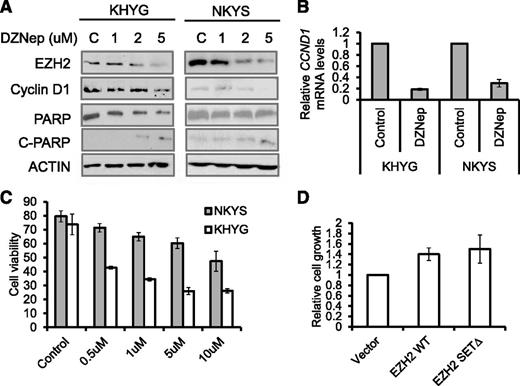
The effects seen with EZH2 knockdown suggest that EZH2 may be a therapeutic target in NKTL. We next explored the use of a compound capable of depleting PRC2 components called DZNep.31 As reported previously,31-34 DZNep effectively and dose-dependently reduced cellular levels of EZH2 in KHYG1 and NKYS, resulting in apoptosis as detected by PARP cleavage using western blot and Annexin V analysis by flow cytometry in a dose-dependent manner (Figure 6A,C). At the same time, CCND1 is downregulated by DZNep at both the mRNA and protein levels (Figure 6A-B). In line with the diminished CCND1, NKYS treated with DZNep showed a substantial reduction in their proliferation rate, as illustrated by a 40% decrease in their BrdU incorporation (supplemental Figure 10). Thus, DZNep was able to phenocopy the effects of EZH2 knockdown on cell growth and CCND1 expression.
DZNep inhibits cell growth and induces apoptosis in NKTL tumor cells. (A) Western blot analysis of NK tumor cells exposed to increasing concentrations of DZNep showed a dose-dependent decrease in EZH2 protein, a decrease in CCND1 protein, and PARP cleavage in response to DZNep treatment. Cells were treated with indicated concentration of DZNep for 48 hours. Actin was used as a loading control. (B) Reduction of CCND1 mRNAs in DZNep-treated cells. The RNA harvested from the cells at 48 hours after treatment with DZNep at 5 µM, reverse-transcribed, subjected to qPCR by using primers specific for CCND1. (C) Quantification of cell viability in KHYG and NKYS cells treated with DZNep. Data are mean ±standard deviation of 3 independent experiments. (D) The rescued effects by EZH2 or EZH2Δ overexpression on DZNep-induced cell growth inhibition. Control plasmid or EZH2-expressing plasmids were cotransfected with pMAX-GFP to NKYS cells. Cells were then cultured in the absence or presence of DZNep at 10 µM for 48 hours. The percentage of GFP+ live cells was accessed by a Tali image-based cytometer.
DZNep inhibits cell growth and induces apoptosis in NKTL tumor cells. (A) Western blot analysis of NK tumor cells exposed to increasing concentrations of DZNep showed a dose-dependent decrease in EZH2 protein, a decrease in CCND1 protein, and PARP cleavage in response to DZNep treatment. Cells were treated with indicated concentration of DZNep for 48 hours. Actin was used as a loading control. (B) Reduction of CCND1 mRNAs in DZNep-treated cells. The RNA harvested from the cells at 48 hours after treatment with DZNep at 5 µM, reverse-transcribed, subjected to qPCR by using primers specific for CCND1. (C) Quantification of cell viability in KHYG and NKYS cells treated with DZNep. Data are mean ±standard deviation of 3 independent experiments. (D) The rescued effects by EZH2 or EZH2Δ overexpression on DZNep-induced cell growth inhibition. Control plasmid or EZH2-expressing plasmids were cotransfected with pMAX-GFP to NKYS cells. Cells were then cultured in the absence or presence of DZNep at 10 µM for 48 hours. The percentage of GFP+ live cells was accessed by a Tali image-based cytometer.
We next explored whether depletion of EZH2 is responsible for the cell growth inhibition and apoptosis observed by DZNep treatment in malignant NK cells. It is worth noting that DZNep depleted endogenous EZH2 protein but had no significant effect on exogenous EZH2 WT and EZH2 SETΔ (supplemental Figure 11). We observed a decrease in the DZNep-induced inhibition of cell growth in NKYS cells expressing both exogenous EZH2 WT and EZH2 SETΔ, compared with cells transfected with empty vector (Figure 6D). Although DZNep is not a specific inhibitor of EZH2 and may affect other molecules,35 these findings indicate that DZNep-induced loss of cell viability in NKTL cells is, in large part, the result of a decrease in EZH2 levels.
Lastly, we evaluated the effects of DZNep on cell growth in other NK cell lines compared with normal NK cells. The MTS assay demonstrated that all of the additional 4 cell lines responded to DZNep treatment, whereas DZNep had minimal effects on normal NK cells (supplemental Figure 12A). Interestingly, the sensitivity of different cell lines to DZNep treatment seems to be related to EZH2 expression levels (supplemental Figure 12B).
Discussion
The data presented here show that EZH2 is aberrantly overexpressed in NKTL and that this is linked to Myc-mediated repression of miRNAs, such as miR26 and miR101 that normally target and inhibit EZH2 expression. This regulatory network demonstrated in NKTL further strengthens the recently proposed model that MYC may stimulate EZH2 overexpression by repression of its negative regulatory miRNAs.9 Importantly, EZH2 overexpression is functionally important in NKTL, and the ability of EZH2 to promote proliferation in NKTL does not require its histone methyltransferase activity. Thus, for the first time our findings have uncovered a crucial and novel role for EZH2 in control of cell proliferation. Given our findings, it is likely that this unconventional role of EZH2 seen in NTKL may represent a more general feature that may be also operational in other malignancies.
The best-understood mechanism by which EZH2 exerts its oncogenic function is to induce gene repression through its effect on chromatin via its histone methyltransferase activity that requires the SET domain of EZH2. However, there has been emerging evidence implying EZH2 functions that are not compatible with the transcription repression model. The oncogenic function of EZH2 has been attributed to the silencing of tumor suppressor genes such as ARF,36 p57KIP2,37 FBXO32,38 p27,33 and BRCA1.39 Bracken and colleagues demonstrated that the downregulation of EZH2 expression by RNAi did not increase the expression of these negative regulators of the cell cycle.27 On the other hand, there was a significant decrease in the positive regulators of cell proliferation such as G1/S-expressed cyclins in EZH2 knockdown cells.27 These observations suggest that EZH2 is required for activation or maintenance of the activated state of certain genes in proliferating cells. The first evidence indicating a role of EZH2 in transcriptional activation was provided by Shi and colleagues40 in which EZH2 interacts directly with estrogen receptor and β-catenin, functionally enhancing gene transactivation in the estrogen and Wnt pathways. In a more recent study, we demonstrated that EZH2 in aggressive breast cancer cells can positively modulate the NF-κB target gene by forming a ternary complex with RelA and RelB.30 Notably, both studies demonstrated that this transcriptional activation activity is independent of the EZH2 SET domain and H3K27me3, although neither study has provided a phenotypic demonstration of such a histone methyltransferase–independent function of EZH2. Our results here clearly demonstrate that EZH2 enhances proliferation of malignant NK cells without requiring its enzyme activity, which highlights a mechanism not well recognized for EZH2 function.
Recent reports increasingly suggest an important role of EZH2 in a wide range of hematologic malignancies, yet its exact tumorigenic role and mechanism and the pathways it deregulates seem to vary in different malignancies. For example, we recently showed that EZH2 enzymatic function in acute myeloid leukemia is important and may be involved in deregulating metabolic pathway genes.34 On the other hand, in the current study in NKTL, the nonenzymatic mechanism seems to be important in driving cell cycle progression by activating CCND1 expression. The understanding of how EZH2 works as an oncogene in different cancers has important therapeutic implications. EZH2 is currently thought to be important in oncogenesis through its enzymatic activity; hence, inhibitors in development are mostly targeting EZH2 enzymatic activity. Our study demonstrates that the oncogenic function of EZH2 may not always be dependent on its enzymatic activities. Therefore, small-molecule drugs that block EZH2 enzymatic activity will not work in these situations. This implies a need to review current therapeutic strategies. In view of the dual function of EZH2 in transcription activation and epigenetic repression, ideally there is a need to identify tumors where EZH2 is predominantly acting through its enzymatic function as a histone methyltransferase, inhibiting the protective role of tumor suppressor genes, as well as tumors where EZH2 is predominantly acting through activation of genes involved in other oncogenic pathways. This will ensure that the appropriate therapeutic strategy can be applied. Conversely, a compound that downregulates EZH2 protein may be effective in both scenarios. Therefore, therapeutic compounds that lead to EZH2 protein degradation rather than specific enzymatic inhibitors may be advantageous.
The proproliferative properties of EZH2 in NKTL support the rationale for using EZH2 inhibitors in the treatment of NKTL. Because targeting of EZH2 is an active area of drug development at present, there is great potential for the development of better treatment modalities. This is especially important for aggressive cancers, such as NKTL, for which no effective curative treatment is currently available.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof. Daniel G. Tenen and Prof. H. Phillip Koeffler for their helpful suggestions.
W.-J.C. was supported by the National Medical Research Council Clinician Scientist Investigator Award. This work is supported in part by the Singapore National Research Foundation and the Ministry of Education under the Research Center of Excellence Program to W.-J.C. S.-B.N. was supported by the National University Health System Clinician Scientist Program Award.
Authorship
Contribution: J.Y. and W.-J.C. conceived and designed the study, analyzed and interpreted the data, and wrote the paper; Q.Y. provided vital reagents and interpreted findings; S.-B.N. provided clinical samples, performed IHC scoring, interpreted the data, and wrote the paper; S.W. performed IHC scoring; J.Y., J.L.-S.T., B.L., T.L.K., J.T., V.S., S.-C.L., C.B., and S.-N.C. performed experiments; G.H. performed bioinformatics analysis; and N.S. contributed the cell lines.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wee-Joo Chng, Department of Hematology-Oncology, National University Cancer Institute of Singapore, National University Health System, 1E, Kent Ridge Rd, Singapore 119228; e-mail: mdccwj@nus.edu.sg.
References
Author notes
J.Y. and S.-B.N. contributed equally to this study.