Key Points
FOXP1 is downregulated in germinal centers, inversely to BCL6, whereby it regulates a network of genes, half of which are also BCL6 targets.
In transgenic mice, constitutive FOXP1 expression impairs GC formation and function, which might contribute to B-cell lymphomagenesis.
Abstract
B-cell maturation and germinal center (GC) formation are dependent on the interplay between BCL6 and other transcriptional regulators. FOXP1 is a transcription factor that regulates early B-cell development, but whether it plays a role in mature B cells is unknown. Analysis of human tonsillar B-cell subpopulations revealed that FOXP1 shows the opposite expression pattern to BCL6, suggesting that FOXP1 regulates the transition from resting follicular B cell to activated GC B cell. Chromatin immunoprecipitation-on-chip and gene expression assays on B cells indicated that FOXP1 acts as a transcriptional activator and repressor of genes involved in the GC reaction, half of which are also BCL6 targets. To study FOXP1 function in vivo, we developed transgenic mice expressing human FOXP1 in lymphoid cells. These mice exhibited irregular formation of splenic GCs, showing a modest increase in naïve and marginal-zone B cells and a significant decrease in GC B cells. Furthermore, aberrant expression of FOXP1 impaired transcription of noncoding γ1 germline transcripts and inhibited efficient class switching to the immunoglobulin G1 isotype. These studies show that FOXP1 is physiologically downregulated in GC B cells and that aberrant expression of FOXP1 impairs mechanisms triggered by B-cell activation, potentially contributing to B-cell lymphomagenesis.
Introduction
The activation of lymphocytes following an antigen encounter is essential for the generation of a broad repertoire of high-affinity protective antibodies. After T-cell–mediated costimulation in secondary lymphoid organs, activated B cells are confronted with important fate decisions. Either they stay in specialized extrafollicular sites, differentiating into short-lived antibody-secreting plasma cells or adopting an early memory phenotype, or they migrate inside lymphoid follicles and form specialized dynamic structures termed germinal centers (GCs).1-3 The mechanisms responsible for these cell-fate decisions at the onset of GC formation are not completely understood. One of the main proteins required by activated B cells to form GCs is BCL6, a transcriptional regulator that controls DNA damage and apoptotic responses and mediates B-cell activation and differentiation.4,5 In the absence of BCL6, high-affinity antibodies cannot be produced and GC formation is abrogated.6,7 Another transcription factor, FOXP1 (forkhead box protein P1), is emerging not only as an essential factor in early pro/pre–B-cell development,8,9 but also as a recurrently deregulated protein highly expressed in a variety of B-cell lymphomas.10-16 Furthermore, FOXP1 has been reported to have a role coordinating transitions between cell proliferation and differentiation in multiple biological contexts including myocytes,17,18 neurons,19-21 monocytes and macrophages,22 T cells,23,24 and stem cells.25,26 Therefore, it is plausible that FOXP1 may also play a role at the transitional stages of mature B-cell differentiation.
Deletion of FOXP1 leads to embryonic lethality.18 Analysis of mice in which Foxp1 was deleted in B lymphocytes revealed that FOXP1 controls the pro-B to pre–B-cell transition within the bone marrow through binding to the ERAG enhancer, thereby regulating the expression of Rag1 and Rag2 and the V-(D)-J [variable-(diversity)-joining] recombination of the immunoglobulin heavy chain gene.8 Conversely, overexpression of FOXP1, either through the chromosomal t(3;14)(p14.1;q32) leading to FOXP1-IGH fusion or associated to trisomy of chromosome 3, has been observed predominantly in mucosa-associated lymphoid tissue (MALT) lymphoma and diffuse large B-cell lymphoma (DLBCL)11-13,16,27,28 and is linked to a poor prognosis.14,15 It has been suggested that FOXP1 expression can promote transformation of MALT lymphoma to DLBCL,10,29 but the gene-regulatory networks targeted by FOXP1 in lymphoma or mature B cells are as yet unknown. Overexpression of human FOXP1 protein in the murine monocyte/macrophage lineage impaired monocyte-to-macrophage maturation in the spleen, reduced the expression of specific surface receptors, and deregulated general macrophage functions.22 Therefore, we asked whether a similar overexpression strategy might help to decipher the putative role of FOXP1 in mature B-cell development.
Recently, FOXP1 has been shown to function in mature naïve T-cell activation.23 This observation, together with the marked expression of FOXP1 in numerous mature B-cell lymphomas and its involvement in multiple cell lineages as a coordinator of developmental transitions, led us to suggest a role for FOXP1 at a more mature stage of B-cell development. We explore here the hypothesis that regulation of FOXP1 at the naïve-to-GC transition is required during naïve B-cell activation and GC reaction.
Methods
Human primary samples and cell lines
Nontumoral fresh human tonsils were obtained with the approval of the institutional review boards in accordance with the Declaration of Helsinki. The human GC-like DLBCL cell line OCI-Ly1 was included in the study. Primary antibodies, primers, and small interfering RNAs (siRNAs) are listed in supplemental Table 3.
ChIP-on-chip
Array-based chromatin immunoprecipitation (ChIP-on-chip) assays were performed in OCI-Ly1 cells as previously described.30 Data were deposited in the Gene Expression Omnibus (accession number GSE44243).
Computational analyses
To identify FOXP1 target genes, we computed the log ratio between the probe intensities of the ChIP product and input as previously described.30,31 De novo motif analysis was performed using the FIRE motif discovery program.32 Large gene expression data sets published previously33 (Gene Expression Omnibus series GSE25639) were also used.
Mouse generation and characterization
Transgenic mice overexpressing the human FOXP1 complementary DNA (NM_032682.5) under the control of Eμ enhancer and SR-α mouse promoter were developed (www.genoway.com). Transgene integration, immunization protocols, flow cytometry assays, and methods for class-switch recombination (CSR) and somatic hypermutation (SHM) analyses were performed as previously described34-36 (supplemental “Methods”). All experiments were conducted with protocols approved by the Ethical Committee of Animal Experimentation of the University of Navarra.
Full methodology information is provided in supplemental “Methods”.
Results
FOXP1 expression is downregulated during the normal GC reaction and shows an inverse correlation with BCL6 expression
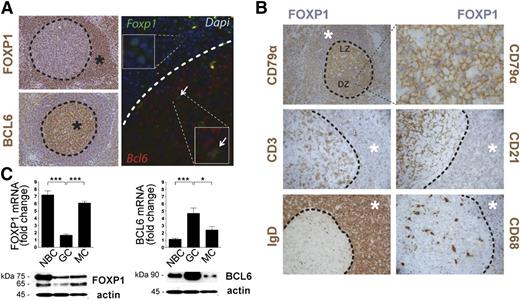
Initial work studying the expression of FOXP1 showed that it is broadly expressed in many normal adult tissues,37-39 including secondary lymphoid follicles.38 To further investigate the expression pattern of FOXP1 in lymphoid tissue, we examined human tonsillar reactive lymphoid follicles by immunohistochemistry (IHC) and immunofluorescence. Nuclear expression of FOXP1 was more abundant in cells surrounding the GC but appeared markedly downregulated inside the GC, with only a small subset of cells expressing FOXP1 (Figure 1A). Conversely, BCL6 exhibited the reciprocal expression pattern to FOXP1, being detected in the nuclei of GC B lymphocytes but not in the cells surrounding the GC. In fact, dual immunofluorescence studies with FOXP1 and BCL6 antibodies detected coexpression of these proteins in only a small fraction of GC cells (Figure 1A), perhaps corresponding to activated B cells that recently entered the GC transitioning from a FOXP1- to a BCL6-mediated transcriptional program. Double IHC analysis using antibodies for FOXP1 and for surface markers of different B-lymphocyte subpopulations (CD79α+ and IgD+), T cells (CD3+), macrophages (CD68+), and follicular/dendritic cells (CD21+) confirmed that FOXP1 was mainly expressed in naïve B cells and that the small subset of FOXP1-expressing cells within the GC corresponded to CD79α+ B lymphocytes in the dark zone of GCs (Figure 1B).
Inverse correlation of FOXP1 and BCL6 expression in GC. (A) Representative images of human tonsil reactive sections examined by IHC (left panels) using anti-FOXP1 and anti-BCL6 antibodies (brown). Also, FOXP1 (green) and BCL6 (red) expression were examined by immunofluorescence (right panels) in human tonsils. * denotes areas of positive signal. Arrows indicate cells that coexpress FOXP1 and BCL6 at the onset of the GC. (B) Double IHC in human tonsil sections for FOXP1 (blue) and other surface markers, including CD79α+ or IgD+ B cells, CD3+ T cells, CD21+ follicular dendritic cells, and CD68+ macrophages (brown). * denotes areas of positive FOXP1 signal (blue). (C) qRT-PCR (top) and immunoblot (bottom) analysis of FOXP1 and BCL6 expression from naïve (NBC), GC, and memory (MC) B cells isolated from human tonsils. qRT-PCR gene expression is shown relative to GAPDH. Bars show mean ± standard error of the mean (SEM) from at least 6 human tonsils. Significance was determined by 2-tailed unpaired Student t tests. Representative western blots from 2 independent experiments are shown. NBC, naïve B cells; MC, memory B cells; mRNA, messenger RNA.
Inverse correlation of FOXP1 and BCL6 expression in GC. (A) Representative images of human tonsil reactive sections examined by IHC (left panels) using anti-FOXP1 and anti-BCL6 antibodies (brown). Also, FOXP1 (green) and BCL6 (red) expression were examined by immunofluorescence (right panels) in human tonsils. * denotes areas of positive signal. Arrows indicate cells that coexpress FOXP1 and BCL6 at the onset of the GC. (B) Double IHC in human tonsil sections for FOXP1 (blue) and other surface markers, including CD79α+ or IgD+ B cells, CD3+ T cells, CD21+ follicular dendritic cells, and CD68+ macrophages (brown). * denotes areas of positive FOXP1 signal (blue). (C) qRT-PCR (top) and immunoblot (bottom) analysis of FOXP1 and BCL6 expression from naïve (NBC), GC, and memory (MC) B cells isolated from human tonsils. qRT-PCR gene expression is shown relative to GAPDH. Bars show mean ± standard error of the mean (SEM) from at least 6 human tonsils. Significance was determined by 2-tailed unpaired Student t tests. Representative western blots from 2 independent experiments are shown. NBC, naïve B cells; MC, memory B cells; mRNA, messenger RNA.
Quantitative measurement of the expression of FOXP1 and BCL6 in isolated B-cell subpopulations at the RNA and protein levels confirmed the higher expression of FOXP1 in pre-GC immunoglobulin (Ig) D+ naïve B cells and post-GC CD27+ memory cells, whereas BCL6 was primarily expressed in CD71+ GC B cells (Figure 1C). FOXP1 has several alternatively spliced isoforms, and our immunoblot analysis in primary human cells (Figure 1C) confirmed the previous observation that whereas the smaller ∼65-kDa isoforms can be detected, the full-length FOXP1 protein of 75 kDa is the predominant isoform in normal tonsils.40
FOXP1 binds genes involved in the GC reaction, half of which are also transcriptionally regulated by BCL6
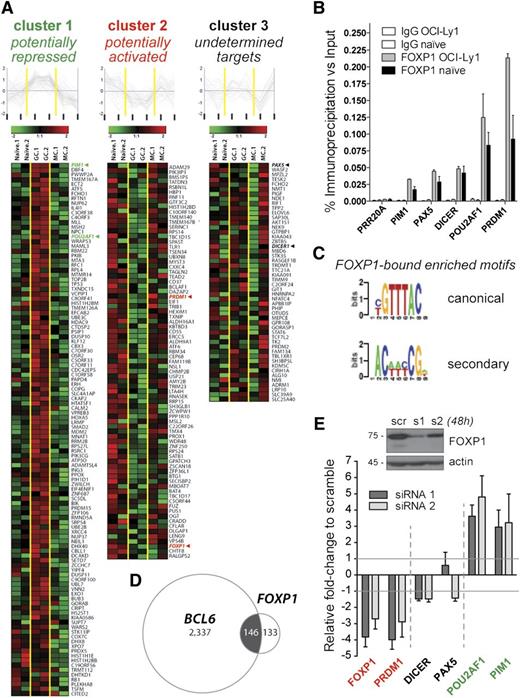
Little is known about transcriptional networks regulated by FOXP1 during the late stages of B-cell development. To identify the target genes of FOXP1 in GC-like cells, we performed ChIP-on-chip of FOXP1 in the human DLBCL cell line OCI-Ly1, which expresses high levels of FOXP1 (supplemental Figure 1A-B). By using a high-stringency overlapping log-ratio approach (see “Methods”), our computational analyses revealed a set of 279 putative FOXP1 target genes (Figure 2A and supplemental Table 1). FOXP1 binding to the promoters of 5 genes of interest (see rationale below) was validated by quantitative ChIP in OCI-Ly1 cells and further confirmed in primary human tonsillar CD19+IgD+ naïve cells (Figure 2B and supplemental Figure 1). The FIRE motif algorithm was used to identify DNA sequences enriched among putative FOXP1 targets in an unbiased manner32 and identified 2 enriched motifs (Figure 2C). One of these motifs corresponded to the canonical GTAAACA motif of the FOXP subfamily and the consensus motif for FOXP1 in human embryonic stem cells (hESCs).26 The second motif [AC(A/T)(A/T)(C/T)CG] did not correspond to known transcription factor binding sites. Notably, the consensus FOXP1 binding sequence and this novel secondary motif were located within FOXP1-bound promoters that were closer to each other than randomly expected (median distance, 205 bp; P = .008), suggesting that both DNA elements may cooperate in driving FOXP1 transcriptional activity. Given that there is an ESC-specific isoform of FOXP1 and not all of the 279 FOXP1-bound genes in B cells overlap with those in hESCs26 or neurons,41 it is reasonable to consider that FOXP1 or different isoforms may regulate distinct transcriptional networks in different cellular contexts.
Identification of a B-cell network of FOXP1 target genes. (A) Heatmaps and line charts representing the top-scoring FOXP1 target genes clustered with respect to their expression profiles in naïve (n = 2), GC (n = 2), and memory (n = 2) human B cells (columns). FOXP1 expression pattern (cluster 2) was included in the analysis for comparison purposes. (B) Validation of FOXP1 occupancy at representative target genes as measured by quantitative ChIP in the OCI-ly1 cell line and in tonsillar naïve B cells. Data are displayed as average fold enrichment relative to the input in 3 independent experiments. (C) FOXP1 canonical and secondary binding motifs detected by the FIRE motif discovery algorithm in our list of FOXP1-bound promoters. (D) Venn diagram showing overlapping between the FOXP1 and BCL6 target genes discovered by our ChIP-on-chip approaches. (E) Transcriptional changes in representative FOXP1 target genes after 48-hour siRNA-mediated knockdown of FOXP1 in OCI-ly1 cells, as evaluated by qRT-PCR in 3 independent experiments. Bars represent gene expression normalized to GAPDH and displayed as fold change relative to scramble-siRNA–treated samples. A representative western blot demonstrating a reduction of FOXP1 protein expression is shown on top.
Identification of a B-cell network of FOXP1 target genes. (A) Heatmaps and line charts representing the top-scoring FOXP1 target genes clustered with respect to their expression profiles in naïve (n = 2), GC (n = 2), and memory (n = 2) human B cells (columns). FOXP1 expression pattern (cluster 2) was included in the analysis for comparison purposes. (B) Validation of FOXP1 occupancy at representative target genes as measured by quantitative ChIP in the OCI-ly1 cell line and in tonsillar naïve B cells. Data are displayed as average fold enrichment relative to the input in 3 independent experiments. (C) FOXP1 canonical and secondary binding motifs detected by the FIRE motif discovery algorithm in our list of FOXP1-bound promoters. (D) Venn diagram showing overlapping between the FOXP1 and BCL6 target genes discovered by our ChIP-on-chip approaches. (E) Transcriptional changes in representative FOXP1 target genes after 48-hour siRNA-mediated knockdown of FOXP1 in OCI-ly1 cells, as evaluated by qRT-PCR in 3 independent experiments. Bars represent gene expression normalized to GAPDH and displayed as fold change relative to scramble-siRNA–treated samples. A representative western blot demonstrating a reduction of FOXP1 protein expression is shown on top.
Next, we sought to determine the functional classes of genes bound by FOXP1 using ingenuity pathway analysis. We found a strong enrichment of cellular pathways related to antigen presentation, cell-mediated immune response, humoral immune response, and cellular and cancer development (supplemental Table 2). Because BCL6 participates in similar pathways in normal B lymphocytes and in DLBCL cells4,30,31 and our results suggested antagonistic functions between BCL6 and FOXP1, we screened for overlapping target genes between both transcription factors by comparing the FOXP1-bound set of genes to those that we previously published for BCL6 using identical ChIP-on-chip assays.30 Among the 24 078 gene promoters analyzed in both ChIP-on-chip microarray experiments (accession number GSE44243), 146 out of the 279 FOXP1 target genes (50%) were also present within the 2483 BCL6 targets (P < 1e-52, hypergeometric test) (Figure 2D). Interestingly, a small set of those joint genes (8%) are known to be targeted by BCL6 in GC B cells but not in DLBCL30 (supplemental Table 1), suggesting a DLBCL-specific FOXP1 binding to certain normal BCL6 targets. Ingenuity pathway analysis of the 146 genes putatively regulated by both FOXP1 and BCL6 revealed enrichment of networks related to hematopoiesis and cancer development as well as genes involved in regulation of cell cycle, cellular growth and proliferation, and DNA replication, recombination, and repair (supplemental Tables 1 and 2). These results indicate that FOXP1 binds to genes involved in GC function and in B-cell lymphoma pathogenesis, many of which are also bound by BCL6. Although the number of FOXP1-bound genes in the OCI-Ly1 cell line is 10-fold lower than the number identified for BCL6,30 we cannot rule out that FOXP1 could bind to additional gene promoters and enhancers or intronic elements, even within nonpromoter regions of BCL6. Supporting this notion, a recent study of neuronal FOXP1 target genes identified a majority of FOXP1-binding sites within intronic regions.41 On the other hand, FOXP1 and BCL6 are rarely coexpressed during the normal GC reaction (Figure 1), and their DNA binding motifs were not significantly closer to each other at joint FOXP1/BCL6 target promoters than expected by chance (P ≥ .189, Wilcoxon test). Furthermore, motifs were not distributed differently in joint targets compared with nonoverlapping and unique targets (P ≥ .53, Wilcoxon test). Therefore, it is plausible that FOXP1 may exert its regulatory function partially in coordination with BCL6, binding to many of the same promoters. However, they are unlikely to bind cooperatively or as a complex to DNA and seem to be engaged in different temporal and spatial contexts.
FOXP1 may function both as activator and repressor of B-cell gene expression
We next explored the regulatory impact of FOXP1 on its target genes. We reasoned that if genes identified by ChIP-on-chip were transcriptionally regulated by FOXP1, coordinated expression between FOXP1 and its targets would be expected during the transition through the GC. To test this hypothesis, we analyzed gene expression profiles of naïve GC cells (centroblasts/centrocytes) and memory B lymphocyte subpopulations isolated from nontumoral human tonsils and compared the expression of FOXP1 and its 279 targets (Figure 2A, GSE25639). A supervised k-means clustering approach (K = 3) was used to explore the separation of the genes into clusters based on their overall expression pattern across naïve-GC memory compartments. Three distinct clusters could be observed, where a high number of candidate targets (∼75%) showed corresponding or opposing associations with FOXP1 expression. Forty-six percent of the FOXP1-bound genes presented opposing expression patterns with FOXP1, suggesting a potential repressor role of FOXP1 in the regulation of these genes (cluster 1 in Figure 2A). Conversely, the expression of 33% of the genes coincided with FOXP1 expression in naïve and/or memory cells but appeared downregulated in GC cells, suggesting potential transcriptional activation of these genes by FOXP1 (cluster 2 in Figure 2A). Finally, a number of FOXP1-bound genes (21%) did not clearly associate with FOXP1 expression, suggesting either sampling heterogeneity or a more complex interplay of transcription factors besides FOXP1 in the regulation of their expression. When the transcript abundance of FOXP1 target genes was examined in existing gene expression profile data sets derived from primary DLBCL cases42 (supplemental Figure 2 and Table 1), most of the genes exhibited a complex interrelationship with FOXP1 levels, while ∼25% of the targets exhibited a positive correlation. Interestingly, some genes switched from an inverse correlation in normal B cells to a positive correlation in DLBCL (supplemental Table 1). As shown in Figure 1C, the ratio of long and short FOXP1 isoforms varies between different populations, and this might also have some effect on FOXP1 target gene expression.
To further explore the transcriptional functions of FOXP1, we selected 5 biologically significant FOXP1 target genes involved in regulating B-cell differentiation (PAX5, PRDM1), B-cell receptor and CD40 lymphocyte signaling (POU2AF1), microRNA processing (DICER), and proto-oncogenic kinase activities during hematopoiesis (PIM1). Most of these genes have also been directly implicated in mature B-cell lymphomagenesis, either by presenting inactivating mutations (PRDM1) or by being targeted by chromosomal translocations (PAX5, POU2AF1, PIM1), genomic amplification (PIM1), or aberrant AID-mediated hypermutation (PIM1, PAX5, and POU2AF1).43,44 To determine its regulatory impact, we used 2 different siRNAs to knock down FOXP1 in OCI-Ly1 cells. FOXP1 knockdown resulted in the upregulation of the 2 potentially FOXP1-repressed genes PIM1 and POU2AF1 (cluster 1) as well as decreased expression of the potentially FOXP1-activated gene PRDM1 (cluster 2) (Figure 2E). Consistent with their assignment to cluster 3, PAX5 or DICER did not show significant differences in expression upon FOXP1 silencing. These results suggest that FOXP1 may exert dual functions as transcriptional activator and repressor in mature B-cell lymphocytes.
Expression of hFOXP1 in mouse B cells does not impair early lymphoid development
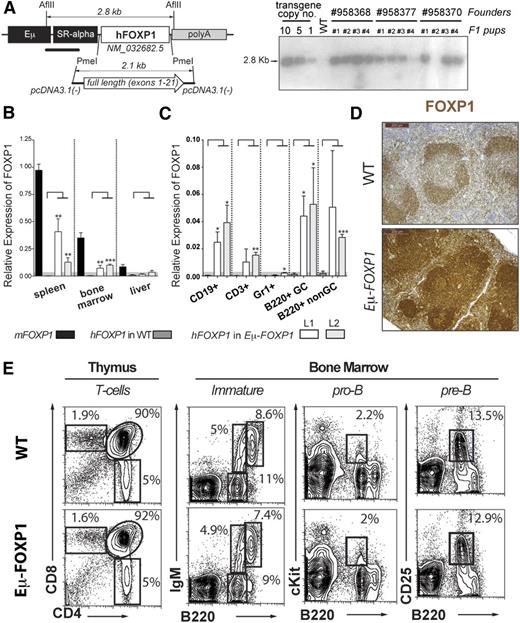
To further explore the function of FOXP1 in B lymphocyte regulation in vivo, we generated C57BL/6-Tg (EµSRα-hFOXP1) transgenic mice by using the immunoglobulin heavy chain enhancer Eµ45 to drive human FOXP1 expression in lymphoid cells (Figure 3A). For simplicity, we refer to these mice as Eµ-FOXP1 from now on. Human and mouse FOXP1 proteins are 96.1% identical and thus most likely share functional attributes. Among independent mouse lines carrying 5 to 10 copies of the human transgene (Figure 3A), 2 lines (L1 and L2) were expanded and characterized. Both Eµ-FOXP1 transgenic lines were obtained in Mendelian numbers, developed normally, and showed a comparable overall phenotype. In addition to endogenous murine FOXP1, ectopic expression of the human FOXP1 messenger RNA was detected in lymphoid organs of Eµ-FOXP1 transgenic mice, including the spleen and bone marrow (Figure 3B). More specifically, human FOXP1 transcripts were observed in splenic CD19+ or B220+ B cells (in both B220+Fas+GL7hi GC and B220+Fas−GL7− non-GC compartments from sheep red blood cell [SRBC]-immunized mice) as well as in thymic CD3+ T cells, but not in splenic Gr1+ granulocytes (Figure 3C). In comparison with wild-type (WT) mice, elevated overall expression of FOXP1 was detected in the splenic lymphoid follicles of Eµ-FOXP1 mice (Figure 3D). These studies show the specific expression of hFOXP1 in the lymphoid lineage of Eµ-FOXP1 mice, thus validating our model for the evaluation of the functional role of FOXP1 in B cells.
Characterization of Eμ-FOXP1 transgenic mice. (A) Illustration of the hFOXP1 transgene construct used for the generation of transgenic mice (left). Southern blot analysis on several transgenic lines to estimate transgene copy number (right). (B) Human and mouse FOXP1 messenger RNA levels were measured by qRT-PCR in spleen, bone marrow, and liver from unimmunized WT and Eμ-FOXP1 mice (n = 3 per mouse line and genotype) and presented relative to endogenous murine FOXP1 (2−ΔΔCt). Significance was determined by 2-tailed unpaired Student t tests. (C) Human FOXP1 expression by qRT-PCR in B CD19+, T CD3+, and granulocyte Gr1+ from unimmunized mice and GC B220+Fas+GL7hi and non-GC B220+Fas−GL7− cells isolated from SRBC-immunized WT and Eμ-FOXP1 mice (n = 3 per mouse line and genotype). Data are presented relative to GAPDH (2−ΔCt). (D) Representative IHC sections from WT and Eμ-FOXP1 mice (line L1) showing the distribution and expression of FOXP1 (brown, sum of human and murine levels) in splenic lymphoid follicles. (E) Representative fluorescence-activated cell sorter (FACS) profiling of the hematopoietic compartments in the thymus and bone marrow of 2-month-old unimmunized WT (n = 3) and Eμ-FOXP1 (n = 3) mice.
Characterization of Eμ-FOXP1 transgenic mice. (A) Illustration of the hFOXP1 transgene construct used for the generation of transgenic mice (left). Southern blot analysis on several transgenic lines to estimate transgene copy number (right). (B) Human and mouse FOXP1 messenger RNA levels were measured by qRT-PCR in spleen, bone marrow, and liver from unimmunized WT and Eμ-FOXP1 mice (n = 3 per mouse line and genotype) and presented relative to endogenous murine FOXP1 (2−ΔΔCt). Significance was determined by 2-tailed unpaired Student t tests. (C) Human FOXP1 expression by qRT-PCR in B CD19+, T CD3+, and granulocyte Gr1+ from unimmunized mice and GC B220+Fas+GL7hi and non-GC B220+Fas−GL7− cells isolated from SRBC-immunized WT and Eμ-FOXP1 mice (n = 3 per mouse line and genotype). Data are presented relative to GAPDH (2−ΔCt). (D) Representative IHC sections from WT and Eμ-FOXP1 mice (line L1) showing the distribution and expression of FOXP1 (brown, sum of human and murine levels) in splenic lymphoid follicles. (E) Representative fluorescence-activated cell sorter (FACS) profiling of the hematopoietic compartments in the thymus and bone marrow of 2-month-old unimmunized WT (n = 3) and Eμ-FOXP1 (n = 3) mice.
Deletion of FOXP1 is known to impair early B-cell development at the pro-B to pre–B-cell transition,8 but the impact of FOXP1 overexpression in the early and late stages of B-cell lymphopoiesis has not been explored. Therefore, we first used flow cytometry to assess the pro-B, pre-B, and immature B-cell populations in the bone marrow of Eµ-FOXP1 mice. A detailed characterization of young unimmunized Eµ-FOXP1 mice revealed largely normal B-cell populations in the bone marrow (Figure 3E). Although FOXP1 has been recently shown to enforce the quiescence of naïve T cells,23 the thymus of Eµ-FOXP1 mice showed normal CD4+ and CD8+ T-cell populations, suggesting no additional gain over the repressor functions that FOXP1 normally exhibits on T-cell proliferation.
Eµ-FOXP1 mice show unbalanced numbers of differentiated B-cell populations and modest impairment of GC formation
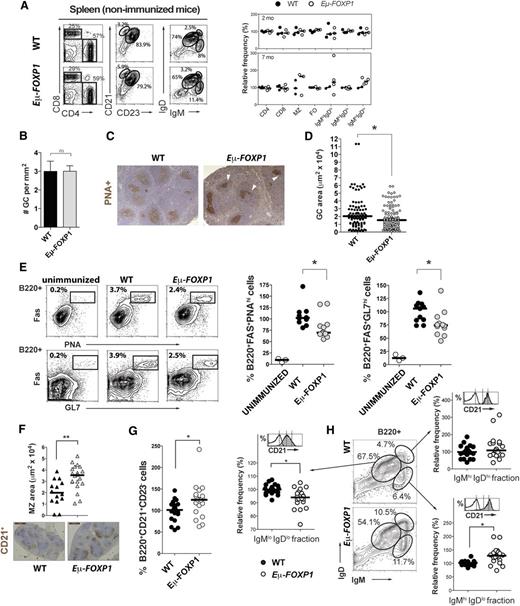
Given the association of FOXP1 overexpression with DLBCL and extranodal marginal zone (MZ)/MALT lymphoma12,16 and its transcriptional regulatory function in naïve B cells, we reasoned that hFOXP1 constitutive expression might disrupt the functions of mature B cells. Accordingly, expansion of mature B cells was detected after a modest upregulation of FOXP1 by knocking down miR-34a in mice.9 To explore in vivo the effect of increased FOXP1 expression in the late stages of B-cell development, cell populations and GC formation were examined by flow cytometry and IHC in WT and transgenic mice (Figure 4). There was no skewing in the mature populations of T or B cells in the spleen of young (2-month-old) unimmunized mice (Figure 4A). After a median follow-up of 12 months, our mice did not develop tumors or any other apparent disease. However, older (7-month-old) transgenic unimmunized mice evidenced tendencies toward a modest expansion of CD21+CD23− MZ cells and IgMhiIgDhi/lo mature B cells (Figure 4A right panel). In order to determine whether the differences in splenic B-cell populations were exacerbated during acute immune responses and whether GC development was impaired by FOXP1 expression, mice were immunized with SRBCs. Two weeks after priming and upon secondary immunization, the formation of a large number of PNA+ GCs was triggered in the spleen of both WT and transgenic mice (average of 2.996 vs 2.998 GCs per mm2, respectively; P = .99) (Figure 4B). However, GCs in transgenic mice showed slightly irregular shapes (Figure 4C) and were of smaller size in comparison with WT mice (median of 1537 vs 2041 μm2, respectively; P = .011) (Figure 4D). Furthermore, in mice expressing human FOXP1, the number of both B220+FAS+PNAhi and B220+FAS+GL7hi GC B cells was significantly reduced compared with controls (Figure 4E; P ≤ .018). Simultaneously, there was a tendency toward an expansion of the CD21+ regions outside the GCs (Figure 4F) and of CD21+CD23− MZ cells (Figure 4G; P = .0102). Furthermore, immunized Eµ-FOXP1 mice showed a significant expansion of splenic IgMhiIgDlo cells (Figure 4H), which is a heterogeneous fraction that may include memory, naïve transitional, and nonrecirculating MZ B cells.46 Indeed, this fraction was enriched in cells expressing high levels of the MZ surface marker CD21 (Figure 4H). On the other hand, the more mature IgMloIgDlo compartment of B cells appeared proportionally reduced. Normally, MZ cells constitute a distinct naïve B-cell lineage that is fully capable of initiating GC formation.47 Altogether, these results suggest a negative impact of FOXP1 during the expansion phase of GC development, where extended FOXP1 expression may alter the efficient differentiation program that promotes GC formation.
Impaired GC formation in Eμ-FOXP1 transgenic mice. (A) Analysis of the hematopoietic compartments in the spleen of unimmunized WT and Eμ-FOXP1 mice. Shown are representative FACS profiles (left) and a scatter plot summarizing the frequency of each cellular population relative to WT (right). Differences between young 2-month-old and older 7-month-old mice (circles) were assayed in 4 independent experiments. (B) The number of PNA+ GCs per mm2 of spleen (mean ± SEM) as measured by IHC from 2-month-old WT (n = 4) and Eμ-FOXP1 (n = 6) mice immunized with SRBCs. (C) Representative IHC staining with the proliferative marker PNA (brown) performed on splenic sections from SRBC-immunized mice. (D) Quantitative analysis of PNA+ surface area corresponding to 94 GCs from WT (n = 4) and 122 GCs from Eμ-FOXP1 (n = 6) 2-month-old immunized mice. (E) Shown are representative FACS profiles of splenic GC B cells after SRBC immunization (left) and the normalized percentage of GC B cells from WT and Eμ-FOXP1 mice (circles) assayed in 4 experiments (right). (F) IHC analysis of MZ CD21+ B cells (brown) in the spleen of WT and Eμ-FOXP1 SRBC-immunized mice (triangles). Shown are average size per mouse of the CD21+ area surrounding GCs (top) and representative IHC images (bottom). (G) Normalized FACS analysis of MZ CD21+CD23− B cells in the spleen from 2- to 8-month-old immunized WT and Eμ-FOXP1 mice (circles) assayed in 7 independent experiments. (H) Representative FACS profile and scatter plots of the normalized relative percentage of splenic B cells expressing different levels of surface IgM and IgD. WT and Eμ-FOXP1 2- to 8-month-old immunized mice (circles) were assayed in 4 independent experiments. Representative histograms of surface CD21 levels are shown for each fraction (in gray) compared with non-B B220− cells (in white). WT (solid lines) and transgenic (dotted lines) histograms were indistinguishable in terms of CD21 expression. The median is shown in all scatter plots. Significance was determined by nonparametric Mann-Whitney U tests.
Impaired GC formation in Eμ-FOXP1 transgenic mice. (A) Analysis of the hematopoietic compartments in the spleen of unimmunized WT and Eμ-FOXP1 mice. Shown are representative FACS profiles (left) and a scatter plot summarizing the frequency of each cellular population relative to WT (right). Differences between young 2-month-old and older 7-month-old mice (circles) were assayed in 4 independent experiments. (B) The number of PNA+ GCs per mm2 of spleen (mean ± SEM) as measured by IHC from 2-month-old WT (n = 4) and Eμ-FOXP1 (n = 6) mice immunized with SRBCs. (C) Representative IHC staining with the proliferative marker PNA (brown) performed on splenic sections from SRBC-immunized mice. (D) Quantitative analysis of PNA+ surface area corresponding to 94 GCs from WT (n = 4) and 122 GCs from Eμ-FOXP1 (n = 6) 2-month-old immunized mice. (E) Shown are representative FACS profiles of splenic GC B cells after SRBC immunization (left) and the normalized percentage of GC B cells from WT and Eμ-FOXP1 mice (circles) assayed in 4 experiments (right). (F) IHC analysis of MZ CD21+ B cells (brown) in the spleen of WT and Eμ-FOXP1 SRBC-immunized mice (triangles). Shown are average size per mouse of the CD21+ area surrounding GCs (top) and representative IHC images (bottom). (G) Normalized FACS analysis of MZ CD21+CD23− B cells in the spleen from 2- to 8-month-old immunized WT and Eμ-FOXP1 mice (circles) assayed in 7 independent experiments. (H) Representative FACS profile and scatter plots of the normalized relative percentage of splenic B cells expressing different levels of surface IgM and IgD. WT and Eμ-FOXP1 2- to 8-month-old immunized mice (circles) were assayed in 4 independent experiments. Representative histograms of surface CD21 levels are shown for each fraction (in gray) compared with non-B B220− cells (in white). WT (solid lines) and transgenic (dotted lines) histograms were indistinguishable in terms of CD21 expression. The median is shown in all scatter plots. Significance was determined by nonparametric Mann-Whitney U tests.
Eμ-FOXP1 transgenic mice exhibit in vivo functional B-cell GC responses
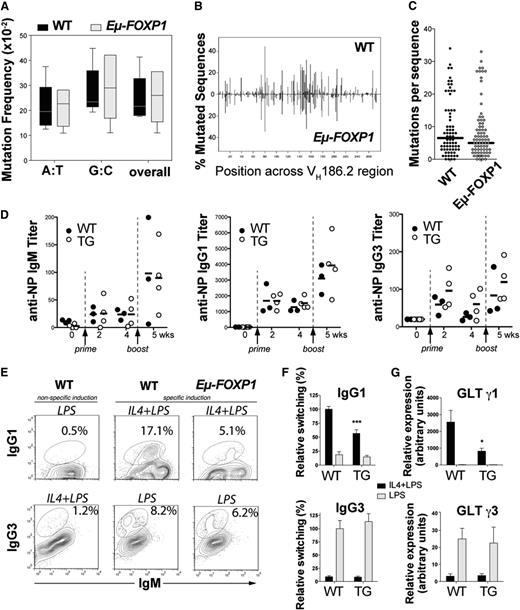
To determine whether there was a functional deficiency of GCs in addition to the effect on cell numbers, we examined the effect of FOXP1 on affinity maturation, which is associated with CSR and SHM of the immunoglobulin loci in GC B cells.48 WT and Eμ-FOXP1 transgenic mice were immunized with the T-dependent antigen 4-hydroxy-3-nitrophenyl-acetyl (NP)36-CGG (chicken γ globulin) in alum and boosted 4 weeks later to elicit secondary antibody responses. Analysis of the canonical NP-related VH186.2 gene indicated that constitutive expression of hFOXP1 in B cells does not have a major impact in the spectrum (Figure 5A) and distribution of SHM (Figure 5B), even though a nonsignificant tendency toward fewer mutations per sequence was observed in Eµ-FOXP1 mice (Figure 5C). The levels of serum anti-NP IgM, IgG1, and IgG3 antibodies were comparable between WT and transgenic mice during primary and secondary immune responses to NP immunization (Figure 5D). Therefore, our results suggest that the smaller GCs observed in immunized Eµ-FOXP1 mice can still support sustained rounds of SHM and CSR to produce normal titers of high-affinity anti-NP antibodies.
Abnormal GC function in Eμ-FOXP1 transgenic mice. (A) Frequencies of unique somatic mutations accumulated in splenic B cells at the VH186.2 region upon NP immunization of 3 WT and 3 transgenic mice. (B) Distribution of all mutations across the sequenced 273-bp-long VH186.2 region. (C) Number of mutations detected in each of the 72 sequences analyzed from WT or the 81 sequences analyzed from Eµ-FOXP1 mice. (D) Humoral immune response of WT (n = 6) and Eµ-FOXP1 (n = 7) mice to immunization with NP(36)-CGG at different times. Serum anti-NP titers and means are shown. (E) A representative FACS profile of ex vivo class switching from IgM to IgG1 and IgG3. Splenocytes from 2-month-old WT and Eµ-FOXP1 mice were stimulated for 72 hours with LPS or IL-4 plus LPS. The percentage of switched cells is indicated in each plot. (F) Bar graphs represent the normalized switching efficiencies of splenic B cells from WT (n = 5) and transgenic (n = 7) mice that were assayed in 4 different experiments. (G) GLTs expressed by WT (n = 4) and Eµ-FOXP1 (n = 7) splenocytes were assayed by qRT-PCR in 3 independent experiments after stimulation for 50 hours with LPS or IL-4 plus LPS, normalized to Igβ levels and represented relative of those of unstimulated WT cells. All bar graphs represent mean ± SEM. Significance was calculated using 2-tailed unpaired Student t tests. TG, transgenic mice; wks, weeks; WT, wild-type mice.
Abnormal GC function in Eμ-FOXP1 transgenic mice. (A) Frequencies of unique somatic mutations accumulated in splenic B cells at the VH186.2 region upon NP immunization of 3 WT and 3 transgenic mice. (B) Distribution of all mutations across the sequenced 273-bp-long VH186.2 region. (C) Number of mutations detected in each of the 72 sequences analyzed from WT or the 81 sequences analyzed from Eµ-FOXP1 mice. (D) Humoral immune response of WT (n = 6) and Eµ-FOXP1 (n = 7) mice to immunization with NP(36)-CGG at different times. Serum anti-NP titers and means are shown. (E) A representative FACS profile of ex vivo class switching from IgM to IgG1 and IgG3. Splenocytes from 2-month-old WT and Eµ-FOXP1 mice were stimulated for 72 hours with LPS or IL-4 plus LPS. The percentage of switched cells is indicated in each plot. (F) Bar graphs represent the normalized switching efficiencies of splenic B cells from WT (n = 5) and transgenic (n = 7) mice that were assayed in 4 different experiments. (G) GLTs expressed by WT (n = 4) and Eµ-FOXP1 (n = 7) splenocytes were assayed by qRT-PCR in 3 independent experiments after stimulation for 50 hours with LPS or IL-4 plus LPS, normalized to Igβ levels and represented relative of those of unstimulated WT cells. All bar graphs represent mean ± SEM. Significance was calculated using 2-tailed unpaired Student t tests. TG, transgenic mice; wks, weeks; WT, wild-type mice.
Abnormal ex vivo CSR in Eμ-FOXP1 splenic B cells
We further investigated ex vivo if hFOXP1 expression could directly impair the molecular mechanisms of isotype switching. Primary splenic B cells from Eµ-FOXP1 and WT mice were stimulated with lipopolysaccharide (LPS) to induce switching from IgM to IgG3 or with LPS plus interleukin 4 (IL-4) to induce switching from IgM to IgG1 (Figure 5E). Although the switching efficiencies to IgG3 were comparable to WT (P = .56), a significant decrease in switching to IgG1 was observed in Eµ-FOXP1 cells (P = .0008) (Figure 5F). Consistently, measurements by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) of noncoding GLTs, which promote switch regions accessibility and precede CSR,49 revealed a significant deficiency in GLTs for γ1 (P = .013) but not in γ3 GLTs expression (P = .84) (Figure 5G). Together, our results suggest that aberrant expression of FOXP1 can suppress γ1 transcription and switching to this isotype, suggesting that constitutive expression of FOXP1 at least partially impairs GC function.
Discussion
Expression of FOXP1 has been linked before to mature B-cell malignancy, but the role of FOXP1 during the late stages of B-cell development has not yet been explored. Here, we provide evidence for a deleterious role of FOXP1 expression in antigen-activated B cells and describe for the first time a FOXP1-bound network of genes involved in B-cell function. We show that FOXP1 is normally downregulated in mature GC B cells at the follicles of secondary lymphoid organs, showing an inverse correlation with BCL6. Consistently, FOXP1 was more abundant in pre-GC IgD+-naïve and CD79α+ B-cell areas surrounding the GCs. Thus, the inverse expression pattern of FOXP1 and BCL6 suggests that these proteins might play antagonistic roles in regulating the GC reaction. Indeed, we have identified a set of 279 FOXP1 target genes in a GC-like human lymphoma cell line, half of which have also been described before as BCL6 targets. Therefore, it is plausible that FOXP1 can impinge on some of BCL6 and GC functions, giving an explanation for their need to be physiologically engaged in different temporal and spatial contexts. On the contrary, this compartmentalization appears disrupted in some DLBCL lymphomas and cell lines such as OCI-Ly1. While care should be taken when comparing purified B-cell populations and heterogeneous DLBCL biopsies, we identified changes in the association of FOXP1 levels and its targets between normal and malignant cells, with a set of genes switching from a FOXP1-related repressed state to an activated one, supporting a potential role of FOXP1 deregulation in lymphomagenesis.
We further identified 2 enriched DNA motifs at FOXP1-bound gene promoters. We corroborated in B cells the preference for FOXP1 binding to the canonical GTAAACA motif, which had been ascribed before to the FOXP subfamily and was recently found in FOXP1-bound genes in hESCs.26 We also identified a second enriched motif [AC(A/T)(A/T)(C/T)CG] that might cooperate in FOXP1 function in B cells. Indeed, modulation of DNA recognition was shown to have profound consequences in FOXP1 functions controlling either pluripotency maintenance or differentiation transcriptional networks in ESCs.26 Furthermore, by exploring a few of our FOXP1 target genes in B cells, we show that FOXP1 may exert both activator and repressor functions. In agreement with these results, a repressor function of FOXP1 was previously shown in T cells,37 while activation functions were linked to FOXP1 activity during early B-cell development.8
To better understand the functional relevance of the downregulation of FOXP1 in GC B cells, we generated transgenic mice that constitutively express human FOXP1 throughout B-cell development. Using this transgenic model, we show that ectopic expression of hFOXP1 does not have a major impact in B-cell development up to the GC stage. However, these mice exhibited abnormal GC formation that resulted in a modest decrease of GC B lymphocytes (B220+Fas+PNAhi/GL7hi) and an increase of naïve and MZ B cells (B220+IgMhiCD21hi), suggesting that FOXP1 downregulation is required for appropriate transit of B cells through the GC. Interestingly, it has been shown that FOXP1 exerts a negative control of T-cell proliferation, preventing naïve T cells from gaining an effector phenotype.23 Therefore, it is plausible that FOXP1 might exhibit an analogous role in promoting quiescence of naïve B cells and hindering, but not completely blocking, the transition of these cells into proliferative GC cells. Furthermore, the ectopic expression of hFOXP1 in ex-vivo–activated B cells impaired the expression of γ1 GLTs at the switch regions and resulted in CSR deficiencies, supporting the notion that aberrant expression of FOXP1 can impact normal GC B-cell functions. Analogously, the B-cell transcription factor Ikaros has also been shown to specifically repress γ2a/γ2b GLTs, suppressing the accessibility of the AID enzyme to the switch regions and influencing the range of isotypes targeted by CSR.35 Our data also suggest that FOXP1 may have functions in post-GC B cells, for instance by transcriptionally activating PRDM1 and thus promoting plasmacytic differentiation. In the Eμ-FOXP1 mice, however, we did not observe increased numbers of plasmatic precursor cells or mature plasma cells (data not shown).
Altogether, our data support the notion that inappropriate expression of FOXP1 can hinder GC formation and function and, therefore, downregulation of FOXP1 is required during normal GC reactions. The functional overlap between FOXP1 and BCL6 provides for the first time a framework to understand their transcriptional crosstalk during physiological humoral responses and to delineate their impact in lymphomagenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs M. D. Scharff and S. N. Wontakal for the critical reading of this manuscript and Dr J. Adams (Walter and Eliza Hall Institute, Melbourne, Australia) for providing the EμSRα vector. This work was supported by grants from the Spanish Ministries of Science, Innovation and Health, Fondo de Investigaciones Sanitarias FIS-PI12/00202 and RD12/0036/0063 (J.A.M.-C.), and PI10/00109 (I.P.-R.), by grant by NCI-R01 CA104348, the Burroughs Wellcome Foundation, the Chemotheraphy and the Raymond and Beverley Sackler Center for Physical and Biomedical Sciences (A.M.), by the Incorpora-Torres Quevedo Program PTQ-11-04774 from the Spanish Ministry of Economy and Competitiveness (S.R.), by a Universidad Cardenal Herrera CEU Santander-Copernicus Program grant (I.P.-R.), and by postdoctoral fellowships from the Foundation for Applied Medical Research (A.S. and E.F.R.) and the Spanish Ministry of Health, Instituto de Salud Carlos III and Fondo de Investigaciones Sanitarias FIS (J.I.M.-F., M.A.A., and L.F.).
Authorship
Contribution: A.S., J.I.M.-F., and S.R. designed and performed research, analyzed and interpreted data, and wrote the manuscript; K.L.B., M.A.A., O.E., R.S., L.F., V.F., E.F.R., L.D.S., and X.S. performed research and revised the manuscript; I.P.-R. analyzed and interpreted data and revised the manuscript; and A.M. and J.A.M.-C. conceived the study, designed research, analyzed and interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jose A. Martinez-Climent, Division of Oncology, Center for Applied Medical Research, University of Navarra, Avda Pio XII, 55, 31008 Pamplona, Spain; e-mail: jamcliment@unav.es; and Ari Melnick, Division of Hematology/Oncology, Department of Medicine, Weill Cornell Medical College, 1300 York Ave, New York, NY 10065; e-mail: amm2014@med.cornell.edu.
References
Author notes
A.S., J.I.M.-F., and S.R. contributed equally to this study as first authors. A.M. and J.A.M.-C. contributed equally to this study as senior authors.