Key Points
High CRM1 expression was associated with short survival of AML patients.
CRM1 inhibitor KPT-185 induces apoptosis mainly in a p53-dependent manner, whereas inhibition of proliferation was p53 independent.
Chromosomal region maintenance 1 (CRM1) is a nuclear export receptor recognizing proteins bearing a leucine-rich nuclear export signal. CRM1 is involved in nuclear export of tumor suppressors such as p53. We investigated the prognostic significance of CRM1 in acute myeloid leukemia (AML) and effects of a novel small-molecule selective inhibitor of CRM1. CRM1 protein expression was determined in 511 newly diagnosed AML patients and was correlated with mouse double minute 2 (MDM2) and p53 levels. High CRM1 expression was associated with short survival of patients and remained an adverse prognostic factor in multivariate analysis. CRM1 inhibitor KPT-185 induced mainly full-length p53 and apoptosis in a p53-dependent manner, whereas inhibition of proliferation was p53 independent. Patient samples with p53 mutations showed low sensitivity to KPT-185. Nuclear retention of p53 induced by CRM1 inhibition synergized with increased levels of p53 induced by MDM2 inhibition in apoptosis induction. KPT-185 and Nutlin-3a, alone and in combination, induced synergistic apoptosis in patient-derived CD34+/CD38– AML, but not in normal progenitor cells. Data suggest that CRM1 exerts an antiapoptotic function and is highly prognostic in AML. We propose a novel combinatorial approach for the therapy of AML, aimed at maximal activation of p53-mediated apoptosis by concomitant MDM2 and CRM1 inhibition.
Introduction
The tumor suppressor p53 is activated in response to malignancy-associated stress signals and transcriptionally regulates genes involved in DNA repair, growth arrest, and apoptosis. Cellular mechanisms for the accumulation, stabilization, and deployment of p53 as a potent transcription factor have been postulated to be imperative for preventing the growth of abnormal or damaged cells.1 p53 loss can promote development of acute myeloid leukemia (AML).2,3
Cellular levels of p53 are critically regulated by mouse double minute 2 (MDM2). MDM2 is a p53-specific E3 ubiquitin ligase, which promotes p53 degradation. MDM2 is frequently overexpressed in AML.4,,-7 The selective MDM2 antagonist Nutlin-3a binds to MDM2 in the p53-binding pocket, disrupts MDM2-p53 interaction, and increases both nuclear and cytoplasmic p53 levels.7,-9 Nutlin-3a induces p53-mediated apoptosis in leukemia cells.7,9,,-12 The clinical analog RG7112 has been shown to activate p53 signaling and induce apoptosis and clinical responses in patients with hematologic malignancies.13,14
p53 is shuttled between the nucleus and the cytoplasm.1,15 Exportin 1 (chromosomal region maintenance 1 [CRM1]) is a member of nuclear export receptors recognizing proteins bearing a leucine-rich nuclear export signal.16 CRM1 is involved in nuclear export of a number of proteins including p53, p21, p27, p73, nucleophosmin-1 (NPM1), protein phosphatase 2, forkhead box protein O3, β-catenin/antigen-presenting cell, topoisomerase II, and nuclear factor κB/inhibitory nuclear factor κB.16,17 CRM1 overexpression has been associated with poor prognosis of solid cancers.18,,-21 CRM1 expression has not yet been investigated in AML. Karyopharm Therapeutics has developed novel, potent, and irreversible small-molecule selective inhibitors of CRM1, selective inhibitors of nuclear export (SINEs). SINEs selectively bind to Cys528 of CRM1, thereby inhibiting CRM1 binding to its target proteins.22 SINEs have been shown to induce apoptosis and block proliferation in malignant cell lines, including pancreas, colon, and breast cancer, as well as leukemias.22,-24 SINEs have shown minimal toxicities in normal human cells including hematopoietic cells.22,-24
Methods
Reagents
The selective CRM1 inhibitor KPT-185 and its inactive trans-enantiomer KPT-301 were synthesized at Karyopharm (Natick, MA). The selective small-molecule antagonist of MDM2, Nutlin-3a,8 was purchased from Cayman Chemical Company (Ann Arbor, MI).
Cell culture
Cells were cultured in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum. OCI-AML-3, MOLM-13, and MV4;11 have wild-type p53; whereas NB4, HL-60, KG-1, THP-1, and U937 have defective (deleted or missense mutated) p53.7,25 Cell lines were harvested in log-phase growth, seeded at a density of 2 × 105 cells per mL and exposed to compounds. Heparinized peripheral blood samples were obtained from AML patients after informed consent, according to institutional guidelines per the Declaration of Helsinki. Human umbilical cord blood was collected according to institutional guidelines. Mononuclear cells were purified by density-gradient centrifugation, and nonadherent cells were resuspended at a density of 1 × 106 cells per mL. Cell viability was evaluated by triplicate counts of trypan blue dye-excluding cells.
Apoptosis analysis
Evaluation of apoptosis by the annexin V–propidium iodide binding assay was performed.7 The extent of apoptosis was quantified as percentage of annexin V–positive cells, and the extent of drug-specific apoptosis was assessed by the following formula: % specific apoptosis = (test – control) × 100 / (100 – control).9
Western blot analysis
Western blot analysis was performed using the Odyssey imaging system (LI-COR Biosciences, Lincoln, NE). The following antibodies were used: mouse monoclonal anti-p53 (DO-1; Santa Cruz Biotechnology, Santa Cruz, CA); rabbit polyclonal anti-p53 (FL-393; Santa Cruz Biotechnology); sheep polyclonal anti-p53 (SAPU; Enzo Life Sciences, Ann Arbor, MI); rabbit polyclonal anti-CRM1 (Santa Cruz Biotechnology); mouse monoclonal anti-p21 (EMD Biosciences, San Diego, CA); mouse monoclonal anti-MDM2 (Santa Cruz Biotechnology); and mouse monoclonal anti–β-actin (Sigma Chemical Co., St. Louis, MO).
Quantitation of intracellular p53 protein by flow cytometry
Cells were fixed and permeabilized with 100% methanol, incubated with fluorescein isothiocyanate–conjugated antibody against p53 or its isotype control (BD Biosciences, San Jose, CA), and analyzed.9
Immunofluorescence and confocal microscopy
Cells were fixed and permeabilized with 100% methanol and incubated with rabbit polyclonal anti-p53 (Santa Cruz Biotechnology) and mouse monoclonal anti–cytochrome c oxidase IV (Molecular Probes, Eugene, OR), followed by Alexa Fluor 488 and 647 secondary antibodies (Molecular Probes). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole. Images were acquired using an Olympus DSU spinning disk confocal microscope.
Gene expression analysis
Messenger RNA expression levels were quantified using TaqMan gene expression assays (TP53I3, Hs00153280_m1; GDF15, Hs00171132_m1; MDM2, Hs00242813_m1; PUMA, Hs00248075_m1; ZMAT3, Hs00536976_m1; p21, Hs00355782_m1; GAPDH, Hs99999905_m1; Applied Biosystems, Foster City, CA) on a 7900HT Fast Real-Time PCR System. Relative quantification between different samples was determined according to the 2–ΔΔCt.
Reverse-phase protein array
Peripheral blood and marrow specimens were collected from 511 patients with newly diagnosed AML evaluated at The University of Texas M.D. Anderson Cancer Center. Normal marrow CD34+ cells from healthy donors aged 18 to 55 were obtained from AllCells (Emeryville, CA). The demographics and clinical characteristics of the patient population have been previously reported.26,27 The methodology and validation of the proteomic profiling technique have been described previously.27,28 A strictly validated CRM1 antibody (H-300, Santa Cruz Biotechnology) was used. The same Reverse-protein protein array (RPPA) was also probed with antibodies against 207 other proteins (listed in supplemental Table 1; see the Blood Web site), as part of a broader proteomic profiling study.
Mutation analysis
Statistical analysis
The statistical analysis was performed using the 2-tailed Student t test. Statistical significance was considered when P < .05. Unless otherwise indicated, average values were expressed as mean ± standard deviation (SD). Synergism, additive effects, and antagonism were assessed as previously described. The combination index (CI), a numerical description of combinatorial effects, was calculated using the more stringent statistical assumption of mutually nonexclusive modes of action. When CI = 1, this equation represents the conservation isobologram and indicates additive effects. CI values <1.0 indicate a more than expected additive effect (synergism), whereas CI values >1.0 indicate antagonism.30
Statistical analysis of RPPA data was performed as previously described.27 Comparison of CRM1 levels between paired samples was done using the paired t test. Associations between CRM1 levels and categorical clinical variables were assessed in the R software program (Version 2.8.0), using standard t tests, linear regression, or mixed-effects linear models. Associations between the protein level and continuous variables were assessed using Pearson and Spearman correlation and linear regression analysis. For correlation of CRM1 expression with that of other proteins, we accounted for multiple testing using a Bonferroni correction and accepted any proteins with a Pearson correlation |R| ≥ 0.2 and P < .0001. The Kaplan-Meier method was used to generate survival curves. A Cox proportional hazards regression model was used to investigate association with survival with protein levels as categorized variables using Statistica Version 10 software (StatSoft, Tulsa, OK). Patients with missing data were excluded from the analysis. Samples were acquired during routine diagnostic assessments in accordance with the regulations and protocols (Laboratory 01-473) approved by the investigational review board of The University of Texas M.D. Anderson Cancer Center. Informed consent was obtained in accordance with the Declaration of Helsinki. Samples were analyzed under a review board–approved laboratory protocol (Laboratory 05-0654).
Results
CRM1 expression has prognostic impact on AML
CRM1 expression was investigated in patient AML samples using an RPPA. CRM1 was variably expressed over a 32-fold range in samples from patients with AML. The CRM1 levels were not significantly different between AML and normal bone marrow–derived CD34+ cells (P = .53; 511 AML and 21 normal samples) (supplemental Figure 1A), although 20.5% of new AML cases showed expression above and 12% below that of normal CD34+ cells. In paired samples, CRM1 expression levels were not significantly different between peripheral blood and marrow AML cells (P = .12, n = 140). CRM1 levels were not significantly affected by age (r = 0) or sex (P = .23). Higher levels of CRM1 were associated with higher marrow blast percentages (r = 0.20, P < .00001), white cell counts (r = 0.12, P = .0079), peripheral blood blast percentages (r = 0.21, P < .00001), and absolute peripheral blood blast count (r = 0.17, P = .0002). Expression was lower in those with favorable cytogenetics compared with those with intermediate or unfavorable cytogenetics (P = .029). CRM1 levels were higher in patients with Fms-like tyrosine kinase-3 (FLT3) mutations (P = .003; supplemental Figure 1B) and marginally higher in patients with NPM1 mutations (P = .06; supplemental Figure 1B). As part of a broader proteomic profiling study of AML, this same RPPA was also probed with antibodies against 207 other proteins (supplemental Table 1); of these, 29 proteins were found to be positively (n = 22) or negatively (n = 7) correlated with CRM1 levels (|R| ≥ 0.2, P < .0001) (supplemental Figure 2). Several proteins that showed correlation with CRM1 expression are components of protein kinase B (AKT/PKB) signaling, including AKT, its upstream phosphatidylinositol-3 kinase p85 (PI3Kp85) and phospho-phosphatase and tensin homolog (phospho-PTEN), and downstream phospho-BCL2-associated agonist of cell death (phospho-BAD) (Ser112, Ser136) and 14-3-3 ζ.
We then investigated the correlation of CRM1 level with p53 and MDM2 levels. In 2-way correlations, MDM2 and p53 levels were inversely correlated (r = −0.17, P = .00007), but CRM1 levels did not correlate with MDM2 levels (r = −0.002, P = .96) or with p53 levels (r = −0.047, P = .29). However in 3-way correlation (using distance weighted least squares), there was a clear interaction, with p53 levels being highest when CRM1 was high and MDM2 levels were low (Figure 1A), raising the possibility CRM1 functions as a complementary mechanism of p53 suppression to MDM2. Overall survival progressively worsened as CRM1 levels increased, with median survival of 66 weeks for those with CRM1 expression in the lowest third, 47 weeks for the middle third, and 37 weeks in the highest third (Figure 1B); there was a statistically significant difference among these groups (P = .007). Similar trends were seen among patients with unfavorable cytogenetics (median survival in low, middle, and high thirds of 41, 39, and 24 weeks, respectively; P = .06). CRM1 did not affect remission duration (P = .33). We also investigated if CRM1 expression is associated with time to remission attainment. CRM1 expression levels were not significantly different among 3 groups with different times to complete remission (<46 days, 46–90 days, or >90 days) (P = .17). A Cox proportional hazards model was performed evaluating for factors that were independent predictors of overall survival. Starting with 13 variables that were univariate predictors in the data set, a stepwise analysis was conducted until only significant (P < .05) variables remained. This was followed by sequential add back of all previously removed variables one by one until a final model with only significant variables remained. The final model contained 6 variables: age, cytogenetics (ie, favorable and unfavorable), white blood cell count, serum albumin level, and high CRM1 level, as shown in Table 1. High CRM1 levels were determined to be an independent predictor of overall survival in both univariate and multivariate analysis.
Increased CRM1 expression predicts for poor survival in AML patients. (A) Three-dimensional surface plot of p53 against CRM1 and MDM2. There is a relationship between the 3 components (P = .0002), and the contour blots, projections of a three-dimensional surface onto a two-dimensional plane, demonstrate that CRM1 (positive correlation) and MDM2 level (negative correlation) interactively modulate p53 levels. The color scale refers to the expression (on the log 2 scale like the rest of the data) of the z-axis component (ie, p53). The scale of p53 ranges from <−0.5 to +2.5. The plot was generated using Statistica software. (B) Kaplan-Meier curves and results of multivariate analysis for overall survival in patients with AML. The final model of multivariate analysis includes 6 variables, and high CRM1 expression is an independent predictor of overall survival in AML.
Increased CRM1 expression predicts for poor survival in AML patients. (A) Three-dimensional surface plot of p53 against CRM1 and MDM2. There is a relationship between the 3 components (P = .0002), and the contour blots, projections of a three-dimensional surface onto a two-dimensional plane, demonstrate that CRM1 (positive correlation) and MDM2 level (negative correlation) interactively modulate p53 levels. The color scale refers to the expression (on the log 2 scale like the rest of the data) of the z-axis component (ie, p53). The scale of p53 ranges from <−0.5 to +2.5. The plot was generated using Statistica software. (B) Kaplan-Meier curves and results of multivariate analysis for overall survival in patients with AML. The final model of multivariate analysis includes 6 variables, and high CRM1 expression is an independent predictor of overall survival in AML.
Multivariate analysis for overall survival (n = 415)
| Variables . | Parameter estimate . | Standard error . | χ2 . | P value . | 95% Lower CL . | 95% Upper CL . | Hazard ratio . | 95% Hazard ratio lower CL . | 95% Hazard ratio upper CL . |
|---|---|---|---|---|---|---|---|---|---|
| Age at diagnosis | 0.039136 | 0.004583 | 72.93462 | .000000 | 0.03015 | 0.048117 | 1.039912 | 1.030613 | 1.049294 |
| Albumin | –0.231112 | 0.080496 | 8.24322 | .004090 | –0.38888 | –0.073343 | 0.793651 | 0.677815 | 0.929282 |
| White blood cell count | 0.005187 | 0.001211 | 18.35753 | .000018 | 0.00281 | 0.007559 | 1.005200 | 1.002818 | 1.007588 |
| Favorable cytogenetics | –0.706299 | 0.336488 | 4.40593 | .035814 | –1.36580 | –0.046794 | 0.493467 | 0.255176 | 0.954284 |
| Unfavorable cytogenetics | 0.612691 | 0.117951 | 26.98225 | .000000 | 0.38151 | 0.843871 | 1.845390 | 1.464495 | 2.325351 |
| High CRM1 | 0.237605 | 0.117512 | 4.08833 | .043180 | 0.00729 | 0.467925 | 1.268209 | 1.007312 | 1.596678 |
| Variables . | Parameter estimate . | Standard error . | χ2 . | P value . | 95% Lower CL . | 95% Upper CL . | Hazard ratio . | 95% Hazard ratio lower CL . | 95% Hazard ratio upper CL . |
|---|---|---|---|---|---|---|---|---|---|
| Age at diagnosis | 0.039136 | 0.004583 | 72.93462 | .000000 | 0.03015 | 0.048117 | 1.039912 | 1.030613 | 1.049294 |
| Albumin | –0.231112 | 0.080496 | 8.24322 | .004090 | –0.38888 | –0.073343 | 0.793651 | 0.677815 | 0.929282 |
| White blood cell count | 0.005187 | 0.001211 | 18.35753 | .000018 | 0.00281 | 0.007559 | 1.005200 | 1.002818 | 1.007588 |
| Favorable cytogenetics | –0.706299 | 0.336488 | 4.40593 | .035814 | –1.36580 | –0.046794 | 0.493467 | 0.255176 | 0.954284 |
| Unfavorable cytogenetics | 0.612691 | 0.117951 | 26.98225 | .000000 | 0.38151 | 0.843871 | 1.845390 | 1.464495 | 2.325351 |
| High CRM1 | 0.237605 | 0.117512 | 4.08833 | .043180 | 0.00729 | 0.467925 | 1.268209 | 1.007312 | 1.596678 |
CL, confidence limit.
p53 is a major determinant of CRM1 inhibition-induced apoptosis in AML
We examined the effect of the CRM1 inhibitor KPT-185 on the viability of AML cell lines. Apoptosis induction by KPT-185 was much more prominent in p53 wild-type cells than in p53-defective cells. ED50 values (effective concentration inducing 50% killing as measured by annexin V positivity) were 150, 90, and 85 nM in p53 wild-type OCI-AML, MOLM-13, and MV4;11 cells; whereas they were >1000 nM in 4 of the 5 p53 mutant cell lines (Figure 2A). KPT-301, an inactive trans-enantiomer of KPT-185, did not exhibit antiproliferative or apoptosis-inducing activities, confirming the selective inhibition of CRM1 by KPT-185 (data not shown). The selective MDM2 inhibitor Nutlin-3a served as positive control for wild-type p53-mediated apoptosis induction (Figure 2A, white bars).8,31 We also investigated the effect of KPT-185 on cell proliferation. IC50 values (concentration at which cell growth is inhibited by 50%) of KPT-185 for wild-type p53 cell lines ranged from 27 to 38 nM, only modestly lower than the range for p53 mutant cell lines (48-112 nM) (Figure 2A), suggesting that KPT-185 potently inhibits cell growth of AML cells, irrespective of p53 functional status.
The CRM1 inhibitor KPT-185 inhibits the growth of AML cell lines through cell cycle arrest and apoptosis induction. (A) AML cell lines with wild-type p53 (OCI-AML-3, MOLM-13, and MV4;11) or mutant p53 (NB4, HL-60, KG-1, THP-1, and U937) cells were incubated with 100, 200, or 500 nM for 72 hours, and the annexin V–positive fractions were measured by flow cytometry (black bars). Cells were treated in parallel with 5 µM Nutlin-3a (white bars). Results are expressed as mean ± SD. (B) Lentivirally transduced wild-type p53 AML cells (virus encoding either control shRNA [shC] or p53-specific shRNA [shp53]) were incubated with 0, 10, 20, 50, 100, 200, 500, or 1000 nM of KPT-185 for 72 hours, and the numbers of viable cells and annexin V–positive fractions were measured. *P < .05; **P < .01; ***P < .001.
The CRM1 inhibitor KPT-185 inhibits the growth of AML cell lines through cell cycle arrest and apoptosis induction. (A) AML cell lines with wild-type p53 (OCI-AML-3, MOLM-13, and MV4;11) or mutant p53 (NB4, HL-60, KG-1, THP-1, and U937) cells were incubated with 100, 200, or 500 nM for 72 hours, and the annexin V–positive fractions were measured by flow cytometry (black bars). Cells were treated in parallel with 5 µM Nutlin-3a (white bars). Results are expressed as mean ± SD. (B) Lentivirally transduced wild-type p53 AML cells (virus encoding either control shRNA [shC] or p53-specific shRNA [shp53]) were incubated with 0, 10, 20, 50, 100, 200, 500, or 1000 nM of KPT-185 for 72 hours, and the numbers of viable cells and annexin V–positive fractions were measured. *P < .05; **P < .01; ***P < .001.
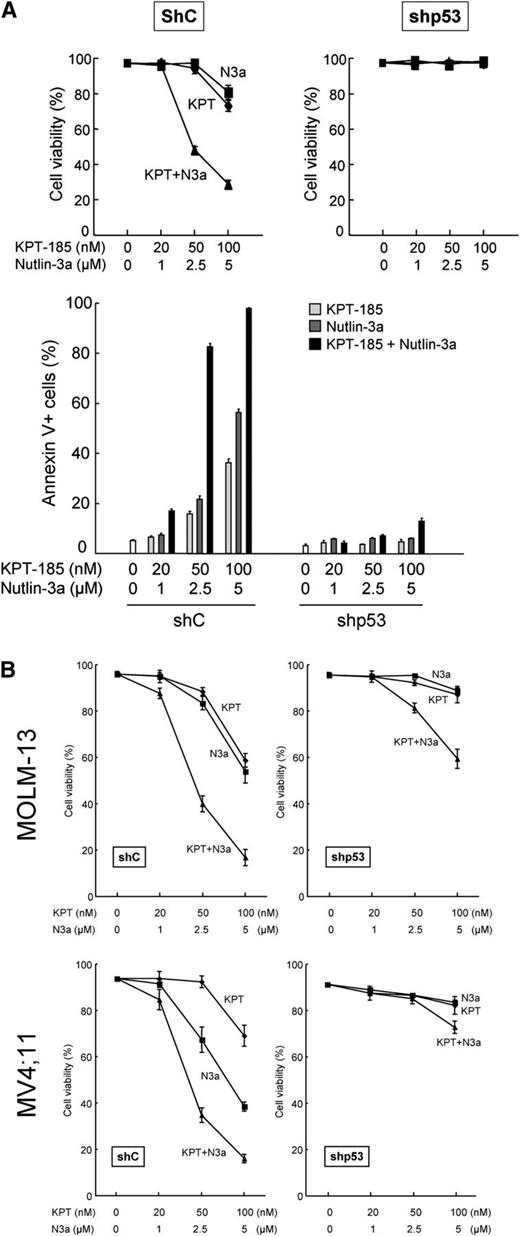
To further define the observed cell growth inhibition and apoptosis induced by KPT-185, we investigated the effect of stable p53 knockdown in 3 p53 wild-type AML lines. p53-specific short hairpin RNA (shRNA) reduced p53 basal levels by 90% to 95% but did not significantly affect CRM1 expression (supplemental Figure 3). At lower doses (≤20 nM) of KPT-185 in all lines, the proportion of viable cells remained high, but cell numbers were substantially decreased (by ∼40% at 20 nM) (Figure 2B, upper and middle panels). This cytostatic effect was p53 independent, in that there was little difference between cell numbers with scrambled or p53-specific shRNA. At higher doses, KPT-185 decreased cell viability and increased the percentage of annexin V–binding cells, which was less prominent in p53 knockdown cells than in control cells (Figure 2B, all panels). These data indicate that the KPT-185 depends on p53 for apoptosis induction, but not for cytostatic effects.
KPT-185 induces p53 response in AML
Inhibition of CRM1 is expected to accumulate p53 in the nucleus. Because it has been reported that p53 degradation occurs most efficiently on cytoplasmic proteasomes,32 nuclear retention of p53 may result in increased cellular levels. CRM1 inhibition by KPT-185 increased cellular p53 protein levels (Figure 3) and activated p53 target genes TP53I3, GDF15, MDM2, PUMA, ZMAT3, and p21 (data not shown). KPT-185 treatment did not affect p53 messenger RNA levels, even at 1000 nM, excluding the possibility that CRM1 inhibition leads to gene induction of TP53 (data not shown).
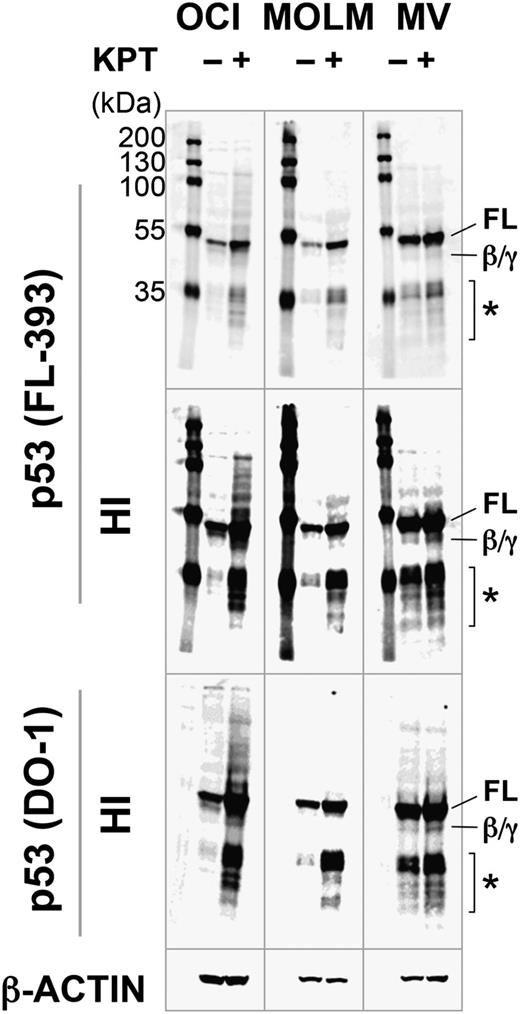
KPT-185 induces full-length p53. OCI-AML-3 (OCI), MOLM-13 (MOLM), and MV4;11 (MV) cells were incubated with 100 nM KPT-185 for 10 hours, and expression of p53 isoforms was determined. KPT-185 increased the levels of full-length p53 (FL) in all cell lines. p53β/γ (β/γ) was barely detected in KPT-treated OCI-AML-3 and MV4;11 cells. Asterisk (*) indicates p53 fragments from full-length p53 or p53β/γ isoforms. Results are representative of 3 independent experiments. HI, high-intensity image.
KPT-185 induces full-length p53. OCI-AML-3 (OCI), MOLM-13 (MOLM), and MV4;11 (MV) cells were incubated with 100 nM KPT-185 for 10 hours, and expression of p53 isoforms was determined. KPT-185 increased the levels of full-length p53 (FL) in all cell lines. p53β/γ (β/γ) was barely detected in KPT-treated OCI-AML-3 and MV4;11 cells. Asterisk (*) indicates p53 fragments from full-length p53 or p53β/γ isoforms. Results are representative of 3 independent experiments. HI, high-intensity image.
We next investigated if KPT-185 increases the levels of specific p53 isoforms in p53 wild-type AML lines. The rabbit polyclonal FL-393 and sheep polyclonal SAPU antibodies recognize all 9 p53 isoforms (full-length p53 [FLp53], p53β/γ, Δ133p53α/β/γ, Δ40p53α/β/γ), and the mouse monoclonal DO-1 antibody detects FLp53, p53β, and p53γ.33 It has been reported that AML cells express FLp53 and p53β/γ.34 In addition to FLp53, FL-393 and DO-1 detected p53β/γ expression in OCI-AML-3 and MV4;11 cells after KPT-185 treatment (Figure 3). Induction of p53β/γ expression was much less prominent than that of FLp53. MOLM-13 cells exclusively expressed FLp53. SAPU did not sufficiently detect p53 isoforms except for FLp53. None of the cells expressed ΔNp53 isoforms (Δ133p53 and Δ40p53). These data suggest that the inhibition of CRM1 promotes the induction of FLp53.
KPT-185 synergizes with the MDM2 inhibitor Nutlin-3a to induce p53 and apoptosis in AML
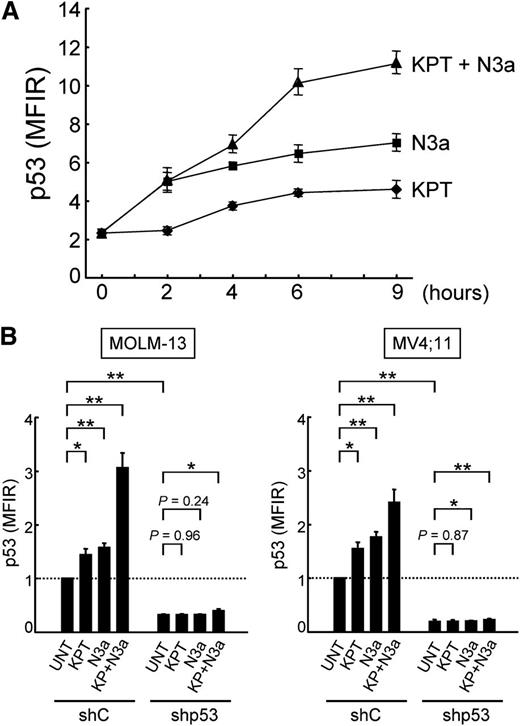
Nutlin-3a increases total cellular levels of p53 in both nucleus and cytoplasm,7,9 whereas CRM1 inhibition restores the nuclear function of p53 by relocation of p53 to the nucleus. Because the mode of action of CRM1 inhibitors is distinct from that of MDM2 inhibitors, we investigated if the KPT-185/Nutlin-3a combination synergistically induces profound p53 response and cell death in AML cells. Treatment of OCI-AML-3 cells with KPT-185 or Nutlin-3a caused a time-dependent increase in p53 levels (Figure 4A). The KPT-185/Nutlin-3a combination resulted in more pronounced p53 induction than the individual agents. Increased p53 induction by the combination was also observed in MOLM-13 and MV4;11 cells (Figure 4B). To investigate if the synergistic p53 induction of p53 by KPT-185 and Nutlin-3a leads to synergistic apoptosis induction, OCI-AML-3 cells expressing scrambled or p53-specific shRNA were treated with KPT-185 and Nutlin-3a, either as individual agents or in combination. As shown in Figure 5A, the synergistic cell death effect was dependent on functional p53 status. This p53-dependent synergistic cell death effect was also found in MOLM-13 and MV4;11 cells (Figure 5B).
KPT-185 and Nutlin-3a synergistically induce p53. (A) OCI-AML-3 cells were treated with 50 nM KPT-185 and/or 2.5 µM Nutlin-3a. KPT-185 synergizes with Nutlin-3a to induce p53. p53 expression levels were expressed as mean fluorescence intensity ratio (MFIR) calculated by the following formula: MFIR = (MFI for anti-p53 antibody)/(MFI for isotypic control). (B) MOLM-13 and MV4;11 cells expressing control shRNA (shC) or p53-specific shRNA (shp53) were treated with 50 nM KPT-185 and/or 2.5 µM Nutlin-3a for 6 hours. Results are expressed as fold change (mean ± SD) relative to the MFIR value in untreated negative control shRNA-expressing cells. Similar results were obtained in 2 other independent experiments. *P < .05; **P < .01; ***P < .001.
KPT-185 and Nutlin-3a synergistically induce p53. (A) OCI-AML-3 cells were treated with 50 nM KPT-185 and/or 2.5 µM Nutlin-3a. KPT-185 synergizes with Nutlin-3a to induce p53. p53 expression levels were expressed as mean fluorescence intensity ratio (MFIR) calculated by the following formula: MFIR = (MFI for anti-p53 antibody)/(MFI for isotypic control). (B) MOLM-13 and MV4;11 cells expressing control shRNA (shC) or p53-specific shRNA (shp53) were treated with 50 nM KPT-185 and/or 2.5 µM Nutlin-3a for 6 hours. Results are expressed as fold change (mean ± SD) relative to the MFIR value in untreated negative control shRNA-expressing cells. Similar results were obtained in 2 other independent experiments. *P < .05; **P < .01; ***P < .001.
The KPT-185/Nutlin-3a combination induces p53-mediated cell death. (A) OCI-AML-3 cells expressing control shRNA (shC) or p53-specific shRNA (shp53) were treated with the indicated concentrations of KPT-185 or Nutlin-3a for 72 hours, either as individual agents or in combination. Cell viability was determined by trypan blue exclusion method. Annexin V–positive fractions were measured by flow cytometry. (B) MOLM-13 and MV4;11 cells expressing control shRNA (shC) or p53-specific shRNA (shp53) were treated with the indicated concentrations of KPT-185 or Nutlin-3a for 72 hours, either as individual agents or in combination. Cell viability was determined by trypan blue exclusion method. Comparable results were obtained in 2 other independent experiments.
The KPT-185/Nutlin-3a combination induces p53-mediated cell death. (A) OCI-AML-3 cells expressing control shRNA (shC) or p53-specific shRNA (shp53) were treated with the indicated concentrations of KPT-185 or Nutlin-3a for 72 hours, either as individual agents or in combination. Cell viability was determined by trypan blue exclusion method. Annexin V–positive fractions were measured by flow cytometry. (B) MOLM-13 and MV4;11 cells expressing control shRNA (shC) or p53-specific shRNA (shp53) were treated with the indicated concentrations of KPT-185 or Nutlin-3a for 72 hours, either as individual agents or in combination. Cell viability was determined by trypan blue exclusion method. Comparable results were obtained in 2 other independent experiments.
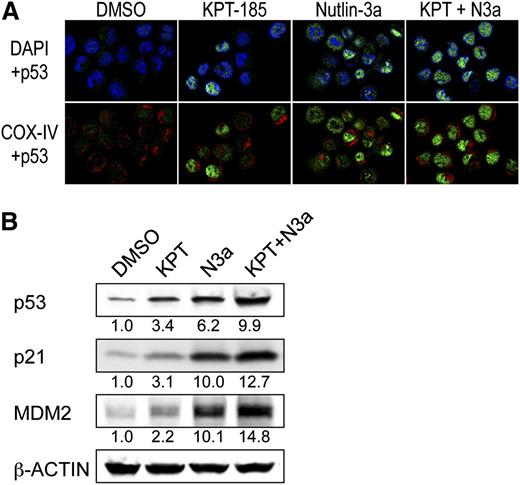
Intracellular localization of p53 was determined using confocal microscopy in OCI-AML-3 cells that have been shown to preferentially express cytoplasmic p53.7 p53 was barely detected in control cells (Figure 6A). After KPT-185 treatment, ∼70% of the cells showed nuclear accumulation of p53. Nutlin-3a induced cytoplasmic, cytoplasmic and nuclear, or nuclear accumulation of p53 in individual cells. When they were combined, almost all cells showed intense nuclear accumulation of p53. In accordance with the synergistic induction of nuclear p53 (Figures 5A and 6A), the KPT-185/Nutlin-3a combination induced profound p53 responses (Figure 6B).
The KPT-185/Nutlin-3a combination enhances nuclear function of p53. (A) The KPT-185/Nutlin-3a combination accumulates p53 into the nucleus of AML cells. OCI-AML-3 cells were treated for 6 hours with 100 nM KPT-185 and 2.5 µM Nutlin-3a. Cells were stained for p53 (green) and mitochondrial marker protein cytochrome c oxidase IV (COX-IV, red) and visualized by confocal microscopy. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole(blue). (B) Expression of p53 target proteins in OCI-AML-3 cells, which were treated with 50 nM KPT-185 and/or 2.5 µM Nutlin-3a for 12 hours. Intensity of the immunoblot signals was quantified, and the relative intensity compared with β-actin was calculated. Results are representative of 3 independent experiments.
The KPT-185/Nutlin-3a combination enhances nuclear function of p53. (A) The KPT-185/Nutlin-3a combination accumulates p53 into the nucleus of AML cells. OCI-AML-3 cells were treated for 6 hours with 100 nM KPT-185 and 2.5 µM Nutlin-3a. Cells were stained for p53 (green) and mitochondrial marker protein cytochrome c oxidase IV (COX-IV, red) and visualized by confocal microscopy. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole(blue). (B) Expression of p53 target proteins in OCI-AML-3 cells, which were treated with 50 nM KPT-185 and/or 2.5 µM Nutlin-3a for 12 hours. Intensity of the immunoblot signals was quantified, and the relative intensity compared with β-actin was calculated. Results are representative of 3 independent experiments.
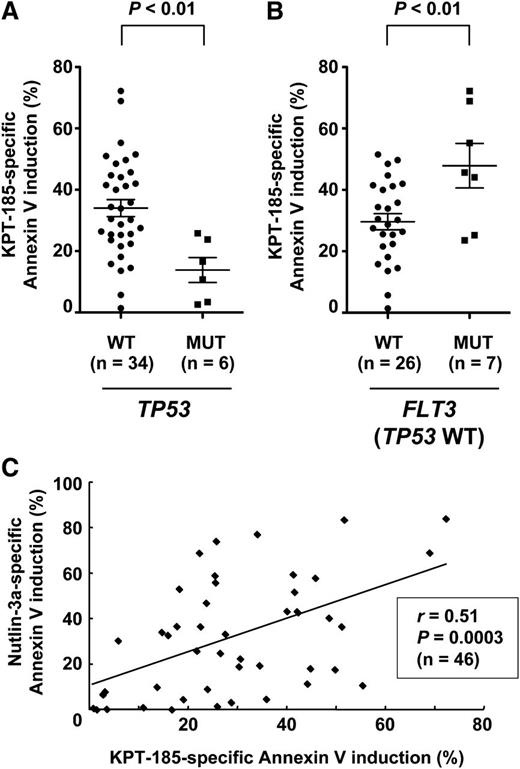
Patient AML cells with mutant p53 are less sensitive to KPT-185
We investigated if p53 mutational status affects cell susceptibility to KPT-185 in primary AML (n = 46; supplemental Table 2). p53 status was available for 40 samples. Six cases had mutant p53, and they were significantly less sensitive to KPT-185 compared with those with wild-type p53 (P < .01) (Figure 7A). FLT3 mutations were associated with increased KPT-185 sensitivity (P < .05), and the difference was more significant in wild-type p53 samples (P = .01) (Figure 7B). Sensitivity to KPT-185 appeared to be independent of NPM1 mutations (27.1 ± 2.6% vs 34.6 ± 6.9%, P = .23). Because the cellular outcome of p53 activation is defined by variations in intrinsic downstream effectors of p53 activation,35 we correlated the extent of apoptosis induced by KPT-185 with that induced by Nutlin-3a. The extent of apoptosis induced by KPT-185 positively correlated with that induced by Nutlin-3a (Figure 7C).
p53 status affects KPT-185 sensitivity in primary AML cells. Primary AML samples were treated for 48 hours with 800 nM KPT-185 and 8 µM Nutlin-3a, and the annexin V–positive fractions were measured by flow cytometry. (A) AML samples with mutant p53 were less sensitive to KPT-185 compared with those with wild-type p53. (B) FLT3 mutations were associated with increased KPT-185 sensitivity in wild-type p53 samples. Results are expressed as mean ± standard error of the mean. (C) The extent of apoptosis induced by KPT-185 positively correlates with that induced by Nutlin-3a.
p53 status affects KPT-185 sensitivity in primary AML cells. Primary AML samples were treated for 48 hours with 800 nM KPT-185 and 8 µM Nutlin-3a, and the annexin V–positive fractions were measured by flow cytometry. (A) AML samples with mutant p53 were less sensitive to KPT-185 compared with those with wild-type p53. (B) FLT3 mutations were associated with increased KPT-185 sensitivity in wild-type p53 samples. Results are expressed as mean ± standard error of the mean. (C) The extent of apoptosis induced by KPT-185 positively correlates with that induced by Nutlin-3a.
CRM1 inhibition by KPT-185 enhances Nutlin-induced p53-mediated apoptosis in primary AML cells but not in normal hematopoietic cells
We examined the combined apoptotic effect of KPT-185 and Nutlin-3a on primary cells (nos. 1-10 in supplemental Table 2). Both KPT-185 and Nutlin-3a induced apoptosis in CD34+/CD38– leukemia stem/progenitor cell populations as effectively as in bulk cells (supplemental Figure 4A), suggesting high sensitivity of progenitor cells to CRM1 and MDM2 inhibition. KPT-185 and Nutlin-3a synergized to induce apoptosis both in bulk and CD34+/CD38– cells. Based on the range of apoptotic effects (45.6% to 79.2% in the combination treatment group), the CI values for ED50 and ED75 (effective concentration inducing 75% killing as measured by annexin V positivity) were considered to directly reflect their combination effect. The CI values were 0.26 (bulk) and 0.30 (CD34+/CD38–) for ED50 and 0.93 (bulk) and 0.46 (CD34+/CD38–) for ED75, indicating a highly synergistic interaction. The synergistic effect was not observed in normal hematopoietic progenitor cells (supplemental Figure 4B). Normal CD34+/CD38– cells were significantly less sensitive to KPT-185 and Nutlin-3a than CD34+/CD38– AML cells (5.6 ± 2.5% vs 32.8 ± 6.7% at 800 nM KPT-185, P < .05; 6.9 ± 2.6% vs 59.8 ± 9.9% at 8 µM Nutlin-3a, P < .01; 8.5 ± 2.8% vs 79.2 ± 4.5% in the combination, P < .0001) (supplemental Figure 4B).
Finally, we investigated if expression of p53 isoforms affects AML cell sensitivity to KPT-185. Primary AML samples (n = 24) variously expressed FLp53 and p53βγ, as determined by western blotting. Consistent with a recent study,34 ΔNp53 isoforms were not detected. Total p53 levels in patient samples relative to those of OCI-AML-3 ranged from 0 to 4.76 (median 0.10). There was no significant correlation between relative p53 level and the percentage of specific annexin V (r = −0.13, P = .53). p53βγ/FLp53 protein ratio ranged from 0 to1.04 (median 0.32). Again, significant correlation was not detected between p53βγ/FLp53 ratio and the percentage of specific annexin V (r = −0.05, P = .82). KPT-185 treatment increased FLp53 levels, but the effect on p53βγ was minimal (n = 5; data not shown).
Discussion
CRM1 expression in AML has not been investigated to date. We profiled CRM1 expression in 511 newly diagnosed AML patients and found that CRM1 expression has a prognostic impact in AML. High CRM1 levels were observed to be an independent predictor of overall survival in both univariate and multivariate analysis. Higher levels of CRM1 were significantly associated with higher marrow blast percentages, white cell counts, peripheral blood blast percentages, and absolute peripheral blood blast count. Expression was significantly lower in those with favorable cytogenetics compared with those with intermediate or unfavorable cytogenetics. Inferior overall survival in a patient group with high CRM1 expression provides additional prognostic definition and rationale for therapeutic targeting of CRM1 in AML.
CRM1 expression was higher in AML with FLT3 mutations, which have been associated with poor survival in AML.36,37 Importantly, CRM1 inhibition by SINEs downregulates FLT3 at the posttranscriptional level in AML cells.24 Because FLT3/internal tandem duplication (ITD) AML cells are susceptible to p53-mediated cell death,38 SINEs that inhibit CRM1 and activate p53 may be beneficial in treating FLT3-mutated AML. Indeed, FLT3 mutations were associated with increased sensitivity to KPT-185 in patient samples, although the sample size was small. High CRM1 levels appeared to be associated with activated AKT signaling. Increased levels of phosphorylated death agonist BAD and 14-3-3 ζ may promote binding of BAD to 14-3-3 ζ to prevent an association between BAD with B-cell CLL/lymphoma 2 (BCL-2) and BCL2-like 1 (BCL-XL), conferring antiapoptotic properties to AML cells. Hence, the role of AKT in the regulation of CRM1 warrants further characterization.
Because MDM2 inhibition itself has not been sufficient to accumulate p53 in the nucleus in AML,7 we investigated if dual inhibition of MDM2 and CRM1 activates the p53-mediated transcription program. The combination of Nutlin-3a and KPT-185 not only increased cellular p53 levels but also accumulated p53 in the nucleus, resulting in synergistic apoptosis induction in AML. Considering that p53 is inactivated in the vast majority of human cancers, either through mutation, increased interactions with its negative regulator MDM2, or cytoplasmic sequestration, the potential effectiveness of the combination strategy could be relevant not only in AML but also in various cancers with wild-type p53. Furthermore, the limited cytotoxicity against normal hematopoietic cells suggests a therapeutic window for the combination.
KPT-185 exhibited potent antiproliferative effects in a p53-independent manner. Etchin et al22 have reported that KPT-185 exhibits p53-independent antileukemic activity in AML cell lines, using an adenosine triphosphate–based assay. Nonspecific proliferation assays like MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-dimethyltetrazolium bromide], MTS [3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium], or adenosine triphosphate–based assays are unable to distinguish effects on cell death or cell growth, and the observed antileukemic activity may be attributable to antiproliferative effects of KPT-185. We found that the p53 status was the major determinant of apoptotic response to KPT-185 in AML cell lines and in primary cells. Induction of apoptosis has been shown to be a key determinant of drug response in AML.39 The hypothesis that p53 status affects clinical response to KPT-SINEs could be validated in patients undergoing clinical trials with KPT-330 (NCT01607892).
One of the AML-specific abnormalities, mutant NPM1, is mediated by CRM1.17 NPM1 mutations create a nuclear export signal motif and disrupt tryptophans at the NPM1 C terminus, leading to CRM1-dependent cytoplasmic accumulation of mutant NPM1. OCI-AML-3 cells carrying the NPM1 mutation have been reported to exhibit cytoplasmic accumulation of NPM1, and cytoplasmic NPM1 has been shown to be retained in the nucleus following CRM1 inhibition.24 Our finding that p53 knockdown substantially blocked KPT-185–induced apoptosis in OCI-AML-3 cells (74.2 ± 2.2% vs 5.4 ± 0.3% at 200 nM KPT-185, P < .001) suggests that CRM1 inhibition induces apoptosis independent of NPM1 mutation status. Alternatively, nuclear accumulation of mutant NPM1 may induce apoptosis through p53 activation. Because nucleoplasmic NPM1 increases p53 stability through interaction with p53 and MDM2,40,41 cytoplasmic accumulation of mutant NPM1 can negatively regulate p53. In this setting, CRM1 inhibition could reactivate p53 and induce apoptosis in a p53-dependent manner. Further studies are required to clarify how NPM1 mutations interfere with the antileukemia effect of CRM1 inhibitors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Elizabeth Shpall of Department of Stem Cell Transplantation & Cellular TherapyACC for providing samples, and Drs Numsen Hail Jr and Peter Ruvolo for valuable discussions.
This work was supported in part by grants from National Institutes of Health Lymphoma Specialized Program of Research Excellence (CA136411), P01 “The Therapy of AML” (CA55164), Leukemia SPORE (CA100632), Cancer Center Support Grant (CA16672), the Paul and Mary Haas Chair in Genetics (M.A.), and by the Japan Leukemia Research Fund (K.K.).
Authorship
Contribution: K.K. initiated, designed, and performed the research, analyzed the data, and wrote the manuscript; V.R., S.D., A.D., M.Z., Y.H.Q., and J.K.B. performed the research; R.E.D., Z.W., K.R.C., N.Z., and S.M.K. analyzed the data; H.K. provided primary patients’ samples and contributed to discussion; S.S. and M.K. developed novel small-molecule inhibitors of CRM1; and M.A. initiated and supervised the research, analyzed the data, and edited the paper.
Conflict-of-interest disclosure: M.A. has sponsored clinical research agreements with Hoffmann-La Roche and received research support by an unrestricted gift from Karyopharm Therapeutics. S.S. and M.K. are employees of Karyopharm Therapeutics, a clinical stage biopharmaceutical company that develops selective inhibitors of nuclear export-targeted therapeutics. The remaining authors declare no competing financial interests.
Correspondence: Kensuke Kojima, Section of Molecular Hematology and Therapy, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 448, Houston, TX 77030; e-mail: kkojima@mdanderson.org; and Michael Andreeff, Section of Molecular Hematology and Therapy, The University of Texas MD Anderson Cancer Center, 1515 Holcombe Blvd, Unit 448, Houston, TX 77030; e-mail: mandreef@mdanderson.org.


![Figure 2. The CRM1 inhibitor KPT-185 inhibits the growth of AML cell lines through cell cycle arrest and apoptosis induction. (A) AML cell lines with wild-type p53 (OCI-AML-3, MOLM-13, and MV4;11) or mutant p53 (NB4, HL-60, KG-1, THP-1, and U937) cells were incubated with 100, 200, or 500 nM for 72 hours, and the annexin V–positive fractions were measured by flow cytometry (black bars). Cells were treated in parallel with 5 µM Nutlin-3a (white bars). Results are expressed as mean ± SD. (B) Lentivirally transduced wild-type p53 AML cells (virus encoding either control shRNA [shC] or p53-specific shRNA [shp53]) were incubated with 0, 10, 20, 50, 100, 200, 500, or 1000 nM of KPT-185 for 72 hours, and the numbers of viable cells and annexin V–positive fractions were measured. *P < .05; **P < .01; ***P < .001.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/20/10.1182_blood-2012-08-447581/2/m_4166f2.jpeg?Expires=1765889658&Signature=K09Wb31igixqOqCPJUpu5Bi2mXziCkMVR29rlLwIvTnlbWEpFSlppra4z8VaMxfSNv2GHdeQcSpR3VOTizxLBKESbzU2uCViZKQI6hIQ9kczqSg9HWqipLuq6k2kr78VHOZPjjsBT2~WCrfJ39oDyS2qgI6ySM0v~XxxCCb2~98or3ZSY4O7uLpK-~KaZRRKYmVfX0TuzxWGZdUEkjDOWjID8wslC1IQrlAVwV954-tju2tTKoqxc~-d1Ql2ex8SMhdKw6NvArF0k~3T7755UjEWCsh0cpu16mAOtt1RbwLF83g-BUGTDaxWfBVQIf8yc9XfrtV5RsEmzzAJpryY8A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)