Key Points
A focused RNAi screen identifies Dhx9 as a regulator of ABT-737 sensitivity in Eµ-myc/Bcl-2 lymphomas.
Dhx9 suppression activates an apoptotic signal through the Chk1/p53 replicative stress pathway in Myc-driven cells.
Abstract
ABT-737 is a promising chemotherapeutic agent that promotes apoptosis by acting as a selective BH3 mimetic to neutralize Bcl-2–like family members. One shortcoming with its use is that Mcl-1, a member of the Bcl-2 family, is poorly inhibited by ABT-737 and thus is a major cause of resistance. We performed a short hairpin RNA (shRNA)-based drop-out screen to identify novel genes and pathways that could reverse resistance to ABT-737 treatment in Eµ-myc/Bcl-2 lymphoma cells engineered to rely on endogenous Mcl-1 for survival. Several drug-sensitive shRNAs were identified that were selectively depleted in the presence of ABT-737. Of these, 2 independent shRNAs targeting the RNA/DNA helicase Dhx9 were found to sensitize lymphomas to ABT-737 to an extent comparable to control Mcl-1 shRNAs. Although Dhx9 suppression sensitized both mouse and human cells to ABT-737 treatment, it did so without altering MCL-1 levels. Rather, loss of Dhx9 appeared to activate a p53-dependent apoptotic program, through aggravation of replicative stress, which was found to be both necessary and sufficient for the ABT-737–shDhx9 synthetic lethal relationship.
Introduction
Activation of the intrinsic apoptotic pathway is largely dictated by the Bcl-2 family of proteins, a structurally diverse group of proteins whose members share a common homologous BH (Bcl-2 Homolog) domain through which they interact. Functionally, these proteins can be subcategorized into 3 groups: 1 prosurvival and 2 proapoptotic. Members of the prosurvival group (which includes Bcl-2, Bcl-XL, Mcl-1, A1, and Bcl-W) prevent mitochondrial outer membrane permeabilization (MOMP; the rate-limiting step of programmed cell death) caused by the formation of oligomeric pores composed of the proapoptotic multiple-BH3 proteins BAX and BAK. The so-called “BH3-only” family of proteins (BAD, BID, BIM, BMF, HRK, NOXA, and PUMA) is the upstream initiator of MOMP and is activated by diverse stress signals ranging from growth factor and nutrient deprivation to DNA damage and organellar stress.1 Although the precise mechanism by which this is accomplished has been contentious over the years, more recent data suggest a model where the combination of both direct activation of BAX/BAK oligomerization and indirect sequestration of Bcl-2–like proteins by the BH3-only proteins is required to trigger MOMP and irreversible cell death.2
Persistent intracellular stress is a common and driving feature of cancer progression that stimulates the intrinsic apoptotic pathway. Thus, evading apoptosis through genetic mutation is considered a hallmark of cancer malignancy. Defective apoptosis not only allows for aberrant (and rare) tumorigenic cells to persist in the tissue but can also lead to resistance to conventional chemotherapy. In fact, most conventional chemotherapeutic agents that are cytotoxic to tumor cells act through activation of ≥1 BH3 proteins.3 Many of these agents will therefore fail as therapy because many aggressive malignant tumors already harbor the necessary genetic alterations to overcome their mechanism of action (eg, Bcl-2 translocations), and therefore, the critical link between death signaling and BH3 activation/MOMP is often disrupted. There has thus been much interest in the development of drugs that can act more directly on the apoptotic machinery (downstream of the oncogenic lesion) to lead to a better therapeutic outcome. One such class is the BH3 mimetics, which are small molecules that act to mimic the interactions between native BH3-only proteins and antiapoptotic mitochondrial proteins.4 The best described drug among them is ABT-737 (and its orally available analog ABT-263 [Navitoclax]), which elicits a predictable MOMP response when administered in vitro or in vivo.5 Although ABT-737 has been shown to be effective as single-agent therapy in several clinically relevant settings, many cancers are refractory to ABT-737 treatment alone,6,7 and this is most likely because of ABT-737’s unique mechanism of action: although it binds to BCL-2, BCL-XL, and BCL-W with high affinity (in a low nanomolar range), it only weakly binds to MCL-1 and A1. Thus, cells engineered to overexpress Mcl-1 are universally resistant to ABT-737 treatment,8 whereas human lymphomas cultured in the presence of low doses of ABT-737 acquire resistance primarily through transcriptional up-regulation of Mcl-1 (and A1).9,10 Importantly, targeting Mcl-1, either by inhibiting its expression or by inactivating the protein, can dramatically improve the effectiveness of ABT-737 intervention.8 Further underscoring its clinical relevance, Mcl-1 has recently been found to be one of the most amplified genes across a wide spectrum of human tumor types.11 Although Mcl-1 is unlikely to be the sole determinant of ABT-737 sensitivity, it is thought to be a major cause of resistance to ABT-737 treatment and could ultimately determine both the clinical outcome and the effectiveness of ABT-737 therapy.
To identify genetic modifiers of ABT-737 sensitivity, we specifically modeled Mcl-1–conferred resistance in the Eμ-myc mouse model, where Myc expression is driven by the IgH enhancer (Eμ), and mice develop pre-B/B-cell lymphomas.12 The Eμ-myc mouse has been extensively used to characterize the functional role of apoptotic regulators in tumorigenesis, as well as in modeling resistance to cancer therapy.13 We performed a focused RNA-interference (RNAi)-based negative selection screen to identify drug-sensitive short hairpin RNAs (shRNAs) and report here that suppression of Dhx9, a multifunctional predominantly nuclear RNA/DNA helicase, is synthetic lethal with ABT-737 and overcomes Mcl-1–mediated resistance. Loss of Dhx9 improved ABT-737 sensitivity by intensifying oncogene-induced replicative stress and triggering induction of the p53 apoptotic program.
Materials and methods
Synthetic lethal RNAi screen
A custom shRNA library targeting the translation machinery, translation control signaling pathways, and RNA helicase family members was designed using miR30-adapted BIOPREDsi predictions (6 shRNAs per gene) and constructed by polymerase chain reaction—cloning a pool of oligonucleotides synthesized on 55k customized arrays (Agilent Technologies). Pools of shRNA polymerase chain reaction fragments were cloned into the MSCV/LTR/mIR30/SV40/green fluorescent protein (GFP; MLS) retrovirus vector, and following sequence verification, libraries were found to contain 4 to 6 shRNAs per gene. Pools were then prepared and spiked with positive (MLS/Mcl-1.1792) and negative control (MLS/Rluc.713) shRNAs at 0.03% of the total pool. These pools of shRNAs (350-1000) were transduced into Arf−/−Eμ-myc/Bcl-2 lymphomas using conditions that predominantly lead to a single retroviral integration and >1000 copies of each shRNA in the infected population (<10% transduction efficiency). Transduced cells were allowed to recover for 2 days, at which point they were split into 2 groups in media containing vehicle or 600 nM ABT-737 (considered T = 0 [T0]). Lymphomas were passaged every 2 days (1:3) and harvested at T0 and T10. Library representation was maintained by keeping >1000-fold more GFP+ cells per shRNA at each passage.
In vitro fitness assays
Lymphomas of indicated genotypes were transduced by a single infection with retrovirus expressing the indicated shRNA in MLS. Lymphomas were plated onto feeder layers at a density of 2.5 × 105 cells/mL, and cells were propagated in the presence of either vehicle (0.1% dimethylsulfoxide) or ABT-737 (600 nM) and passaged every 2 to 3 days at a 1:3 split. The percent GFP+ population was measured on the indicated days (∼5 × 104 cells analyzed per data point). To discriminate live from dead cells, lymphomas were stained with Propidium Iodide (PI), and both forward and side scatter and PI measurements were taken using a Guava Easycyte. Cells exhibiting PI staining and reduced forward scatter were excluded from analysis.
Cell cycle analysis
Cell cycle was analyzed using ethanol fixation/acid denaturation/PI staining.14 For S-phase transition assays, lymphomas were synchronized at the G1/S border using a double thymidine block. Briefly, lymphomas were treated with 2 mM thymidine for 16 hours. Thymidine was then removed by washing cells 3 times in B-cell media (BCM) followed by continued culturing for an additional 8 hours, at which point thymidine was added again for 16 hours. Lymphomas were then washed 3 times in prewarmed BCM and released into media containing 10 μM 5-bromo-2'-deoxyuridine (BrdU) for 30 minutes. Cells were washed 3 times in prewarmed BCM and chased in BrdU-free BCM. Cells (∼250 000) were collected at the indicated time points, washed with phosphate-buffered saline (PBS) twice, fixed in ethanol, and stored at −20°C until further processing. Lymphomas were treated with 0.5% Triton X-100/2N HCl for 30 minutes with end-over-end incubation at room temperature to denature genomic DNA. Cells were neutralized with 1 M sodium borate, pH 8.5, washed several times with 1% bovine serum albumin/0.5% Triton X-100 in PBS, and incubated with a 1:100 dilution of anti-BrdU antibodies conjugated to Alexa-647 for 30 minutes at room temperature. Cells were then washed 3 times with PBS and resuspended in 500 μL of PBS containing 5 μg/mL PI. BrdU+ lymphomas were then gated and tracked as they progressed through S phase.
Please see the supplemental Materials and Methods on the Blood website for additional information.
Results
Modeling Mcl-1–dependent ABT-737 resistance
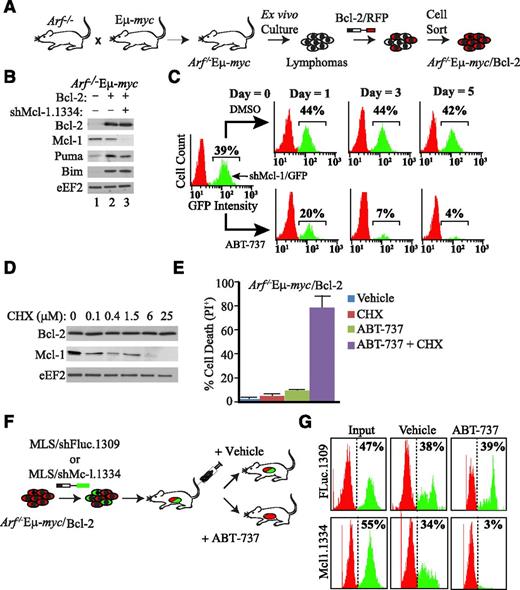
We chose to perform an RNAi-based drop-out screen in the Eμ-myc mouse model to identify apoptotic regulators capable of reversing ABT-737 resistance in an Mcl-1–dependent model. In most Eµ-myc mouse lymphoma lines that we tested, shRNAs targeting Mcl-1 were poorly tolerated—lymphomas expressing these shRNAs were rapidly depleted (supplemental Figure 1A), likely because of the key prosurvival role of Mcl-1 in the hematopoietic compartment.15,16 In the context of RNAi-based negative selection screens, where we wished to identify shRNAs phenocopying loss of Mcl-1, this limits the ability to differentiate lethal shRNAs that influence only Mcl-1 function from lethal shRNAs acting via alternative nonapoptotic mechanisms (eg, growth arrest). To circumvent these technical difficulties, we ectopically expressed Bcl-2 in Arf−/−Eµ-myc lymphomas such that shRNAs targeting Mcl-1 were tolerated and showed minimal loss after 8 days in culture (Figure 1A-B; supplemental Figure 1A). Importantly, shRNAs targeting essential genes (eg, ribosomal protein L15 [rpL15]) were readily depleted in cells overexpressing Bcl-2 (supplemental Figure 1B). Given the heterogeneity of apoptotic lesions found in spontaneous Eµ-myc lymphomas,17,18 we chose to take advantage of Eμ-myc lymphomas derived on the Arf−/− background, because loss of Arf alleviates the selective pressure of Myc-driven lymphomas to inactivate the apoptotic machinery, and thus, these will retain a consistent apoptotic response following standard chemotherapy.19 To confirm that resistance to ABT-737 could be conferred by endogenous Mcl-1 in Arf−/−Eµ-myc/Bcl-2 cells, we inhibited Mcl-1 expression with either RNAi or using the translation inhibitor cycloheximide (CHX)—conditions that dramatically reduce MCL-1 protein levels and elicit apoptosis in parental Arf−/−Eµ-myc lymphomas (supplemental Figure 1A,C).20,21 Importantly, Mcl-1 inhibition in Arf−/−Eµ-myc/Bcl-2 lymphomas showed a synthetic lethal relationship with ABT-737 both ex vivo (Figure 1C-E) and in vivo (Figure 1F-G). Furthermore, increasing MCL-1 levels desensitized lymphomas to ABT-737 (supplemental Figure 2). Therefore, engineered Arf−/−Eµ-myc/Bcl-2 cells are tolerant to ABT-737 insofar as MCL-1 activity is maintained, establishing a model of ABT-737 resistance mediated by endogenous Mcl-1.
Resistance to ABT-737 is reversed on suppression of Mcl-1 in the Eμ-myc model. (A) Schematic diagram illustrating derivation of the ABT-737–responsive Arf−/−Eμ-myc/Bcl-2 tumor model. (B) Immunoblot analysis of selective Bcl-2–related family members in extracts from Arf−/−Eμ-myc/Bcl-2 lymphomas transduced with MLS expressing shMcl1.1334 and/or the Bcl-2 cDNA. (C) Knockdown of Mcl-1 is synthetic lethal with ABT-737 in Arf−/−Eμ-myc/Bcl-2 lymphomas ex vivo. Flow cytometry analysis of Arf−/−Eμ-myc/Bcl-2 lymphomas infected with MLS/shMcl-1.1334 and cultured in the presence of vehicle or 600 nM ABT-737 for the indicated time periods. (D) Western blot analysis indicating that inhibition of protein synthesis results in rapid depletion of MCL-1. Arf−/−Eμ-myc/Bcl-2 lymphomas were exposed to the indicated CHX (concentration) for 12 hours, at which point cell extracts were prepared and probed by western blot for the indicated proteins. (E) Loss of MCL-1 protein levels by blocking protein synthesis is synthetic lethal with ABT-737 in Arf−/−Eμ-myc/Bcl-2 lymphomas. Cells were treated with 600 nM ABT-737 and 25 μM CHX for 16 hours, and cell death was quantitated by PI staining. Cell death was determined by PI staining. n = 3; errors bars represent ± standard error of the mean. (F) Schematic representation of experimental procedure used to assess ABT-737 sensitivity in vivo in Arf−/−Eμ-myc/Bcl-2 lymphomas. (G) Suppression of Mcl-1 is synthetic lethal with ABT-737 in Arf−/−Eμ-myc/Bcl-2 lymphomas in vivo. Following infection of Arf−/−Eμ-myc/Bcl-2 (RFP+) with FLuc.1309 or Mcl-1.1334 (GFP+), cells were injected into syngeneic C57BL/6 mice. ABT-737 treatment was initiated 2 days following delivery of cells and continued for 7 days, at which point lymphomas were harvested and analyzed by flow cytometry.
Resistance to ABT-737 is reversed on suppression of Mcl-1 in the Eμ-myc model. (A) Schematic diagram illustrating derivation of the ABT-737–responsive Arf−/−Eμ-myc/Bcl-2 tumor model. (B) Immunoblot analysis of selective Bcl-2–related family members in extracts from Arf−/−Eμ-myc/Bcl-2 lymphomas transduced with MLS expressing shMcl1.1334 and/or the Bcl-2 cDNA. (C) Knockdown of Mcl-1 is synthetic lethal with ABT-737 in Arf−/−Eμ-myc/Bcl-2 lymphomas ex vivo. Flow cytometry analysis of Arf−/−Eμ-myc/Bcl-2 lymphomas infected with MLS/shMcl-1.1334 and cultured in the presence of vehicle or 600 nM ABT-737 for the indicated time periods. (D) Western blot analysis indicating that inhibition of protein synthesis results in rapid depletion of MCL-1. Arf−/−Eμ-myc/Bcl-2 lymphomas were exposed to the indicated CHX (concentration) for 12 hours, at which point cell extracts were prepared and probed by western blot for the indicated proteins. (E) Loss of MCL-1 protein levels by blocking protein synthesis is synthetic lethal with ABT-737 in Arf−/−Eμ-myc/Bcl-2 lymphomas. Cells were treated with 600 nM ABT-737 and 25 μM CHX for 16 hours, and cell death was quantitated by PI staining. Cell death was determined by PI staining. n = 3; errors bars represent ± standard error of the mean. (F) Schematic representation of experimental procedure used to assess ABT-737 sensitivity in vivo in Arf−/−Eμ-myc/Bcl-2 lymphomas. (G) Suppression of Mcl-1 is synthetic lethal with ABT-737 in Arf−/−Eμ-myc/Bcl-2 lymphomas in vivo. Following infection of Arf−/−Eμ-myc/Bcl-2 (RFP+) with FLuc.1309 or Mcl-1.1334 (GFP+), cells were injected into syngeneic C57BL/6 mice. ABT-737 treatment was initiated 2 days following delivery of cells and continued for 7 days, at which point lymphomas were harvested and analyzed by flow cytometry.
RNAi-based synthetic lethal screen to identify genetic modifiers of ABT-737 sensitivity
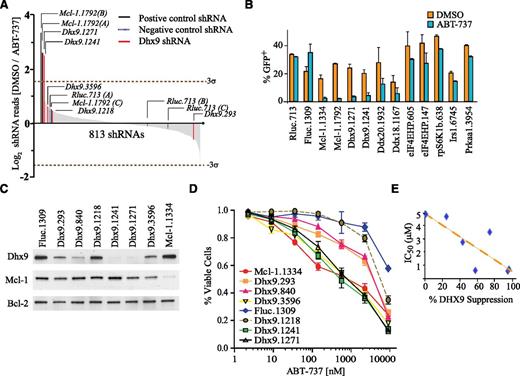
Because inactivation of Mcl-1 is an effective approach to resensitizing lymphomas to ABT-737 (Figure 1C-G) and endogenous Mcl-1 is under translational control in Eμ-myc lymphomas,20,22 we generated a custom miR30-based shRNA library targeting known components of the protein synthesis apparatus. This library included shRNAs directed to amino acyl-tRNA synthetases, large and small ribosomal proteins, initiation factors, elongation factors, termination factors, RNA helicases, and components of signaling pathways known to regulate protein synthesis (supplemental Figure 3A; supplemental Table 1). The library consisted of 1810 unique shRNAs (targeting 299 genes including controls) cloned into MLS, a miR30-based retroviral vector that coexpresses GFP (supplemental Figure 3B). The library was split into 3 pools, and these were transduced into Arf−/−Eμ-myc/Bcl-2 lymphomas (supplemental Figure 4). Infected cells were cultured for an additional 10 days (T10) in the presence of vehicle or ABT-737, and changes in shRNA representation were monitored by deep sequencing of shRNA guide strands amplified from genomic DNA. Each experimental time point was performed on 3 independent biological replicates, and correlation coefficients between replicates ranged from 0.7 to 0.96 (supplemental Table 2). In total, 1330 shRNAs (73% of library) were detectable above background in all 3 biological replicates at T0 (supplemental Table 3). By T10, 517 shRNAs were depleted from vehicle-treated cells (supplemental Table 3; supplemental Figure 5). Among the straight lethal shRNAs, 79% were shRNAs directed to genes encoding core components of the translational machinery—not surprising given our stringent selection criteria and the essential nature of the core protein synthetic apparatus.23 Of the remaining shRNAs, 10 shRNAs were synthetic lethal with ABT-737 (>2.5-fold depletion; Figure 2A; supplemental Table 3). These included 2 independent shRNAs targeting Dhx9 (Dhx9.1241 and Dhx9.1271), 6 unique shRNAs targeting eIF4E-HP, rpS6K, Ddx18, Ddx20, Irs1, and Prkaa1, and 2 positive control shRNAs targeting Mcl-1. These shRNAs were validated on a one-by-one basis and in independent Arf−/−Eμ-myc/Bcl-2 lymphoma lines (Figure 2B). Both Dhx9 shRNAs were depleted greater than fivefold in both the initial screen and in secondary validation assays, and thus Dhx9 was selected for more extensive evaluation (Figure 2B; supplemental Table 3). We compared the dose-response curves of a series of Arf−/−Eμ-myc/Bcl-2 cells stably expressing Dhx9 shRNAs (sorted for GFP), which covered a wide range of knockdown efficiencies (Figure 2C) and found that the more potent the shRNA (Dhx9.1241, Dhx9.1271, and Dhx9.3596), the greater the sensitization to ABT-737 (∼5- to 10-fold reduction at the EC50; Figure 2C-E). This was not a generalized sensitization to all chemotherapies, because there was very little change in sensitivity to DNA damaging therapies, such as etoposide (supplemental Figure 6). We further confirmed that Dhx9 knockdown specifically sensitized lymphomas to ABT-737 by monitoring the fitness of cells expressing Dhx9 shRNAs over time in a competition assay that showed selective depletion of Dhx9 shRNA expressing cells (GFP+) only in the presence of ABT-737 but not dimethylsulfoxide (supplemental Figure 7). Although knockdown efficiency for each Dhx9 shRNA correlated well with their sensitivity to ABT-737 (Figure 2E), surprisingly, there was a poor correlation between MCL-1 protein levels and Dhx9 knockdown efficiency (Figure 2C). This suggested that the synthetic lethal relationship between loss of Dhx9 and treatment with ABT-737 in Arf−/−Eμ-myc/Bcl-2 lymphomas was either independent of Mcl-1 gene function or was through inactivation of MCL-1 antiapoptotic activity.
Dhx9 is a genetic suppressor of ABT-737 sensitivity. (A) A pooled synthetic lethal shRNA screen in Myc-driven lymphomas depicting changes in representation of 813 informative shRNAs during 10 days of culture. shRNA depletion was calculated as the relative abundance in vehicle (dimethylsulfoxide)-treated cells divided by the relative abundance in ABT-737–treated cells. Values are plotted as the average of triplicate values in descending order. The dotted line represents 3 standard deviations from the population mean. Parenthesis denotes control shRNA pool location. (B) Percentage of GFP-positive Arf−/−Eμ-myc/Bcl-2 lymphoma cells cultured in the presence or absence of 600 nM ABT-737 for 8 days after transduction with MLS constructs expressing the indicated shRNAs. Error bars are standard error of the mean; n = 3. (C) Western blot form whole cell lysates of Arf−/−Eμ-myc/Bcl-2 lymphomas infected with the indicated shRNAs. (D) Viability of Arf−/−Eμ-myc/Bcl-2 cells expressing the indicated shRNAs. The fraction of PI-negative cells was determined 16 hours after exposure to increasing concentrations of ABT-737 and set relative to vehicle-treated cells. Error bars are standard error of the mean; n = 3. (E) Scatter plot comparing the Dhx9 knockdown to ABT-737 sensitivity (R2 = 0.66; P = .025; calculated using a paired Student t test).
Dhx9 is a genetic suppressor of ABT-737 sensitivity. (A) A pooled synthetic lethal shRNA screen in Myc-driven lymphomas depicting changes in representation of 813 informative shRNAs during 10 days of culture. shRNA depletion was calculated as the relative abundance in vehicle (dimethylsulfoxide)-treated cells divided by the relative abundance in ABT-737–treated cells. Values are plotted as the average of triplicate values in descending order. The dotted line represents 3 standard deviations from the population mean. Parenthesis denotes control shRNA pool location. (B) Percentage of GFP-positive Arf−/−Eμ-myc/Bcl-2 lymphoma cells cultured in the presence or absence of 600 nM ABT-737 for 8 days after transduction with MLS constructs expressing the indicated shRNAs. Error bars are standard error of the mean; n = 3. (C) Western blot form whole cell lysates of Arf−/−Eμ-myc/Bcl-2 lymphomas infected with the indicated shRNAs. (D) Viability of Arf−/−Eμ-myc/Bcl-2 cells expressing the indicated shRNAs. The fraction of PI-negative cells was determined 16 hours after exposure to increasing concentrations of ABT-737 and set relative to vehicle-treated cells. Error bars are standard error of the mean; n = 3. (E) Scatter plot comparing the Dhx9 knockdown to ABT-737 sensitivity (R2 = 0.66; P = .025; calculated using a paired Student t test).
Dhx9 knockdown activates a p53 transcriptional program.
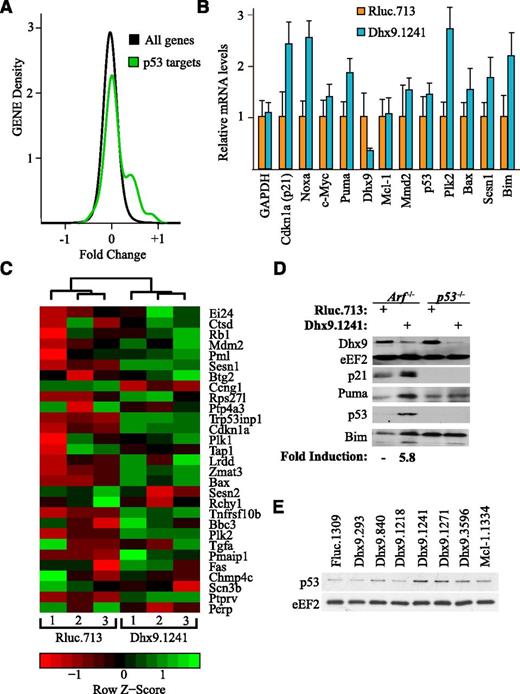
To gain insight into the synthetic lethal relationship between Dhx9 suppression and ABT-737 sensitization, we performed gene expression analysis on Arf−/−Eμ-myc/Bcl-2 lymphomas expressing either control Rluc.713 or the more potent Dhx9 shRNA, Dhx9.1241. After applying the random variance model 2-tailed Student t test and adjusting the P values for multiple testing using the Benjamini and Hochberg false discovery rate method, we identified 552 genes that were differentially expressed (false discovery rate <0.05) on silencing of Dhx9 (supplemental Table 4). We initially used gene set enrichment analysis but failed to uncover any molecular signatures induced by Dhx9 knockdown. We noticed that among the most statistically significant differentially expressed genes (supplemental Table 4), 7 were bona fide p53 transcriptional targets. Thus, we tested whether a validated set of p53 target genes24 was elevated relative to all genes on Dhx9 silencing. Indeed, expression of a subset of p53 target genes was significantly induced on Dhx9 suppression (Figure 3A-B). These results were validated by quantitative reverse transcription-polymerase chain reaction (Figure 3C). Additionally, we confirmed that on knockdown of Dhx9, p53 protein levels were elevated (approximately sixfold), as well as the protein levels of the p53 transcriptional targets p21 and PUMA (Figure 3D). There was a strong correlation between Dhx9 knockdown and p53 induction, with the most potent Dhx9 shRNAs (Dhx9.1241 and Dhx9.1271) yielding the highest levels of p53 induction (Figure 3E; supplemental Figure 8A). Moreover, the levels of p53 induction correlated with the degree to which these shRNAs sensitized lymphomas to ABT-737 (supplemental Figure 8B). Therefore, silencing of Dhx9 induces p53-dependent transcription, and this correlates with ABT-737 sensitivity.
Dhx9 suppression activates proapoptotic p53 signaling. (A) p53 transcriptional targets are elevated on Dhx9 suppression. The distribution of fold changes between Arf−/−Eµ-myc/Bcl2 lymphomas expressing Rluc.713 or Dhx9.1241 was compared for p53 target genes24 and non-p53 target genes. n = 3; Kolmogorov-Smirnov test, P = .0005. (B) Validation of microarray results by quantifying p53 transcriptional targets by quantitative reverse transcription-polymerase chain reaction. Values are set relative to β-actin. Error bars represent ± standard error of the mean; n = 6. (C) Heat map and clustering analysis of p53 targets in Arf−/−Eμ-myc/Bcl-2 lymphomas expressing Rluc.713 or Dhx9.1241. Cells were harvested 5 days after infection, and 3 independent biological replicates were used for each shRNA. The expression relative to per gene mean and standard deviation (z-score) is rendered in a red-black-green pseudo color scheme for p53 targets.24 Hierarchical clustering indicates p53 transcriptional target efficiently segregate lymphomas expressing Rluc.713 from those expressing Dhx9.1241. (D) Immunoblot analysis of cell extracts prepared from Arf−/−Eμ-myc/Bcl-2 and p53−/−Eμ-myc/Bcl-2 lymphoma cells. Cells were infected with MLS constructs expressing the indicated shRNAs (top) and probed with antibodies to the protein targets indicated to the left. Quantitation of the average fold p53 induction in response to Dhx9 suppression from 3 independent experiments. (E) Immunoblot analysis of cell extracts prepared from Arf−/−Eμ-myc/Bcl-2 lymphoma cells infected with MLS constructs expressing the indicated shRNAs.
Dhx9 suppression activates proapoptotic p53 signaling. (A) p53 transcriptional targets are elevated on Dhx9 suppression. The distribution of fold changes between Arf−/−Eµ-myc/Bcl2 lymphomas expressing Rluc.713 or Dhx9.1241 was compared for p53 target genes24 and non-p53 target genes. n = 3; Kolmogorov-Smirnov test, P = .0005. (B) Validation of microarray results by quantifying p53 transcriptional targets by quantitative reverse transcription-polymerase chain reaction. Values are set relative to β-actin. Error bars represent ± standard error of the mean; n = 6. (C) Heat map and clustering analysis of p53 targets in Arf−/−Eμ-myc/Bcl-2 lymphomas expressing Rluc.713 or Dhx9.1241. Cells were harvested 5 days after infection, and 3 independent biological replicates were used for each shRNA. The expression relative to per gene mean and standard deviation (z-score) is rendered in a red-black-green pseudo color scheme for p53 targets.24 Hierarchical clustering indicates p53 transcriptional target efficiently segregate lymphomas expressing Rluc.713 from those expressing Dhx9.1241. (D) Immunoblot analysis of cell extracts prepared from Arf−/−Eμ-myc/Bcl-2 and p53−/−Eμ-myc/Bcl-2 lymphoma cells. Cells were infected with MLS constructs expressing the indicated shRNAs (top) and probed with antibodies to the protein targets indicated to the left. Quantitation of the average fold p53 induction in response to Dhx9 suppression from 3 independent experiments. (E) Immunoblot analysis of cell extracts prepared from Arf−/−Eμ-myc/Bcl-2 lymphoma cells infected with MLS constructs expressing the indicated shRNAs.
p53 is essential for the synthetic lethal relationship between ABT-737 and Dhx9 suppression.
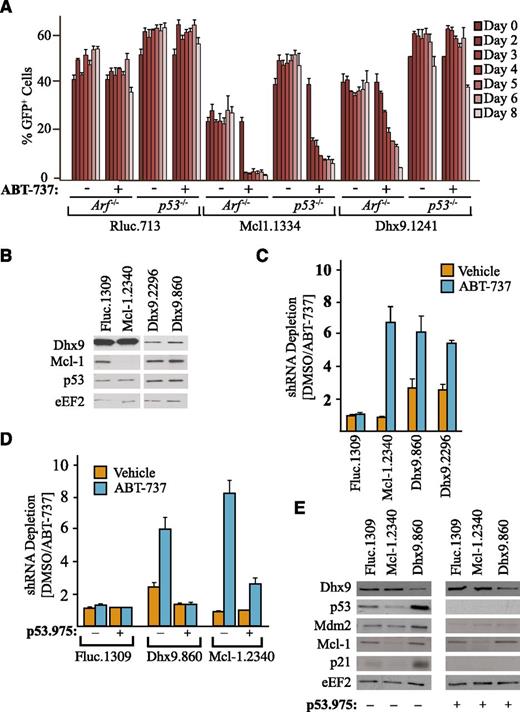
To determine whether induction of p53 was necessary for the synthetic lethal relationship between Dhx9 shRNAs and ABT-737 in Arf−/−Eμ-myc/Bcl-2 lymphomas, we performed similar competition experiments as above (supplemental Figure 7), except in p53−/−Eμ-myc/Bcl-2 lymphomas (Figure 4A). In contrast to Arf−/−Eμ-myc/Bcl-2 cells, Dhx9.1241 was not significantly depleted in the presence of ABT-737 in p53−/−Eμ-myc/Bcl-2 lymphomas, unlike Mcl-1.1334, which depleted regardless of p53 (Figure 4A). We also tested whether Nutlin-3, a specific inhibitor of the p53/MDM-2 interaction that directly increases p53 levels in the absence of genotoxic stress,25 could synergize with ABT-737 in killing Arf−/−Eμ-myc/Bcl-2. The combination of Nutlin-3 with ABT-737 potently killed Arf−/−Eμ-myc/Bcl-2 lymphomas, and this was completely abolished in the absence of p53 (supplemental Figure 9A-B).
Synergy between Dhx9 suppression and ABT-737 is p53 dependent. (A) Arf−/−Eμ-myc/Bcl-2 or p53−/−Eμ-myc/Bcl-2 lymphomas were infected with MLS constructs expressing the indicated shRNAs and cultured in the presence of media containing vehicle or ABT-737, and the percent GFP+ cells was determined over an 8-day period. Error bars represent ± standard error of the mean; n = 4. (B) Immunoblot of cell extracts prepared from U2OS cells that had been infected with lentiviral constructs expressing the indicated shRNAs and harvested 3 days after infection. The ratio of shRNA abundance in the dimethylsulfoxide control-treated lymphomas was set relative to ABT-737–treated samples. (C) U2OS cells were infected with lentiviruses expressing the indicated shRNAs, and the percent GFP+ cell population after 2 days of culture in the presence of vehicle or ABT-737 was determined. The ratio of shRNA abundance in the dimethylsulfoxide control-treated lymphomas was set relative to ABT-737–treated samples. Error bars represent standard error of the mean; n = 2. (D) Competition assays were performed as in panel C except using U2OS cells that had been previously transduced with a lentivirus expressing p53.975. Error bars represent mean ± standard error of the mean; n = 3. (E) Immunoblot of extracts prepared from U2OS cells infected with lentivirus expressing the indicated shRNAs and harvested 4 days after infection.
Synergy between Dhx9 suppression and ABT-737 is p53 dependent. (A) Arf−/−Eμ-myc/Bcl-2 or p53−/−Eμ-myc/Bcl-2 lymphomas were infected with MLS constructs expressing the indicated shRNAs and cultured in the presence of media containing vehicle or ABT-737, and the percent GFP+ cells was determined over an 8-day period. Error bars represent ± standard error of the mean; n = 4. (B) Immunoblot of cell extracts prepared from U2OS cells that had been infected with lentiviral constructs expressing the indicated shRNAs and harvested 3 days after infection. The ratio of shRNA abundance in the dimethylsulfoxide control-treated lymphomas was set relative to ABT-737–treated samples. (C) U2OS cells were infected with lentiviruses expressing the indicated shRNAs, and the percent GFP+ cell population after 2 days of culture in the presence of vehicle or ABT-737 was determined. The ratio of shRNA abundance in the dimethylsulfoxide control-treated lymphomas was set relative to ABT-737–treated samples. Error bars represent standard error of the mean; n = 2. (D) Competition assays were performed as in panel C except using U2OS cells that had been previously transduced with a lentivirus expressing p53.975. Error bars represent mean ± standard error of the mean; n = 3. (E) Immunoblot of extracts prepared from U2OS cells infected with lentivirus expressing the indicated shRNAs and harvested 4 days after infection.
To extend the relevance of our observations to human cancer, we generated shRNAs against human Dhx9 and tested these in U2OS osteosarcoma cells (which have retained a functional p53 pathway). As in the Eμ-myc lymphomas, shRNAs targeting Dhx9 in U2OS cells also activated p53 (Figure 4B) and were synergistically depleted by ABT-737 (Figure 4C). This effect was also entirely dependent on activation of p53, as knockdown of p53 completely abolished this relationship between loss of Dhx9 and ABT-737 (Figure 4D-E). Interestingly, even in the absence of ABT-737, Dhx9 shRNAs had reduced fitness and were selected against over time relative to both Fluc and Mcl-1 shRNAs in U2OS cells (Figure 4C-D, approximate two- to threefold reduction). This is consistent with previous observations demonstrating that induction of p53 in U2OS cells causes predominantly growth arrest and limited apoptosis.25 Thus, p53 appears both necessary and sufficient for the sensitization to ABT-737 treatment on suppression of Dhx9—a feature conserved in mouse and human tumor cells.
The very design of the Arf−/−Eμ-myc/Bcl-2 lymphomas allows us to infer that shRNAs against Dhx9 are not essential for growth or proliferation of Eμ-myc lymphomas, given that they are maintained in the absence of ABT-737 treatment. Conversely, this implies that knockdown of Dhx9, without the protection afforded by Bcl-2, could be lethal on its own, most likely because of apoptosis. To formally test this, we measured the fitness of the Dhx9.1241, Rluc.713, and Mcl-1.1334 hairpins using competition assays in absence of drug in parental Arf−/−Eμ-myc, as well as p53−/−Eμ-myc lymphoma cells (supplemental Figure 10A-C). Both Mcl-1.1334 and Dhx9.1241 were rapidly depleted in Arf−/−Eμ-myc cells; however, in the Bcl-2–expressing cells or p53−/−Eμ-myc cells, Dhx9.1241 was no longer rapidly depleted. This suggests that the net effect of activation of p53-dependent transcription caused by suppression of Dhx9 is an apoptotic mechanism.
Noxa is required for ABT-737 sensitization by Dhx9 suppression.
We observed an ∼2.5-fold increase in the mRNA levels of the proapoptotic p53 transcriptional targets Noxa, Puma, and Bim on knockdown of Dhx9 (Figure 3C), which all could potentially inactivate MCL-1.2 Although each of these BH3-only proteins can antagonize MCL-1 antiapoptotic function, they differ in their overall binding profiles: PUMA and BIM can bind strongly to all Bcl-2–like family members (both ABT-737 sensitive and insensitive), whereas NOXA can only bind MCL-1 and A1.26 Given that in some contexts, such as chronic lymphocytic leukemia, where the amount of BIM released from a BIM/BCL2 complex on ABT-737 treatment is an indicator of sensitivity,27 we tested how BIM might influence the ability of Dhx9 suppression to sensitize lymphomas to ABT-737. Knockdown of Bim increased the ABT-737 IC50 equally in the presence of shFLuc or shDhx9, thus failing to selectively prevent Dhx9 shRNAs from sensitizing lymphomas to ABT-737 (supplemental Figure 11). To determine whether either Noxa or Puma was essential for Dhx9-mediated ABT-737 sensitization, we measured the ability of ABT-737 to selectively deplete Dhx9.1241 shRNA in Arf−/−Eμ-myc/Bcl-2, p53−/−Eμ-myc/Bcl-2 Puma−/−Eμ-myc/Bcl-2, and Noxa−/−Eμ-myc/Bcl-2 lymphomas, relative to the control Rluc.713 shRNA (supplemental Figure 12A). In the absence of Noxa, ABT-737 failed to enhance elimination of Dhx9.1241-expressing lymphomas, whereas loss of Puma only mildly impacted ABT-737–mediated depletion of Dhx9.1241-expressing lymphomas (supplemental Figure 12A). These results were confirmed in sorted Dhx9.1241 Noxa−/−Eμ-myc/Bcl-2 cells (supplemental Figure 12B) and, importantly, were not caused by differences in knockdown efficiencies or in the levels of p53 induction (supplemental Figure 12C). Thus, it appears that in the presence of excess Bcl-2, the p53/Noxa apoptotic arm is critical for the synthetic lethal association between Dhx9 shRNAs and ABT-737.
Given the role of Noxa in overcoming resistance to ABT-737 and its selectivity for MCL-1, we also suppressed Dhx9 in Arf−/−Eμ-myc lymphomas overexpressing Mcl-1 instead of Bcl-2. In contrast to Bcl-2–overexpressing lymphomas, in Arf−/−Eμ-myc/Mcl-1 lymphomas, Dhx9 shRNAs were depleted over time (supplemental Figure 13). Thus, targeting Dhx9 can be exploited to overcome Mcl-1–mediated survival.
Targeting Dhx9 to sensitize Myc-dependent cells to ABT-737
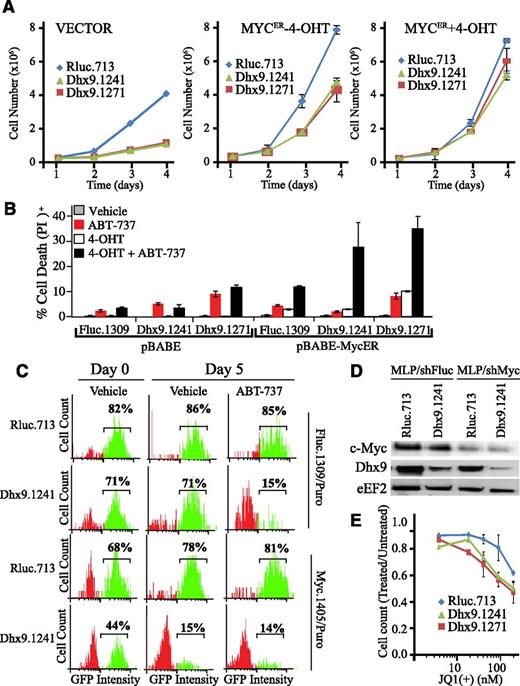
Given that our screen was performed in the context of constitutive c-Myc expression, we asked if Myc could be a genetic modifier of the cellular response to Dhx9 knockdown. In immortalized fibroblasts, in the absence of constitutive Myc expression, Dhx9 suppression induced a pronounced growth defect, in contrast to observations in Eμ-myc lymphomas (Figure 5A; supplemental Figure 10). To test whether Myc could rescue this defect, we generated an inducible Myc-ER fibroblast cell line responsive to 4-hydroxytamoxifen (4-OHT). Even without 4-OHT, there is sufficient Myc-ER activity to partially overcome the growth arrest induced by loss of Dhx9 (Figure 5A). 4-OHT eliminated any growth differences in cells expressing Rluc shRNAs relative to those expressing Dhx9 shRNAs (Figure 5A). Importantly, shRNAs targeting Dhx9 only strongly sensitize Myc-ER cells to ABT-737 on full Myc activation (+4-OHT; Figure 5B). This was also the case in U2OS/MycER cells, where activation of Myc was required for Dhx9 knockdown to induce maximal ABT-737 killing (supplemental Figure 14).
Loss of Dhx9 acts in concert with Myc to enhance ABT-737 sensitivity. (A) Growth curves of NIH3T3, NIH3T3/Myc-ER(−4-OHT), and NIH3T3/Myc-ER(+4-OHT) fibroblasts expressing the indicated shRNAs. (B) Cell death was measured in NIH3T3 cells expressing pBABE or pBABE-MycER, as well as the indicated shRNAs. Infected cells were sorted by flow cytometry, exposed to 250 nM 4-OHT for 24 hours and then to vehicle or ABT-737 for an additional 12 hours. Cells were harvested, stained with PI, and analyzed by flow cytometry. Error bars represent standard error of the mean; n = 3. (C) Competition assay of control (shRluc) or Dhx9 shRNAs in the context of either normal Myc levels or Myc suppression in Arf−/−Eμ-myc/Bcl-2 lymphomas. Lymphomas were infected with the indicated shRNAs, selected with puromycin, and cultured in the presence of media containing vehicle or ABT-737. The percent GFP+ cells was determined over a 5-day period. (D) Immunoblot analysis of extracts prepared from Arf−/−Eμ-myc/Bcl-2 cells that had been infected with retroviral vectors expressing the indicated shRNAs and puromycin selected and sorted for GFP+ lymphomas. (E) Arf−/−Eμ-myc/Bcl-2 lymphomas were infected with the indicated shRNAs and sorted with flow cytometry for GFP+. Lymphomas were plated at 100 000 cells per well and allowed to grow an additional 2 days. Cells were then collected and counted. Error bars represent standard error of the mean; n = 3.
Loss of Dhx9 acts in concert with Myc to enhance ABT-737 sensitivity. (A) Growth curves of NIH3T3, NIH3T3/Myc-ER(−4-OHT), and NIH3T3/Myc-ER(+4-OHT) fibroblasts expressing the indicated shRNAs. (B) Cell death was measured in NIH3T3 cells expressing pBABE or pBABE-MycER, as well as the indicated shRNAs. Infected cells were sorted by flow cytometry, exposed to 250 nM 4-OHT for 24 hours and then to vehicle or ABT-737 for an additional 12 hours. Cells were harvested, stained with PI, and analyzed by flow cytometry. Error bars represent standard error of the mean; n = 3. (C) Competition assay of control (shRluc) or Dhx9 shRNAs in the context of either normal Myc levels or Myc suppression in Arf−/−Eμ-myc/Bcl-2 lymphomas. Lymphomas were infected with the indicated shRNAs, selected with puromycin, and cultured in the presence of media containing vehicle or ABT-737. The percent GFP+ cells was determined over a 5-day period. (D) Immunoblot analysis of extracts prepared from Arf−/−Eμ-myc/Bcl-2 cells that had been infected with retroviral vectors expressing the indicated shRNAs and puromycin selected and sorted for GFP+ lymphomas. (E) Arf−/−Eμ-myc/Bcl-2 lymphomas were infected with the indicated shRNAs and sorted with flow cytometry for GFP+. Lymphomas were plated at 100 000 cells per well and allowed to grow an additional 2 days. Cells were then collected and counted. Error bars represent standard error of the mean; n = 3.
We also generated Arf−/−Eμ-myc/Bcl-2 expressing either shRNAs targeting Fluc or c-Myc that we partially transduced with GFP+ shRNA vectors expressing shRNAs targeting Rluc or Dhx9. On partial knockdown of Myc (Figure 5C), Dhx9 shRNAs weredepleted in the absence of ABT-737, and ABT-737 failed to select against Dhx9 shRNA-expressing cells (Figure 5D). To confirm these results, we used JQ1(+), a BET bromodomain inhibitor that targets Myc transcription. At the doses tested (up to 200 nM), 48 hours of JQ1(+) treatment did not trigger cell death in Arf−/−Eμ-myc/Bcl-2 lymphomas, likely because of excess Bcl-2, but did reduce growth (Figure 5E; supplemental Figure 15). Notably, lymphomas with reduced Dhx9 levels were more susceptible to JQ1(+)-induced growth arrest despite no detectable increase in cell death (Figure 5E). The combination of JQ1(+) and ABT-737 induced cell death, regardless of DHX9 levels, consistent with the dependency of Myc-driven lymphomas on c-Myc for continued survival and growth (supplemental Figure 15).28 Therefore, elevated Myc activity is necessary for Dhx9 knockdown to sensitize cells to ABT-737.
Dhx9 knockdown activates p53 through replicative stress
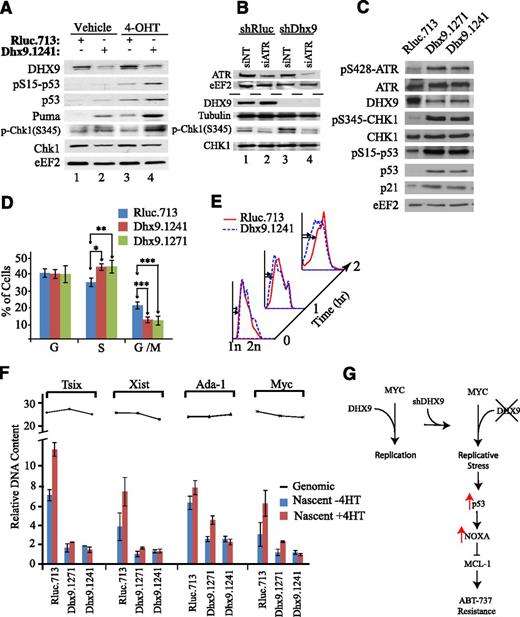
Induction of p53 is required for Dhx9 to alter ABT-737 sensitivity (Figure 4), and importantly, constitutive Myc activity alters the cellular response to Dhx9 knockdown and subsequent p53 induction (Figure 5). Constitutive Myc can overcome growth arrest associated with replication deficiencies,29,30 yet this activates a potent p53 apoptotic program.31 Given the association of Dhx9 with the replication machinery, we investigated S-phase checkpoint signaling in cells expressing either Rluc or Dhx9 hairpins. Knockdown of Dhx9 in Myc-ER cells increased Chk1 S345 phosphorylation, an event that could be prevented by ATR suppression (Figure 6A-B). In Myc-ER fibroblasts (−4-OHT), loss of Dhx9 weakly elevated p53, phospho-p53, and downstream p53 targets (Figure 6A). The combination of Dhx9 suppression and Myc activation potently stimulated the phosphorylation of p53, with a corresponding increase in p53 levels and its downstream targets (Figure 6A), as well as increasing cell death (Figure 5B). In lymphomas, knockdown of Dhx9 increased Chk1 S345 phosphorylation, a classic marker of replicative stress, as well as ATR and p53 phosphorylation (Figure 6C), suggesting activation of the S-phase checkpoint pathway. These results highlight the importance of the interplay between Dhx9/Myc/p53 in overcoming resistance to ABT-737.
Loss of Dhx9 induces replicative stress and exacerbates Myc induction of p53. (A) Immunoblot analysis of extracts prepared from MycER/3T3 cells that had been infected with MLS expressing the indicated shRNAs. Cells were harvest 48 hours after exposure to vehicle or 250 nM 4-OHT. (B) Immunoblot analysis of extracts prepared from Myc-ER/3T3 that had been infected with MLS expressing the indicated shRNAs. Cells were then transfected with the indicated siRNA, and 48 hours later, lysates were generated and blotted with the indicated antibodies. (C) Immunoblot analysis of extracts prepared from Arf−/−Eμ-myc/Bcl-2 cells infected with the indicated shRNAs. (D) Cell cycle distribution of Arf−/−Eμ-myc/Bcl-2 lymphomas expressing the indicated shRNAs. Error bars represent standard error of the mean; n = 5; *P = .01; **P = .05, ***P < .01 (calculated using an unpaired Student t test). (E) S-phase progression of Arf−/−Eμ-myc/Bcl-2 expressing the indicated shRNAs on release from a double thymidine block. BrdU+ lymphomas were monitored at the indicated times after release. Cells were stained with PI and analyzed by flow cytometry. (F) Genomic (line graph) and nascent DNA (bar graph) was isolated from MycER/3T3 cells expressing the indicated shRNAs. DNA was harvested 48 hours after exposure to vehicle or 250 nM 4-OHT. The presence of nascent DNA corresponding to Tsix, Xist, Ada-1, and Myc origins was quantitated with quantitative reverse transcription-polymerase chain reaction. Error bars represent standard deviation; n = 3. (G) Schematic representation of a model by which Dhx9 knockdown enhances ABT-737 sensitivity.
Loss of Dhx9 induces replicative stress and exacerbates Myc induction of p53. (A) Immunoblot analysis of extracts prepared from MycER/3T3 cells that had been infected with MLS expressing the indicated shRNAs. Cells were harvest 48 hours after exposure to vehicle or 250 nM 4-OHT. (B) Immunoblot analysis of extracts prepared from Myc-ER/3T3 that had been infected with MLS expressing the indicated shRNAs. Cells were then transfected with the indicated siRNA, and 48 hours later, lysates were generated and blotted with the indicated antibodies. (C) Immunoblot analysis of extracts prepared from Arf−/−Eμ-myc/Bcl-2 cells infected with the indicated shRNAs. (D) Cell cycle distribution of Arf−/−Eμ-myc/Bcl-2 lymphomas expressing the indicated shRNAs. Error bars represent standard error of the mean; n = 5; *P = .01; **P = .05, ***P < .01 (calculated using an unpaired Student t test). (E) S-phase progression of Arf−/−Eμ-myc/Bcl-2 expressing the indicated shRNAs on release from a double thymidine block. BrdU+ lymphomas were monitored at the indicated times after release. Cells were stained with PI and analyzed by flow cytometry. (F) Genomic (line graph) and nascent DNA (bar graph) was isolated from MycER/3T3 cells expressing the indicated shRNAs. DNA was harvested 48 hours after exposure to vehicle or 250 nM 4-OHT. The presence of nascent DNA corresponding to Tsix, Xist, Ada-1, and Myc origins was quantitated with quantitative reverse transcription-polymerase chain reaction. Error bars represent standard deviation; n = 3. (G) Schematic representation of a model by which Dhx9 knockdown enhances ABT-737 sensitivity.
In both the lymphoma and Myc-ER fibroblasts, Dhx9 suppression enhances the number of asynchronous cells found in S-phase with a reduction in the number of cells in G2/M, signifying a defect in S-phase (Figure 6D; supplemental Figure 16). Following release from a double thymidine block, Arf−/−Eμ-myc/Bcl-2 lymphomas expressing Dhx.1241 exhibited a delay in transition across early S-phase (Figure 6E). In Myc-ER fibroblasts, loss of Dhx9 reduces the induction of replication-dependent dsDNA breaks induced by camptothecin, but not those induced by γ-IR (replication-independent), indicative of a reduction in DNA replication (supplemental Figure 17A)—this despite more cells present in S-phase on Dhx9 knockdown (supplemental Figure 16). Furthermore, etoposide, which targets replicating cells,32 was less effective at inducing cell death when Dhx9 was suppressed (supplemental Figure 17B). We also measured the activity of several well-characterized mouse origins of DNA replication in Myc-ER fibroblasts.33,34 Dhx9 suppression significantly reduced the frequency of firing at several origins and also blocked the ability of Myc to increase origin activity (Figure 6F).29
To independently confirm replicative stress is sufficient to lead to ABT-737 sensitization, we exposed Arf−/−Eμ-myc/Bcl-2 cells to hydroxyurea, a compound that induces replication stress by altering the dNTP pools available to DNA polymerases. This is sufficient to activate Chk1/p53 signaling (supplemental Figure 18A), as well as sensitize Arf−/−Eμ-myc/Bcl-2 lymphomas to ABT-737 (supplemental Figure 18B).
Last, to address which functions of Dhx9 might be required for ABT-737 resistance, we rescued loss of Dhx9 with a wild-type shDhx9.1271-resistant, human (h)Dhx9 cDNA (supplemental Figure 16). We then tested a series of Dhx9 mutants for their ability to reverse ABT-737 sensitivity, none of which were capable of doing so (supplemental Figure 19). Therefore, loss of Dhx9, in the context of Myc activation, is sufficient to elicit a p53-dependent response priming cells for ABT-737–induced cell death.
Discussion
We performed a focused shRNA screen to identify novel gene products that could overcome Mcl-1–mediated resistance to ABT-737 in an Eμ-myc lymphoma cell line engineered to rely on ectopic Bcl-2 and endogenous Mcl-1 expression for survival. Two independent shRNAs targeting Dhx9 were identified that were as effective as Mcl-1 shRNAs in reversing ABT-737 resistance, but, to our surprise, had no discernible effect on MCL-1 protein levels (Figure 2C). Because of the many and varied cellular functions ascribed to Dhx9,35 we analyzed gene signature pathways that were affected by Dhx9 suppression to obtain insight into the mechanism of Dhx9-mediated ABT-737 sensitization. A validated p53 gene set24 was significantly increased in lymphomas expressing Dhx9 shRNAs (Figure 3A), and hierarchical clustering of experimental samples based on expression profiles of p53 targets was sufficient to differentiate those lymphomas expressing Dhx9 shRNAs from controls (Figure 3B).
Activation of p53 in the Eμ-myc model is an essential component of the chemotherapeutic apoptotic response and is largely mediated downstream through transcriptional activation of the BH3-only proteins PUMA, BIM, and NOXA.36 In our study, we found that only Noxa was necessary for Dhx9 shRNAs to overcome ABT-737 resistance, whereas loss of Puma or Bim had only minimal effects, even though both Puma and Bim were transcriptionally induced on Dhx9 knockdown. Although lymphomas lacking Bim were rendered more resistant to ABT-737, knockdown of Dhx9 remained sufficient to sensitize these lymphomas to ABT-737. Although a minor apoptotic role has been attributed to Noxa in the Eμ-myc model,37 our data are consistent with previous reports demonstrating Noxa induction sensitizes cells to ABT-737.8 One possible explanation is that in our setting, Bcl-2 is sufficiently in excess such that ABT-737 does not release adequate levels of PUMA and/or BIM to antagonize MCL-1, essentially restricting the contribution of Dhx9-induced Puma or Bim to ABT-737 sensitivity. In contrast, Noxa serves a unique function in that BCL-2 cannot engage NOXA, whereas loss of Bim or Puma could be functionally supplanted by each other, such that individual loss of one would have negligible impact on the net amount of BH3-only proteins circulating on ABT-737 treatment. Interestingly, in the case of histone deacetylase inhibitors where increased sensitivity to ABT-737 is achieved through induction of p53, this enhancement also appears solely dependent on Noxa,38 perhaps reflecting a similar phenomenon.
Given the importance of p53 in mediating varied chemotherapeutic responses, the relative paucity of data connecting p53 status with sensitivity to ABT-737 treatment (and other BH3 mimetics) is somewhat surprising, especially because p53-mediated induction of BH3-only proteins has been shown to enhance ABT-737 treatment. Combination therapies with ABT-737 aimed at inducing genotoxic stress pathways requires an intact p53/Noxa-Puma pathway to synergize with ABT-737.18 Taken together, these data suggest that secondary to Mcl-1 expression levels, p53 status could be an important determinant of ABT-737 sensitivity—an issue that may be of clinical significance given the design of trials assessing the efficacy of Navitoclax with genotoxic agents (ie, NCT00868413, NCT01423539, and NCT00878449).
Our data indicate that suppression of Dhx9 widens the therapeutic window of ABT-737 by taking advantage of, and exacerbating, a preactivated replicative stress response pathway. In fact, oncogene-driven replicative stress is a common feature of a wide variety of tumor types, and exploiting it for therapeutic gain, especially in the context of Myc, has previously been used with some success. Treatment of Eμ-myc lymphomas with novel Chk1 inhibitors dramatically induces p53 signaling, leading to robust DNA damage and pronounced apoptosis; a consequence of disrupting the S-phase checkpoint pathway and subsequent DNA repair.31,39 An RNAi screen in MYCN-driven neuroblastoma cell lines revealed that siRNAs against, and pharmacological inhibition of, Chk1 is cytotoxic, underscoring the reliance of these cells on a functional replicative stress response pathway and confirming the sensitivity of Myc-driven tumors to disruption of the S-phase checkpoint pathway.40 Furthermore, in the Eμ-myc mouse model, inactivation of either Wrn or ATR, both of which are essential for recovery from Myc-induced replicative stress, results in a dramatic delay in lymphoma onset and robust activation of p53 signaling.30,41,42 Interestingly, in the context of the Myc oncogene, loss of Dhx9 results in a phenotype that overlaps with that of Wrn defective cells: exacerbation of p53 signaling, activation of S-phase checkpoint signaling, abnormal accumulation of cells in S-phase, and defective transition across the early phases of DNA synthesis.30,41,42 DHX9 has been identified as an interacting partner of WRN, a RecQ DNA helicase whose loss or mutation leads to replicative stress and genomic instability.43-45 DHX9 enhances the 3′-5′ exonuclease activity of WRN in vitro, which is required for its ability to resolve stalled replication forks and permit efficient transit through S-phase.46 Consistent with its role in DNA replication, DHX9 associates with components of the DNA replication machinery (eg, TOP2A, PCNA, and BRCA1) and its helicase specificity toward triple helical structures and DNA:RNA hybrids—two intermediates formed during replication fork stalling.47,48 Knockdown of Dhx9 is therefore sufficient to activate an ATR/Chk1 signaling cascade, leading to accumulation of cells in S-phase, delaying transition across S-phase, and altering origin activity, all suggesting that Dhx9 likely influences the fidelity of DNA replication.
Our data show that targeting Dhx9 only elicits an apoptotic response when combined with constitutive MYC activity. Correspondingly, inactivating Myc diminishes the ability of Dhx9 hairpins to sensitize lymphomas to ABT-737. In the absence of Myc, loss of Dhx9 delays growth and in immortalized fibroblasts is protective against DNA damaging agents. These results suggest targeting Dhx9 in combination with ABT-737 may be useful in specifically targeting highly chemoresistant lymphomas, such as double hit Myc/Bcl-2 lymphomas49 with minimal nonspecific toxicity to nontransformed cells. As well, our results suggest that inhibitors of Chk1 (currently undergoing extensive clinical evaluation), which have proven to be effective as single agents against Myc-driven cancers, may synergize with ABT-737 and perhaps broaden their applicability in cancer therapies. Our study provided novel insight into Dhx9 biology and the role that the replicative stress response pathway plays in ABT-737 sensitization.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Clare Scott (Walter and Eliza Hall Institute) for supplying Puma−/−Eμ-myc and Noxa−/−Eμ-myc lymphomas and Dr Michael Hemann for Bim shRNAs.
J.R.M. was supported by a Cole Foundation Fellowship. O.L. was supported by the Swedish Research Council. This work was supported by the Canadian Institutes of Health Research (MOP-115126 to J.P.), a Townshead/Lamarre Family Foundation grant (to J.P.), and an National Institutes of Health/National Cancer Institute program project grant (CA087497 to S.W.L).
Authorship
Contribution: J.R.M., A.M., and J.P. conceived and designed the study and constructed the shRNA library; J.R.M., A.M., T.L., and D.D.P. performed the experiments; C.M., J.R.M., and J.P. processed and analyzed the Solexa sequencing data; O.L. performed statistical evaluation of microarray data; S.W.L., M.Z-H., H.T., and F.G. provided key reagents and expertise; and J.R.M., A.M., S.W.L., and J.P. prepared the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jerry Pelletier, Department of Biochemistry, McIntyre Medical Sciences Building, Room 810, 3655 Drummond St, McGill University, Montreal, Quebec, Canada H3G 1Y6; e-mail: jerry.pelletier@mcgill.ca.
References
Author notes
J.R.M. and A.M. contributed equally to this work.