Key Points
We observed that SMAD7, a negative regulator of TGF-β receptor-I kinase, is markedly reduced in MDS, and leads to ineffective hematopoiesis.
Increased levels of microRNA-21 are seen in MDS and reduce SMAD7 levels, thus overactivating TGF-β signaling.
Abstract
Myelodysplastic syndromes (MDS) are characterized by ineffective hematopoiesis that leads to peripheral cytopenias. We observed that SMAD7, a negative regulator of transforming growth factor–beta (TGF-β) receptor-I kinase, is markedly reduced in MDS and leads to ineffective hematopoiesis by overactivation of TGF-β signaling. To determine the cause of SMAD7 reduction in MDS, we analyzed the 3′UTR of the gene and determined that it contains a highly conserved putative binding site for microRNA-21. We observed significantly elevated levels of miR-21 in MDS marrow samples when compared with age-matched controls. miR-21 was shown to directly bind to the 3′UTR of SMAD7 and reduce its expression in hematopoietic cells. Next, we tested the role of miR-21 in regulating TGF-β signaling in a TGF-β–overexpressing transgenic mouse model that develops progressive anemia and dysplasia and thus serves as a model of human bone marrow failure. Treatment with a chemically modified miR-21 inhibitor led to significant increases in hematocrit and led to an increase in SMAD7 expression in vivo. Inhibition of miR-21 also led to an increase in erythroid colony formation from primary MDS bone marrow progenitors, demonstrating its ability in stimulating hematopoiesis in vitro. Taken together, these studies demonstrate the role of miR-21 in regulating overactivated TGF-β signaling in MDS.
Introduction
The myelodysplastic syndromes (MDS) are clonal stem cell disorders characterized by cytologic dysplasia and ineffective hematopoiesis.1-3 Although approximately one-third of patients’ disease may progress to acute leukemia, refractory cytopenias are the principal cause of morbidity and mortality in patients with MDS.4 In fact, approximately two-thirds of patients present with lower-risk disease characterized by hypercellular marrows, with increased rates of apoptosis in the progenitor and differentiated cell compartments in the marrow.5-8 Ineffective hematopoiesis arising from abortive maturation leads to peripheral cytopenias. Higher grade or more advanced disease categories are associated with a significant risk of leukemia transformation with a corresponding lower apoptotic index and higher percentage of marrow blasts.
Cytokines play important roles in the regulation of normal hematopoiesis, and a balance between the actions of hematopoietic growth factors and myelosuppressive factors is required for optimal production of different hematopoietic cell lineages. Excess production of inhibitory cytokines amplifies ineffective hematopoiesis inherent to the MDS clone. Transforming growth factor-beta (TGF-β) is a myelosuppressive cytokine that has been implicated in the hematopoietic suppression in MDS. The plasma levels of TGF-β have been reported to be elevated in some9-13 but not all studies14-17 and are supported by greater TGF-β immunohistochemical staining in selected studies. Conflicting data may arise from technical limitations of bone marrow immunohistochemical analyses of a secreted protein, as well as the biological heterogeneity of the disease itself. Thus, we investigated the role of TGF-β in MDS by direct examination of receptor signal activation to conclusively determine its role in the pathogenesis of ineffective hematopoiesis in MDS. We determined that the SMAD2 is upregulated and overactivated in MDS bone marrow progenitors, thereby demonstrating sustained TGF-β signal activation in this disease. Because there is conflicting data about upregulation of extracellular TGF-β levels in MDS, we next sought to determine the molecular basis of TGF-β receptor-I overactivation and subsequent SMAD2 phosphorylation/activation in this disease. We observed that SMAD7, a negative regulator of TGF-β receptor-I kinase, is markedly decreased in MDS and this leads to overactivation of TGF-β signal transduction, even in the absence of increased levels of extracellular TGF-β.18 Thus, we wanted to focus on the molecular alterations that lead to SMAD7 reduction in MDS. In the present study, we were able to show that miR-21 is increased in MDS, binds to the 3′UTR of the SMAD7 gene, and leads to consequent upregulation of the TGF-β pathway in MDS. We were also able to show that inhibition of this microRNA is able to abrogate TGF-β signaling both in vitro and in vivo and stimulates hematopoiesis in MDS.
Materials and methods
Cells lines and reagents
Human CD34+ cells were isolated from the bone marrow of normal or MDS patients after obtaining informed consent in accordance with the Declaration of Helsinki and approval by the institutional review board of Albert Einstein College of Medicine. Bone marrow CD34+ cells from various normal donors were also obtained from AllCells (Emeryville, CA). HS-5 cell line (CRL-11882) was purchased from ATCC (Manassas, VA). TGF-β1 was bought from R&D Systems (Minneapolis, MN). The phos-SMAD2 (S465/467) and actin antibodies were from Cell Signaling Technology (Beverly, MA), and SMAD2 antibody was from Invitrogen (Carlsbad, CA), SMAD7 antibody was from ABcam (Cambridge, MA). Locked nucleic acid (LNA)-modified miR-21 inhibitor was provided by Exiqon (Woburn, MA).
Cell lysis and immunoblotting
Gene expression analysis
Gene expression data were obtained from 183 MDS CD34+ cells and 17 controls.20 Clinical information for the cases was provided by the authors. SMAD7 and SMAD2 expression was represented as box plots using R statistical software.
qPCR for miR-21
Quantitative reverse-transcriptase polymerase chain reaction (RT-PCR) was done for miR-21 and endogenous control, RNU44, on MDS and age-matched control bone marrow mononuclear cells (MNCs) after obtaining informed consent approved by the institutional review board of Moffitt Cancer Center. Reverse-transcriptase reactions and real-time PCR were performed on Applied Biosystems Real-Time PCR instruments (Applied Biosystems, Foster City, CA) according to the manufacturer’s protocols as described elsewhere.21,22
Murine experiments
TGF-β1 transgenic mice expressing a fusion gene (Alb/TGF) consisting of modified porcine TGF-β1 cDNA under the control of the regulatory elements of the mouse albumin gene23 were used under animal institute–approved protocol. Mice were given LNA miR-21 inhibitors (Exiqon) at a dose of 25 mg/kg given intraperitoneally on alternate days for a total of 5 doses. Mismatched oligos were used as controls at similar doses. Blood counts were analyzed by Advia machine (Siemens, Princeton, NJ).
SMAD7 gene reporter assay
HS-5 cells were plated in 6-well plates 24 hours before transfection. Cells were transfected in triplicate with SuperFect (Qiagen, Alameda, CA) according to the manufacturer’s instructions.24 To generate SMAD7 3′UTR mutants containing mutations in the conserved miR-21 binding site, site-directed mutagenesis was performed using the wild-type 3′UTR as the template. In the 3′UTR mutant, the nucleotide sequence complementary to nt 2–5 of miR-21 was mutated to the same sequence as that in miR-21 (from AGCT to TCGA). Luciferase reporters containing wild-type and mutant 3′UTR of SMAD7 gene were used for transfection with Pre-miR miR-21 precursors (Ambion, Invitrogen) or controls (30 nM). The generation of wild-type and mutant constructs has been described previously.25 Cells were harvested 24 hours later in reporter lysis buffer, and luciferase and renilla activities were determined using Dual Luciferase System (Promega, Madison, WI).
miR-21 overexpression
A plasmid-encoding miR-21 and green fluorescent protein (GFP) in the pMIF-cGFP-Zeo lentiviral vector (Systems Biosciences, Mountain View, CA) was introduced into cells by nucleofection. The empty pMIF-cGFP-Zeo vector, encoding only GFP, was used as a negative control.
Hematopoietic progenitor cell assays
Hematopoietic progenitor colony formation was determined by clonogenic assays in methylcellulose, as in our previous studies.19,26 All participants in the study signed informed consent, approved by the institutional review board of Albert Einstein College of Medicine. Granulocyte/macrophage colony-forming units (CFU-GM) and erythroid burst-forming units (BFU-E) from bone marrow samples were scored on day 14 of the culture.
Fluorescence in situ hybridization
After 14 days of methylcellulose culture, cells were isolated from the plates and cytospun on microscopic slides. The cells on the slides were fixed in Carnoy’s solution. The following dual color probes (Abbott Molecular, Abbott Park, IL) were used to detect 7q deletions and 5q deletions: LSI D7S522 (7q31) SpectrumOrange/CEP7 SpectrumGreen, EGR-1 SpectrumOrange/D5S721/D5S23 SpectrumGreen. Fluorescence in situ hybridization was performed according to the manufacturer’s instructions. Cells were counterstained with 4′6-diamidino-2-phenylindole and examined on a Zeiss Axioplan 2 fluorescence microscope (Zeiss, North Chesterfield, VA).
Results
SMAD7 is reduced in bone marrow progenitors in MDS
SMAD7 is a negative regulator of the TGF-β signaling pathway and inhibits the activity of the TGF-β receptor-I kinase. We analyzed the expression of SMAD7 in a large gene expression dataset obtained from bone marrow CD34+–selected samples from MDS patients20 We found that SMAD7 is significantly reduced in these samples when compared with healthy controls (Figure 1, mean log2 expression 8.31 in controls vs 6.32 in MDS samples, false discovery rate < 0.1, Benjamin Hochberg correction multiple testing, Student’s t-test < 0.001). Furthermore, SMAD2, the effector SMAD protein for TGF-β signaling was found to be increased in the same samples (Figure 1B, mean log2 expression 8.4 in controls vs 8.8 in MDS samples, false discovery rate < 0.1, Benjamin Hochberg correction multiple testing, Student’s t-test < 0.001), demonstrating the sustained activation of TGF-β pathway in these samples. Subset analysis revealed striking SMAD7 reduction in all subsets of MDS examined (Figure 1A). Correlation of SMAD7 levels with clinical parameters did not reveal any associations with age, sex, or cytogenetics, revealing it to be a pervasive finding in MDS.
SMAD7 expression is significantly decreased in MDS CD34+ cells. (A) SMAD7 expression in 183 samples of MDS CD34+ cells and 17 healthy controls reveals reduction in all subsets of MDS. (false discovery rate < 0.1, Benjamin Hochberg correction multiple testing). (B) SMAD2, the effector SMAD protein for TGF-β signaling, was found to be increased in the same samples. (C) Heatmaps showing expression values for both genes. (The 5q- patients were a subset of the RA patient cohort.)
SMAD7 expression is significantly decreased in MDS CD34+ cells. (A) SMAD7 expression in 183 samples of MDS CD34+ cells and 17 healthy controls reveals reduction in all subsets of MDS. (false discovery rate < 0.1, Benjamin Hochberg correction multiple testing). (B) SMAD2, the effector SMAD protein for TGF-β signaling, was found to be increased in the same samples. (C) Heatmaps showing expression values for both genes. (The 5q- patients were a subset of the RA patient cohort.)
microRNA-21 is increased in MDS and has a putative binding site on the SMAD7 3′UTR
microRNAs have been implicated in the pathogenesis of cancer27 and have also recently been shown to be dysregulated in cases of MDS.21 To evaluate whether any of these aberrantly expressed microRNAs could target SMAD7, we evaluated the 3′UTR of the transcript and found putative binding sites for various microRNAs (Figure 2A). miR-21 was found to have a highly conserved binding site in the 3′UTR of SMAD7 and was also one of the microRNAs aberrantly expressed in a genome-wide analysis of 41 MDS samples (Figure 2B).21 Thus, we evaluated the expression of miR-21 in primary MDS bone marrow samples (N = 19; 10 low-risk, 9 high-risk MDS, supplemental Table 1) by quantitative RT-PCR and compared it with age-matched controls (N = 13). miR-21 levels were significantly increased in MDS when compared with controls, and the mean expression was more than fourfold higher (Student’s t-test, P = .03) (Figure 2C). Both lower-risk MDS (low and Int-1 International Prognostic Scoring System) and higher-risk MDS (Int-2 and high International Prognostic Scoring System) had significantly higher expression when compared with controls (Student’s t-test, P < .05).
miR-21 has a putative binding site on the SMAD7 3′UTR and is overexpressed in MDS. (A) Targetscan software predicts a strong possibility of miR-21 binding site at position 1122 of the SMAD7 3′UTR with a highly conserved site. (B) Microarray analysis of 41 untreated MDS marrow samples were compared with 10 age-matched controls21 and demonstrate upregulation of miR-21 (mean intensity ± SEM, Student’s t-test, P = .001). (C) Quantitative RT-PCR was done for miR-21 and endogenous control, RNU44, on 19 MDS and 13 age-matched control bone marrow MNCs using the Ambion Taqman miroRNA assay kit. MDS samples had significantly higher expression of miR-21 (*Student’s t-test, P = .04). Both lower- and higher-risk MDS (low-risk MDS, N = 10, P value = .04; high-risk MDS, N = 9, P = .03, Student’s t-test) had higher mean expression when compared with controls.
miR-21 has a putative binding site on the SMAD7 3′UTR and is overexpressed in MDS. (A) Targetscan software predicts a strong possibility of miR-21 binding site at position 1122 of the SMAD7 3′UTR with a highly conserved site. (B) Microarray analysis of 41 untreated MDS marrow samples were compared with 10 age-matched controls21 and demonstrate upregulation of miR-21 (mean intensity ± SEM, Student’s t-test, P = .001). (C) Quantitative RT-PCR was done for miR-21 and endogenous control, RNU44, on 19 MDS and 13 age-matched control bone marrow MNCs using the Ambion Taqman miroRNA assay kit. MDS samples had significantly higher expression of miR-21 (*Student’s t-test, P = .04). Both lower- and higher-risk MDS (low-risk MDS, N = 10, P value = .04; high-risk MDS, N = 9, P = .03, Student’s t-test) had higher mean expression when compared with controls.
miR-21 binds directly to 3′UTR of SMAD7, reduces the expression of SMAD7, and inhibits erythroid colony formation
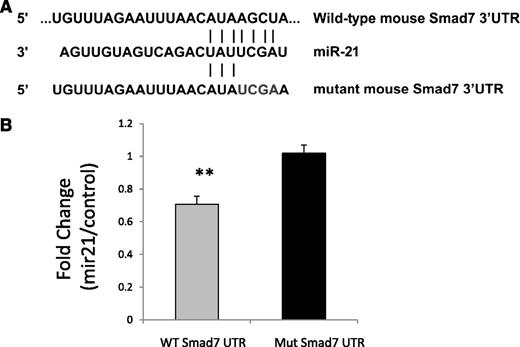
We next wanted to determine whether miR-21 expression can decrease SMAD7 expression and whether this effect is caused by direct interaction of miR-21 with the SMAD7 3′UTR. A luciferase reporter containing the 3′UTR segment containing predicted microRNA interaction sites was used to analyze the effect of miR-21 on SMAD7 gene transcription in bone marrow stromal HS-5 cells. The nucleotide sequence complementary to nts 2–5 of miR-21 binding site in the 3′UTR of SMAD7 was mutated to the same sequence as that in miR-21 (from AGCT to TCGA)25 (Figure 3A). Dual luciferase activity was measured and showed decreased SMAD7 luciferase activity caused by expression of miR-21 compared with cells transfected with control mimics. This was abrogated by mutation in its binding site on the UTR (Student’s t-test, P = .001), demonstrating a direct effect of miR-21 on the 3′UTR of SMAD7 (Figure 3A).
miR-21 binds to SMAD7 3′UTR. (A) The nucleotide sequence complementary to nt 2–5 of miR-21 in the 3′UTR of SMAD7 was mutated to the same sequence as that in miR-21 (from AGCT to TCGA). (B) Luciferase reporters containing wild-type and mutant 3′UTR of the SMAD7 gene were used for transfection with control miRNA or miR-21 precursors into bone marrow stromal HS-5 cells. Dual luciferase activity was measured and shows decreased luciferase activity caused by miR-21 that is abrogated by mutation in its binding site on the UTR (Student’s t-test, P = .001) compared with cells transfected with control mimics.
miR-21 binds to SMAD7 3′UTR. (A) The nucleotide sequence complementary to nt 2–5 of miR-21 in the 3′UTR of SMAD7 was mutated to the same sequence as that in miR-21 (from AGCT to TCGA). (B) Luciferase reporters containing wild-type and mutant 3′UTR of the SMAD7 gene were used for transfection with control miRNA or miR-21 precursors into bone marrow stromal HS-5 cells. Dual luciferase activity was measured and shows decreased luciferase activity caused by miR-21 that is abrogated by mutation in its binding site on the UTR (Student’s t-test, P = .001) compared with cells transfected with control mimics.
Subsequently, the effect of miR-21 on SMAD7 expression was tested by using a chemically modified LNA inhibitor that led to decreased levels of miR-21 transcripts (Figure 4A) when compared with mismatched control. We observed that inhibition of miR-21 led to an increase in SMAD7 protein expression in leukemic cell lines and in primary MDS patient samples (Figure 4B). Inhibition of miR-21 also led to decreased phosphorylation/activation of SMAD2 in a MDS patient sample (Figure 4C). Next, we tested the functional role of miR-21 in regulating the effects of TGF-β on normal hematopoiesis and the role of SMAD7 in regulating these effects. CD34+ cells were transfected with shRNAs against SMAD7 and controls as performed previously,18 sorted for GFP expression, and then treated with TGF-β in the presence and absence of the miR-21 inhibitor. We observed that miR-21 inhibition was able to abrogate the inhibitory responses of TGF-β on erythroid colony formation from primary human CD34+ stem cells that were transfected with scrambled control shRNAs (Figure 4D). This effect was not seen in cells transfected with shRNAs against SMAD7.
Inhibition of miR-21 can abrogate the effects of TGF-β on hematopoietic cells. miR-21 levels were measured in leukemic K562 cells after 48-hour treatment with LNA inhibitor of miR-21 and compared with mismatched control (mm control). miR-21 transcripts were significantly decreased after LNA treatment (50 mM) as measured by quantitative RT-PCR (mean ± SEM of 2 experiments, Student’s t-test, P < .001). (B) SMAD7 protein expression is increased after miR-21 inhibition (50 mM, 48-hour treatment) in HL-60, TF-1 cell lines and in leukocytes from 2 MDS patients, as shown by immunoblotting. Actin was used as a protein-loading control. (C) miR-21 inhibitor treatment leads to decreased p-SMAD2 in MDS leukocytes when compared with mismatched-control LNAs (50 mM). (D) Primary CD34+ progenitors were nucleofected with GFP coexpressing anti-SMAD7 (or scrambled control) shRNA constructs18 and sorted after 24 hours. GFP-positive cells were grown in methylcellulose in the presence and absence of TGF-β (20 ng/mL) and miR-21 inhibitor (50 mM), and erythroid colonies were counted after 14 days. miR-21 inhibitor treatment abrogates the inhibitory effects of TGF-β on erythroid colonies (mean ± SEM of 2 independent experiments, Student’s t-test, P = .04) in cells that were transfected with scrambled shRNA controls. SMAD7 shRNA–transfected progenitors did not demonstrate any reversal of TGF-β–induced inhibition after miR-21 inhibitor treatment. (E) miR-21 and GFP were co-overexpressed in leukemic K562 cells using the pMIF-cGFP-Zeo vector, leading to decreased levels of SMAD7 as detected by immunoblotting when compared with the GFP-only control. (F) Primary CD34+ progenitors were nucleofected with miR-21and GFP or with the GFP-only control and sorted after 24 hours. GFP-positive cells were grown in methylcellulose in the presence of TGF-β (20 ng/mL) and hematopoietic colonies were counted after 14 days. (G) miR-21 overexpression led to decreased erythroid colony formation (mean ± SEM of 2 independent experiments, Student’s t-test, P = .01) and also led to qualitative reductions in colony sizes.
Inhibition of miR-21 can abrogate the effects of TGF-β on hematopoietic cells. miR-21 levels were measured in leukemic K562 cells after 48-hour treatment with LNA inhibitor of miR-21 and compared with mismatched control (mm control). miR-21 transcripts were significantly decreased after LNA treatment (50 mM) as measured by quantitative RT-PCR (mean ± SEM of 2 experiments, Student’s t-test, P < .001). (B) SMAD7 protein expression is increased after miR-21 inhibition (50 mM, 48-hour treatment) in HL-60, TF-1 cell lines and in leukocytes from 2 MDS patients, as shown by immunoblotting. Actin was used as a protein-loading control. (C) miR-21 inhibitor treatment leads to decreased p-SMAD2 in MDS leukocytes when compared with mismatched-control LNAs (50 mM). (D) Primary CD34+ progenitors were nucleofected with GFP coexpressing anti-SMAD7 (or scrambled control) shRNA constructs18 and sorted after 24 hours. GFP-positive cells were grown in methylcellulose in the presence and absence of TGF-β (20 ng/mL) and miR-21 inhibitor (50 mM), and erythroid colonies were counted after 14 days. miR-21 inhibitor treatment abrogates the inhibitory effects of TGF-β on erythroid colonies (mean ± SEM of 2 independent experiments, Student’s t-test, P = .04) in cells that were transfected with scrambled shRNA controls. SMAD7 shRNA–transfected progenitors did not demonstrate any reversal of TGF-β–induced inhibition after miR-21 inhibitor treatment. (E) miR-21 and GFP were co-overexpressed in leukemic K562 cells using the pMIF-cGFP-Zeo vector, leading to decreased levels of SMAD7 as detected by immunoblotting when compared with the GFP-only control. (F) Primary CD34+ progenitors were nucleofected with miR-21and GFP or with the GFP-only control and sorted after 24 hours. GFP-positive cells were grown in methylcellulose in the presence of TGF-β (20 ng/mL) and hematopoietic colonies were counted after 14 days. (G) miR-21 overexpression led to decreased erythroid colony formation (mean ± SEM of 2 independent experiments, Student’s t-test, P = .01) and also led to qualitative reductions in colony sizes.
Furthermore, we also overexpressed miR-21 in separate experiments by using lentiviral vector-encoding miR-21 precursors (pMIF-cGFP-Zeo), along with a control vector that expressed only GFP. Overexpression of miR-21 led to decreased SMAD7 protein levels in leukemic cells (Figure 4E) and led to quantitative and qualitative reduction in erythroid colony formation from primary CD34+ cells (Figure 4F-G). Taken together, these data demonstrate that miR-21 potentiates TGF-β signaling in hematopoietic cells via downregulation of SMAD7.
Inhibition of miR-21 can improve anemia in a model of TGF-β–driven bone marrow failure
To test the role of miR-21 in regulating TGF-β signaling in vivo, we used chemically modified inhibitors of miR-21 and tested them in a transgenic mouse expressing a fusion gene (Alb/TGF) consisting of modified porcine TGF-β cDNA under the control of the regulatory elements of the mouse albumin gene.23 We have shown that these mice constitutively secrete TGF-β, become anemic, and have histologic marrow findings that mimic human MDS, thus serving as an in vivo model of bone marrow failure.28 These mice were randomized into a treatment or placebo group on the basis of pretreatment hemotocrit levels. Blood counts were measured after administration of miR-21 inhibitor or control. miR-21 inhibitor treatment led to increased expression of SMAD7 and inhibition of constitutive SMAD2 activation in murine bone marrow (see Figure 5C). Mice treated with miR-21 inhibitor had a significant increase in hematocrit (Figure 5A) and were able to form more erythroid colonies from marrow-derived cells (Figure 5B) when compared with control (Student’s t-test, P < .05). These results demonstrate the role of miR-21 inhibition in inhibiting TGF-β signaling in vivo.
Treatment with mir21 inhibitor leads to an increase in red blood cells in TGF-β transgenic mice. (A) Mice expressing TGF-β (Alb/TGFβ+) were treated with LNA-modified anti–miR-21 (n = 7) or mismatched placebo controls (n = 6) for 5 doses. Mice treated with anti–miR-21 oligos had a significant increase in hematocrit (HCT, Student’s t-test, P = .03), hemoglobin (Hgb [gm/DL], Student’s t-test, P = .049), and red blood cells (RBCs [×106], Student’s t-test, P = .0005). (B) Equal numbers of bone marrow cells were grown in methylcellulose and demonstrated increased erythroid colony formation in the mice treated with miR-21 inhibitor (n = 5 in each group, Student’s t-test, P = .04. (C) Representative bone marrow biopsy samples from both groups show an increase in SMAD7 levels in the treatment group with a corresponding decrease in phospho-SMAD2.
Treatment with mir21 inhibitor leads to an increase in red blood cells in TGF-β transgenic mice. (A) Mice expressing TGF-β (Alb/TGFβ+) were treated with LNA-modified anti–miR-21 (n = 7) or mismatched placebo controls (n = 6) for 5 doses. Mice treated with anti–miR-21 oligos had a significant increase in hematocrit (HCT, Student’s t-test, P = .03), hemoglobin (Hgb [gm/DL], Student’s t-test, P = .049), and red blood cells (RBCs [×106], Student’s t-test, P = .0005). (B) Equal numbers of bone marrow cells were grown in methylcellulose and demonstrated increased erythroid colony formation in the mice treated with miR-21 inhibitor (n = 5 in each group, Student’s t-test, P = .04. (C) Representative bone marrow biopsy samples from both groups show an increase in SMAD7 levels in the treatment group with a corresponding decrease in phospho-SMAD2.
Inhibition of miR-21 can stimulate MDS hematopoiesis
Finally, we tested the ability of chemically modified (LNA) miR-21 inhibitor in vitro in 5 primary MDS bone marrow samples. These included different subtypes of MDS (Table 1). Mononuclear cells from the bone marrow were grown in methylcellulose with cytokines in the presence of miR-21 inhibitor and control. Treatment with the miR-21 inhibitor resulted in a significant increase in erythroid (BFU-E) colony numbers in all patients (results depicted as means in Figure 6A and Table 1). miR-21 inhibitor treatment did not result in any significant increase in colony formation from healthy controls (Figure 6B). Karyotypic analysis of colonies from 2 MDS samples revealed a reduction in percentage of cells, with cytogenetic alterations in miR-21 inhibitor–treated cells when compared with controls (20% in treated vs 29% in controls for 5q- deletion; 3% in treated vs 11% in controls for 7q- deletion). In addition to confirming the findings from our studies in mice, these results point to the potential stimulatory effect of miR-21 inhibition on MDS hematopoiesis.
MDS patient characteristics and response to miR-21 inhibition
| N . | Age/Sex . | WBC (103/mL) . | Hgb (g/dL) . | Plt (103/mL) . | Subtype . | Cytogenetics . | Colonies post treatment with mismatched control . | Colonies post treatment with miR-21 inhibitor . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| BFU-E . | CFU-GM . | BFU-E . | CFU-GM . | |||||||
| 1 | 63/M | 1.2 | 9 | 14 | RCMD | +11 | 204 | 12 | 259 | 24 |
| 2 | 79/f | 2.9 | 9.6 | 29 | RAEB | −7q | 22 | 4 | 26 | 5 |
| 3 | 68/F | 3.5 | 9.3 | 35 | RAEB | −5q, −18 | 20 | 37 | 49 | 46 |
| 4 | 70/M | 18.2 | 8.5 | 84 | RAEB | Nml | 91 | 93 | 121 | 86 |
| 5 | 69/F | 5.2 | 8.8 | 75 | RCMD | Nml | 62 | 26 | 98 | 25 |
| N . | Age/Sex . | WBC (103/mL) . | Hgb (g/dL) . | Plt (103/mL) . | Subtype . | Cytogenetics . | Colonies post treatment with mismatched control . | Colonies post treatment with miR-21 inhibitor . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| BFU-E . | CFU-GM . | BFU-E . | CFU-GM . | |||||||
| 1 | 63/M | 1.2 | 9 | 14 | RCMD | +11 | 204 | 12 | 259 | 24 |
| 2 | 79/f | 2.9 | 9.6 | 29 | RAEB | −7q | 22 | 4 | 26 | 5 |
| 3 | 68/F | 3.5 | 9.3 | 35 | RAEB | −5q, −18 | 20 | 37 | 49 | 46 |
| 4 | 70/M | 18.2 | 8.5 | 84 | RAEB | Nml | 91 | 93 | 121 | 86 |
| 5 | 69/F | 5.2 | 8.8 | 75 | RCMD | Nml | 62 | 26 | 98 | 25 |
WBC, white blood cells; plt, platelets; Hgb, hemoglobin.
Inhibition of miR-21 stimulates erythropoiesis in MDS. MDS bone marrow–derived MNCs from 5 patients with MDS (A) and 3 controls (B) were plated in methylcellulose and cytokines in the presence and absence of miR-21 inhibitor (50 nM). Colonies were scored at Day 14 and results were expressed as means ± SEM of independent experiments.
Inhibition of miR-21 stimulates erythropoiesis in MDS. MDS bone marrow–derived MNCs from 5 patients with MDS (A) and 3 controls (B) were plated in methylcellulose and cytokines in the presence and absence of miR-21 inhibitor (50 nM). Colonies were scored at Day 14 and results were expressed as means ± SEM of independent experiments.
Discussion
The discovery of effective treatments for MDS has been challenged by the limited insight into molecular pathogenesis of the ineffective hematopoiesis seen in these disorders. Even though TGF-β levels have been shown to be elevated in a subset of MDS cases, our previous work has shown that intracellular signaling is activated in a large proportion of cases. Furthermore, we demonstrated that SMAD7 downregulation may be the key mechanism driving the suppression of hematopoiesis in MDS via overactivation of the TGF-SMAD2 pathway. This would also explain increased TGF-β signal transduction even in the absence of large amounts of circulating TGF-β that have been observed in various studies in MDS patients.14-17 In the present study, we determined the reasons for downregulation of SMAD7 in MDS and observed that miR-21 can bind to the 3′UTR region of the SMAD7 gene, reduce its expression, and potentiate TGF-β signaling in hematopoietic cells. Furthermore, inhibition of miR-21 can stimulate hematopoiesis in vitro and in vivo, thus demonstrating a critical role of this microRNA in regulation of myelosuppresive pathways in MDS.
SMAD7 is an important regulator of TGF-β signaling and its dysregulation has been implicated in many disease and cancer models.29 Genome-wide association studies have shown that polymorphisms in the SMAD7 gene are some of the most significant genetic alterations30 in colorectal cancer. A recent study showed that decreased SMAD7 is also seen in idiopathic pulmonary fibrosis and contributes to TGF-β–mediated fibrosis seen in this disease.25 Our study demonstrates that SMAD7 reduction is seen in a large number of MDS samples and points to its role in hematopoietic failure seen in this disease. The bone marrow at earlier stages of MDS is hypercellular and is characterized by ineffective hematopoiesis affecting both stem and progenitor cells. Because TGF-β is a critical regulator of hematopoiesis and affects both proliferation and differentiation of stem cells,31 an aberrant increase in TGF-β signaling can potentially explain the changes seen in MDS.
There could be many possible reasons for SMAD7 reduction in MDS. Even though the deletion of chromosome 18/18q, where the SMAD7 gene is located, was noticed to be a common deletion in MDS,32 it is only seen in a small proportion of patients with the disease. Furthermore, even though splicing mutations have recently been discovered in MDS,33 it is possible that splicing abnormalities can also affect SMAD7 transcripts, even though this information is not yet available. Because microRNAs are important regulators of genes, we decided to examine miR-mediated degradation of SMAD7 in MDS. We were able to show that miR-21 directly binds to the SMAD7 3′UTR region and regulated TGF-β signaling. miR-21 has been well studied in cancer and has been shown to influence a variety of targets including PTEN, PLAG1, and NFIB among others. A recent study demonstrated that miR-21–mediated degradation of SMAD7 was seen in pulmonary tissue and led to increased TGF-β–mediated fibrosis in idiopathic lung fibrosis.25 We show that this phenomenon can be seen in a hematologic disease and can explain the reduced SMAD7 levels seen in bone marrow progenitors. Our findings also raise the question of why miR-21 is found to be upregulated in MDS. In recent studies, the miR-21 gene locus has been shown to contain STAT3 binding sites.34 Furthermore, STAT3 has also been shown to directly lead to miR-21 upregulation in immune cells.34 Our genomic analysis of hematopoietic stem cells in MDS demonstrated that STAT3 is selectively upregulated in these cells. Thus, it is possible that this leads to miR-21 upregulation seen by us in MDS that in turn enhances TGF-β signaling that leads to ineffective hematopoiesis.
Most importantly, our results demonstrate that the activation of the TGF-β pathway by SMAD7 reduction in MDS can be abrogated by miR-21 inhibition both in vitro and in vivo. Ineffective hematopoiesis and anemia cause most of the morbidity in patients with MDS. Two-thirds of all MDS patients are at the low-risk stage of disease, have a lower chance of the disease progressing to leukemia, and have problems associated primarily with low red blood cell counts. Because TGF-β has been specifically shown to regulate both the early and late stages of erythropoiesis,31 miR-21 can be predicted to affect red cell production in particular, as was seen in the present study. Thus, taken together our findings support the exploration of miR-21 inhibitors in future preclinical and clinical studies in MDS.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by a Leukemia and Lymphoma Society Translational Research grant, the American Cancer Society (RSG-09-037), the Gabrielle Angel foundation, the Immunology and Immunooncology Training Program (T32 CA009173), the Partnership for Cures, the Department of Defense, National Institutes of Health (R01HL082946, R01HL116336), Leukaemia and Lymphoma Research UK, and a Carl Gottschalk Research Scholarship from the American Society of Nephrology.
Authorship
Contribution: T.D.B., L.Z., L.S., R.K., G.C., K.G., R.T., S.G., I.M., T.J., X.J., R.P., and K.B. performed research; Y.Y. analyzed the data and provided statistical support; A.P., J.B., and S.K. contributed samples and gene expression data; U.S., C.S., W.J., G.L., P.K., and A.L. contributed samples and reagents; and T.D.B., M.B., and A.V. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Amit Verma, Chanin 302B, Albert Einstein Cancer Center, 1300 Morris Park Ave, Bronx, NY 10461; e-mail: averma@aecom.yu.edu; or Markus Bitzer, Internal Medicine/Nephrology, University of Michigan, MSRB 2, 1570C, 1150 W. Medical Center Dr, Ann Arbor, MI 48105-5676; e-mail: markusbi@med.umich.edu.
References
Author notes
T.D.B., L.Z., and L.S. contributed equally to this study.




![Figure 5. Treatment with mir21 inhibitor leads to an increase in red blood cells in TGF-β transgenic mice. (A) Mice expressing TGF-β (Alb/TGFβ+) were treated with LNA-modified anti–miR-21 (n = 7) or mismatched placebo controls (n = 6) for 5 doses. Mice treated with anti–miR-21 oligos had a significant increase in hematocrit (HCT, Student’s t-test, P = .03), hemoglobin (Hgb [gm/DL], Student’s t-test, P = .049), and red blood cells (RBCs [×106], Student’s t-test, P = .0005). (B) Equal numbers of bone marrow cells were grown in methylcellulose and demonstrated increased erythroid colony formation in the mice treated with miR-21 inhibitor (n = 5 in each group, Student’s t-test, P = .04. (C) Representative bone marrow biopsy samples from both groups show an increase in SMAD7 levels in the treatment group with a corresponding decrease in phospho-SMAD2.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/15/10.1182_blood-2011-12-397067/4/m_2875f5.jpeg?Expires=1767751429&Signature=DaEeDEf~jW~sbiRa1u-dPTrY2Yl8yZnY6nK0xIkpNjq9pBJG7o3HwYzlybseanDSJ69z11jn-FHpXA89PPyRIyIu31kH3D65McZMtzjoIKKuD5bwwOct7XmjuTc7Qio8ft2gZnbD04ri2G6iXo1P98WxWO20g6sgCIJpEJYTeb1SSUM0AYyc2vgJeU8i8uGv6eL60OmW~fTJhzIyzJkSjVDC84lsGZZdbEN0Uh9TBGc68LPEaxvPSRtTYiJEOsJcF1vKp12UkUuDQlpOdddz~NCgB6KvirKf3JOAMicIKlZ5l6X-fb6wOLj~XJsg~mL3JklMARFDpEMTcSgqjHRinw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal