Key Points
Infant acute lymphoblastic leukemia is sensitive to therapeutic targeting by apoptosis, necoptosis, and autophagy activation whether MLL is rearranged or germline.
The disease-specific form of triple death mode killing by obatoclax overcomes the intrinsic resistance of MLL-rearranged infant acute lymphoblastic to cell death.
Abstract
Survival in infants younger than 1 year who have acute lymphoblastic leukemia (ALL) is inferior whether MLL is rearranged (R) or germline (G). MLL translocations confer chemotherapy resistance, and infants experience excess complications. We characterized in vitro sensitivity to the pan-antiapoptotic BCL-2 family inhibitor obatoclax mesylate in diagnostic leukemia cells from 54 infants with ALL/bilineal acute leukemia because of the role of prosurvival BCL-2 proteins in resistance, their imbalanced expression in infant ALL, and evidence of obatoclax activity with a favorable toxicity profile in early adult leukemia trials. Overall, half maximal effective concentrations (EC50s) were lower than 176 nM (the maximal plasma concentration [Cmax] with recommended adult dose) in 76% of samples, whether in MLL-AF4, MLL-ENL, or other MLL-R or MLL-G subsets, and regardless of patients’ poor prognostic features. However, MLL status and partner genes correlated with EC50. Combined approaches including flow cytometry, Western blot, obatoclax treatment with death pathway inhibition, microarray analyses, and/or electron microscopy indicated a unique killing mechanism involving apoptosis, necroptosis, and autophagy in MLL-AF4 ALL cell lines and primary MLL-R and MLL-G infant ALL cells. This in vitro obatoclax activity and its multiple killing mechanisms across molecular cytogenetic subsets provide a rationale to incorporate a similarly acting compound into combination strategies to combat infant ALL.
Introduction
Chemotherapy resistance and exaggerated age-related vulnerability to complications make infant acute lymphoblastic leukemia (ALL) extremely difficult to cure with intensive modern-era treatments (Z.E.D. et al, manuscript in preparation).1-3 More-efficacious, better-tolerated therapies are urgent. MLL translocations, which occur in 75% of ALL in infants younger than 1 year, are associated with poor outcomes, but survival in MLL-germline (G) cases also is inferior compared with ALL in children younger than 1 year (Z.E.D. et al, manuscript submitted).1,2
The most common MLL translocation in infant ALL, MLL-AF4 (MLLT2, AFF1), confers resistance to cell death.4 Antiapoptotic BCL-2 protein in MLL-rearranged (MLL-R) infant ALL is abundant, and BCL-2 antisense sensitized MLL-AF4 cell lines to die.5 Additional BCL-2 family members (eg, MCL-1, BCL-XL) that downregulate intrinsic apoptosis by forming complexes with proapoptotic BAX, BAK, and BH3-only proteins also promote leukemia cell survival.6 In MLL-R infant ALL, high MCL-1 mRNA expression correlated with in vitro prednisone resistance.7 MLL-AF4 targeting siRNAs decreased BCL-XL expression and increased apoptosis in MLL-AF4 ALL cell lines.8 BCL-XL antisense enhanced etoposide-induced apoptosis in SEM-K2 cells with this translocation in a xenograft model.9
The pan-antiapoptotic BCL-2 family small molecule inhibitor obatoclax mesylate (GeminX Pharmaceuticals, Malvern, PA; now an indirect, wholly owned subsidiary of Teva Pharmaceutical Industries Ltd.) binds the BH3-binding pocket and antagonizes a broad spectrum of prosurvival BCL-2 proteins.6 Obatoclax exhibited preclinical activity and synergy with chemotherapy in various solid tumors, leukemias, and lymphomas (reviewed in Brown and Felix10 ). Obatoclax was well-tolerated with minimal toxicities in early adult trials and, as monotherapy, induced an 8-month complete remission of MLL-R secondary AML.11
Despite MCL-1/BAK and BCL-XL/BAK dimer disruption and induction of apoptosis,12 recent studies suggested heterogeneity and cell type specificity in obatoclax killing mechanisms. Obatoclax killing of Bax−/−/Bak−/− mouse embryo fibroblasts implicated alternative death pathway activation.13 Obatoclax caused death receptor-mediated apoptosis or autophagy in pancreatic carcinoma14 and autophagy-dependent necroptosis when combined with dexamethasone in steroid-resistant pediatric ALL.15
Obatoclax activity and molecular targets have not been reported in detail in infant ALL. In a large sampling of cases, we find that primary infant ALL cells are obatoclax-sensitive and vulnerable to obatoclax-induced cytotoxicity in a MLL partner-gene-dependent manner. Moreover, for the first time, we describe a highly novel triple killing mechanism of obatoclax across primary MLL-R and MLL-G infant ALL cells that uses apoptosis, necroptosis, and autophagy. Both early in vitro activity data from a small number of these cases and the synergy data that we show here justified including a MLL-R stratum in a phase 1 pediatric trial (registered at www.clinicaltrials.gov as #NCT00933985). With the recent company transfer of obatoclax, this clinical trial stopped enrolling, and the MLL-R stratum did not open, but these results provide an increasing rationale to target abnormal cell death regulation in infant ALL with a similarly acting compound.
Materials and methods
The Children’s Hospital of Philadelphia Institutional Review Board approved this research.
Patient samples and cell lines
Cryopreserved pretreatment diagnostic bone marrow, peripheral blood, or apheresis samples from 54 infants (age, 1-365 days; mean, 169 days) with acute leukemia (52 ALL, 2 bilineal) selected on the basis of availability were received deidentified from the Children’s Oncology Group (COG) P9407 trial (n = 48; 47 treatment eligible) or obtained from the Children’s Hospital of Philadelphia (n = 6). WBC counts ranged from 15.3 to 1175 × 103/μL (mean, 452 × 103/μL). Molecular-cytogenetic classification by MLL status and partner genes was described.5,16 An apheresis sample from a 6.5-year-old boy (WBC, 408 × 103/μL) with MLL-AF4 ALL was obtained from the Children’s Hospital of Philadelphia. Mononuclear cells were enriched by Ficoll-Paque (Amersham, Pittsburgh, PA) centrifugation before cryopreservation of diagnostic specimens. Unstimulated peripheral blood mononuclear cells (PBMCs) collected by apheresis from a healthy adult were purchased from the University of Pennsylvania Human Immunology Core and cryopreserved before use. MLL-AF4 ALL cell lines RS4:11 and SEM-K2 were maintained as described.5
MTT assays
Primary leukemia cells/PBMCs were thawed, acclimated briefly, plated at 2 × 106 cells/mL in RPMI-1640 (Invitrogen, Grand Island, NY) with 20% serum substitute (BIT 9500; StemCell Technologies, Vancouver, BC, Canada) and 10 ng/mL interleukin 7 and stem cell factor (R&D Systems, Minneapolis, MN) at 37°C/5% carbon dioxide, and treated for 72 hours with obatoclax (courtesy GeminX Pharmaceuticals). Obatoclax–chemotherapy combinations were evaluated in primary MLL-AF4 ALL cells treated for 72 hours with doxorubicin (ADR), cytosine arabinoside, etoposide, dexamethasone, vincristine (Sigma-Aldrich, St. Louis, MO) or L-asparaginase (Merck, Whitehouse Station, NJ) at increasing concentrations alone or combined with fixed obatoclax doses.
For genetic autophagy inhibition, 5 × 106 log phase SEM-K2 cells were transfected with 1-5 μg Dharmacon (Waltham, MA) ON-TARGETplus BECN1 siRNA #1 (5′-GGAACUCACAGCUCCAUUA-3′; J-010552-06), #2 (5′-CUAAGGAGCUGCCGUUAUA-3′; J-010552-07), or a nontargeting control siRNA (D-001810-01) using a Nucleofector Kit R for Cell Lines, program T16 (Amaxa Biosystems, Allendale, NJ), and the cells were then incubated overnight before plating. Twenty-four hours later (48 hours after nucleofection), the cells were treated with vehicle or obatoclax for 24, 48, or 72 hours for BECN1 Western blot analysis or for 72 hours for MTT [(3–4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide] assays.
Cell lines were plated at 0.5 × 106 cells/mL, acclimated for 1 day, and treated for 72 hours with vehicle, ADR, or obatoclax alone or with 3-methyladenine (3-MA; Sigma-Aldrich), Necrostatin-1 (Nec-1; Sigma-Aldrich), and/or zVAD-fmk (Promega, Madison, WI). MTT assays were performed to ensure that chemical cell death inhibitor exposures were minimally cytotoxic (see supplemental Figure 1A on the Blood website). Primary infant ALL cells, plated as described, were treated with obatoclax combined with inhibitors at minimally cytotoxic concentrations (supplemental Figure 1B).
MTT assays were performed according to instructions. After background signal (media control) subtraction, data were normalized to vehicle for single-agent obatoclax and obatoclax–chemotherapy combinations; to vehicle-treated, siRNA-transfected cells for assays using siRNAs; or to cells treated with inhibitor or inhibitor combinations to account for any toxicity resulting from the inhibitors for assays combining obatoclax with chemical cell death inhibition.
Half maximal effective concentrations (EC50s) of obatoclax in diagnostic infant samples and PBMCs were calculated on the basis of cell survival in MTT assays by generating an inhibitory sigmoid Emax model (1.0 top down to 0.0 bottom, variable slope), using GraphPad Prism (version 4.03; La Jolla, CA). In 2 cases, EC50s could not be obtained with this model and the data were fitted with an inhibitory sigmoid Emax model (top down to bottom, variable slope), using Prism, or with an inhibitory sigmoid Emax model, using WinNonlin version 4.1 (Pharsight, Cary, NC). Box and whisker plots of EC50s in primary infant cases sorted by MLL rearrangement status and partner genes were generated using R statistical software.17 EC50s in MLL-AF4 vs other subgroups and in different clinical subsets were compared by Wilcoxon rank sum test. Survival curves were estimated using the Kaplan-Meier method and standard errors.
For obatoclax–chemotherapy combinations, WinNonlin was used to construct inhibitory sigmoid Emax models and determine single-agent EC50s and Hill coefficients. Combination indices (CIs) were calculated according to Loewe additivity criteria to determine synergy. Three-dimensional scatter plots were generated using the modeling software CombiTool (version 2.001; Bäretswil, Switzerland) to graph CIs of actual drug interactions in MTT assays and predicted Loewe additivity zero-interaction response surfaces.5
A sigmoid Emax model (top down to bottom, variable slope) within Prism was used to calculate EC50s for all cell line assays. Obatoclax–chemical inhibitor combinations were compared in Excel (Microsoft, Redmond, Washington) by t-test with 2-tailed distribution.
Flow cytometry
Cells lines plated at 0.5 × 106 cells/mL and treated 24 hours later with vehicle, ADR, or obatoclax were collected at specified times and prepared for analysis on a BD FACSCalibur (BD Biosciences, Sparks, MD).5 Propidium iodide (PI) uptake, activated caspase 3, and terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assay data were analyzed using CellQuest Pro version 5.2 (BD Biosciences). Cell cycle was modeled in live cells, using ModFit LT (Verity Software House, Topsham, ME). Differences were analyzed using a general linear model procedure with SAS software (version 9.2; Cary, NC) and Tukey’s post hoc pairwise comparisons.
Primary patient cells plated at 2 × 106 cells/mL were treated with vehicle or obatoclax, collected at specified times, and washed twice with 2% fetal bovine serum/phosphate-buffered saline for light scatter analysis on a BD FACSCalibur. Contour plots were generated using CellQuest.
Western blot analyses
Either cell lines were plated at 0.5 × 106 cells/mL, treated 24 hours later with vehicle, ADR, or obatoclax alone or with Nec-1 and/or zVAD-fmk alone or together, and collected at times specified; or siRNA-transfected cells were plated, treated, and collected as described earlier. Protein in whole-cell lysates was measured using a DC protein assay kit (BioRad, Hercules, CA). Proteins were separated on NuPAGE gels (Invitrogen), transferred and probed with antibodies to LC3B (Cell Signaling Technology, Danvers, MA; or gift from Thompson laboratory), p62 (Santa Cruz Biotechnology, Danvers, MA; Abgent, San Diego, CA; or Cell Signaling), PARP (BD Pharmigen, Sparks, MD), BECN1 (Santa Cruz Biotechnology), or ACTB (Sigma-Aldrich). Species-specific horseradish perioxidase–conjugated secondary antibodies were used for signal detection with ECL-plus (Amersham). Primary patient cells plated at 2 × 106 cells/mL were treated with vehicle, obatoclax, or ADR for Western blot analyses.
Microarray analysis
Cell lines were plated at 106 cells/mL; 24 hours later, 10 × 106 cells were treated with vehicle or obatoclax at the 72-hour EC50 or 3 × EC50 for 6 hours (2 replicates/condition). Cells were lysed with TRIzol (Invitrogen), and RNA was isolated and cRNA prepared for U133_Plus_2 (Affymetrix, Santa Clara, CA) microarray chip analysis.18 Differentially expressed genes (analysis of variance, P ≤ .05; >50% up-/downregulation) were identified by comparing mean log2 expression for obatoclax treatment vs vehicle.
Heatmaps were generated by 2-way clustering, using Euclidian distance. Autophagy and necroptosis probesets are not annotated in U133_Plus_2 arrays. Autophagy-related probesets were identified by downloading array gene ontology (GO) annotations from R/Bioconductor (http://www.bioconductor.org) and extracting 60 probeset identifications associated with any GO term descendant of GO:006914 (autophagy) in the GO hierarchy. Necroptosis-associated probesets (n = 294) were retrieved by matching annotations with those in Vandenabeele et al.19
Electron microscopy
SEM-K2 cells plated at 106 cells/mL or primary patient cells plated at 2 × 106 cells/mL were treated with vehicle, ADR, or obatoclax for specified times and washed with warm phosphate-buffered saline. In a primary case, Ficoll-Paque (Amersham) centrifugation was performed a second time after thawing before treatment. Pellets were fixed with prewarmed fixative (2.5% glutaraldehyde, 2% paraformaldehyde in sodium cacodylate) for 1 hour and processed.20 Ultrathin sections were examined with a Jeol (Peabody, MA) JEM-1010 electron microscope fitted with an Advanced Microscopy Techniques (AMT) (Woburn, MA) digital camera with dedicated image acquisition software.
Results
Broad-spectrum single-agent in vitro obatoclax activity across clinical and MLL-R and MLL-G subsets of infant ALL
Table 1 summarizes the clinical covariates and MLL status/partner genes in the 52 infant ALL and 2 infant bilineal acute leukemia cases. High-risk patients were overrepresented, with 33% being 90 days old or younger at diagnosis, 98% having a WBC of 50 × 103/μL or greater, and 87% having MLL-R leukemia (Z.E.D. et al, manuscript submitted).16 The mean obatoclax EC50 in all 54 cases was 136 nM. Obatoclax showed broad-spectrum single-agent in vitro activity across samples from all clinical risk groups, including those from infants 90 days old or younger, and regardless of high WBC or MLL-R vs MLL-G status (Table 1; Figure 1A). The Cmax with the recommended adult phase 2 dose (28 mg/m2 by intravenous infusion administered over 3 hours) was 72.9 ng/mL (176 nM; coefficient of variation 44%),11,21 suggesting that clinically achievable adult exposures could be cytotoxic against infant ALL in vitro. Using a cutoff of EC50 lower than 176 nM, most cases (41/54; 76%) in all 4 molecular cytogenetic subgroups (19/28 MLL-AF4; 10/11 MLL-ENL; 6/8 other MLL-R; and 6/7 MLL-G) were obatoclax sensitive (Figure 1A-B). However, MLL translocation status and partner genes affected EC50. EC50s in MLL-AF4 cases were significantly higher than in the remainder of cases combined (P = .037), as well as in the MLL-ENL subgroup (P = .044; Figure 1B; Table 1).
Association of obatoclax sensitivity in infant ALL/bilineal acute leukemia with clinical covariates and molecular/cytogenetic characteristics
| . | N (%) . | EC50 [nM] range/mean/median . | Wilcoxon P value . |
|---|---|---|---|
| Total | 54 (100) | 10-834/ 136/ 75.5 | |
| Age | .5327 | ||
| ≤90 days | 18 (33) | 10-834/ 152/ 87 | |
| >90 days | 36 (67) | 92-365/ 145/ 86 | |
| Sex | .3561 | ||
| Male | 33 (61) | 10-834/ 143/ 98 | |
| Female | 21 (39) | 22-550/ 126/ 58 | |
| WBC | NA | ||
| ≥50 × 103/μL | 53 (98) | 10-834/ 136/ 75 | |
| <50 × 103/μL | 1 (2) | 163 | |
| MLL Status | .8368; MLL-R vs G | ||
| Rearranged | 47 (87) | 10-834/ 135/ 75 | |
| MLL-AF4(AFF1) | 28 (52) | 26-834/ 168/ 109 | .03774; MLL-AF4 vs MLL-ENL/Other MLL-R/MLL-G .04409; MLL-AF4 vs MLL-ENL |
| MLL-ENL(MLLT1) | 11 (20) | 13-294/ 76/ 67 | |
| Other MLL-R* | 8 (15) | 10-356/ 101/ 41 | |
| Germline | 7 (13) | 31-488/ 145/ 78 | |
| Event** | .3274 | ||
| Yes | 32 (68) | 13-435/ 116/ 100 | |
| No | 15 (32) | 10-488/ 116/ 53 |
| . | N (%) . | EC50 [nM] range/mean/median . | Wilcoxon P value . |
|---|---|---|---|
| Total | 54 (100) | 10-834/ 136/ 75.5 | |
| Age | .5327 | ||
| ≤90 days | 18 (33) | 10-834/ 152/ 87 | |
| >90 days | 36 (67) | 92-365/ 145/ 86 | |
| Sex | .3561 | ||
| Male | 33 (61) | 10-834/ 143/ 98 | |
| Female | 21 (39) | 22-550/ 126/ 58 | |
| WBC | NA | ||
| ≥50 × 103/μL | 53 (98) | 10-834/ 136/ 75 | |
| <50 × 103/μL | 1 (2) | 163 | |
| MLL Status | .8368; MLL-R vs G | ||
| Rearranged | 47 (87) | 10-834/ 135/ 75 | |
| MLL-AF4(AFF1) | 28 (52) | 26-834/ 168/ 109 | .03774; MLL-AF4 vs MLL-ENL/Other MLL-R/MLL-G .04409; MLL-AF4 vs MLL-ENL |
| MLL-ENL(MLLT1) | 11 (20) | 13-294/ 76/ 67 | |
| Other MLL-R* | 8 (15) | 10-356/ 101/ 41 | |
| Germline | 7 (13) | 31-488/ 145/ 78 | |
| Event** | .3274 | ||
| Yes | 32 (68) | 13-435/ 116/ 100 | |
| No | 15 (32) | 10-488/ 116/ 53 |
NA, not applicable
Rearrangements include 2 MLL-AF9 (MLLT3), 1 MLL-EPS15, 1 MLL-ASAH3, and 4 MLL-R, not AF4, ENL, or AF9.
47 COG P9407 treatment-eligible cases; events include induction failure, induction death, relapse, remission death, and second malignancy.
MTT assays showing broad-spectrum single-agent obatoclax cytotoxicity in primary infant ALL. (A) Surviving fraction plots of MTT assay data by MLL subsets in diagnostic leukemia samples from 54 cases of primary infant ALL (n = 52) or infant bilineal acute leukemia (n = 2). Cells were treated for 72 hours with increasing obatoclax concentrations. Experiments were performed 1-3 times per patient sample (3-6 replicates/condition per experiment). (B) Box and whisker plots of EC50 values ascertained by inhibitory sigmoid Emax models of MTT assay data in 54 cases shown in (A). Significant differences between EC50 values were determined by Wilcoxon’s test (Table 1). (C) Kaplan-Meier curves showing similar EFS among infants with pretreatment diagnostic ALL samples in obatoclax-sensitive and obatoclax-resistant categories defined using a 176-nM cutoff. (D) Surviving fraction plot of MTT assay data for PBMCs after 72 hours of treatment with increasing obatoclax concentrations; assay was performed twice (1-3 replicates/condition per experiment).
MTT assays showing broad-spectrum single-agent obatoclax cytotoxicity in primary infant ALL. (A) Surviving fraction plots of MTT assay data by MLL subsets in diagnostic leukemia samples from 54 cases of primary infant ALL (n = 52) or infant bilineal acute leukemia (n = 2). Cells were treated for 72 hours with increasing obatoclax concentrations. Experiments were performed 1-3 times per patient sample (3-6 replicates/condition per experiment). (B) Box and whisker plots of EC50 values ascertained by inhibitory sigmoid Emax models of MTT assay data in 54 cases shown in (A). Significant differences between EC50 values were determined by Wilcoxon’s test (Table 1). (C) Kaplan-Meier curves showing similar EFS among infants with pretreatment diagnostic ALL samples in obatoclax-sensitive and obatoclax-resistant categories defined using a 176-nM cutoff. (D) Surviving fraction plot of MTT assay data for PBMCs after 72 hours of treatment with increasing obatoclax concentrations; assay was performed twice (1-3 replicates/condition per experiment).
There was no difference between obatoclax EC50s in pretreatment diagnostic ALL samples from 32 treatment-eligible patients who experienced events on the COG P9407 trial vs EC50s in 15 who did not (Table 1). This suggests that even in cases where events occur with intensified infant ALL therapy, the cells could be sensitive to obatoclax killing. Conversely, using the 176-nM cutoff to define sensitivity, the Kaplan-Meier plots in Figure 1C indicate similar event-free survival (EFS) among infants with pretreatment diagnostic ALL samples in obatoclax-sensitive (n = 37) and obatoclax-resistant (n = 10) categories.
The obatoclax EC50 in normal subject PBMCs was 805 nM (Figure 1D), suggesting that higher doses than those needed for cytotoxicity against infant ALL cells would be needed for toxicity to occur in normal blood cells.
Extensive in vitro synergy of obatoclax with chemotherapy in primary pediatric MLL-AF4 ALL cells
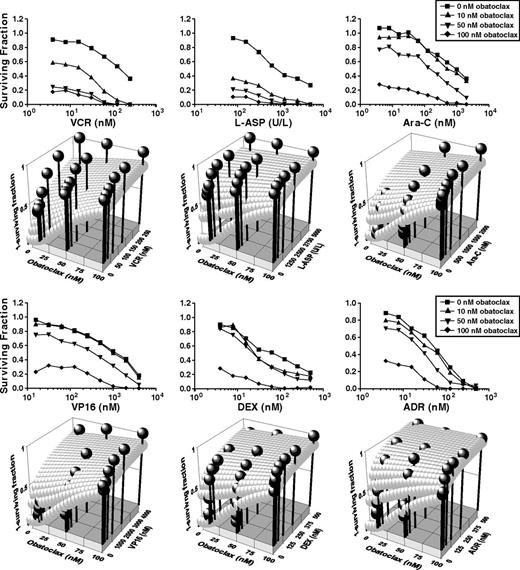
Combination effects of obatoclax with chemotherapies used for infant ALL were determined in MTT assays of primary pediatric MLL-AF4 ALL cells with a single-agent obatoclax EC50 (67 nM), similar to many infant cases. Surviving fraction plots indicated reduced EC50s for all 6 obatoclax–chemotherapy combinations compared with chemotherapy alone (Figure 2, rows 1 and 3). Response surface models of experimental CIs based on Loewe additivity criteria indicated synergy (CI < 1; black spheres above zero interaction plane) at multiple concentrations for every obatoclax–chemotherapy combination tested (Figure 2, rows 2 and 4).
Synergy between obatoclax and chemotherapy against primary pediatric MLL-AF4 ALL. Cells were treated with obatoclax alone, indicated chemotherapy drug alone, or increasing concentrations of chemotherapy drug combined with obatoclax at fixed concentrations. Data from MTT assays performed 72 hours after treatment of each combination are shown in surviving fraction plots (first and third rows) and response surface models (second and fourth rows). Each experimental point represents average of 2 independent experiments (3 replicates/condition per experiment). An inhibitory sigmoid Emax model was used to determine the EC50 and Hill coefficient for single-agent dose responses (obatoclax: EC50 = 67.2 ± 13 nM, Hill coefficient = 1.91 ± 0.60; vincristine: EC50 = 150 ± 35 nM, Hill coefficient = 0.911 ± 0.160; L-aspariginase: EC50 = 958 ± 123 U/L, Hill coefficient = 0.785 ± 0.067; etoposide: EC50 = 1125 ± 210 nM, Hill coefficient = 0.76 ± 0.12; doxorubicin: EC50 = 47.4 ± 5.99 nM, Hill coefficient = 1.13 ± 0.13; cytosine arabinodise: EC50 = 693 ± 119 nM, Hill coefficient = 0.70 ± 0.10; dexamethasone: EC50 = 62.6 ± 8.37 nM, Hill coefficient = 0.64 ± 0.05). Three-dimensional scatter plots show Loewe additivity zero-interaction response surfaces for each obatoclax–chemotherapy combination (gray spheres) derived from single-agent experiments. If actual experimental combination effects (black spheres) are above zero-interaction response surface, combination is synergistic; if below, antagonistic; and if on the response surface, additive. Note synergistic interactions between obatoclax and each chemotherapy agent tested. ADR, doxorubicin; Ara-C, cytosine arabinodise; DEX, dexamethasone; L-ASP, L-aspariginase; VCR, vincristine; VP16, etoposide.
Synergy between obatoclax and chemotherapy against primary pediatric MLL-AF4 ALL. Cells were treated with obatoclax alone, indicated chemotherapy drug alone, or increasing concentrations of chemotherapy drug combined with obatoclax at fixed concentrations. Data from MTT assays performed 72 hours after treatment of each combination are shown in surviving fraction plots (first and third rows) and response surface models (second and fourth rows). Each experimental point represents average of 2 independent experiments (3 replicates/condition per experiment). An inhibitory sigmoid Emax model was used to determine the EC50 and Hill coefficient for single-agent dose responses (obatoclax: EC50 = 67.2 ± 13 nM, Hill coefficient = 1.91 ± 0.60; vincristine: EC50 = 150 ± 35 nM, Hill coefficient = 0.911 ± 0.160; L-aspariginase: EC50 = 958 ± 123 U/L, Hill coefficient = 0.785 ± 0.067; etoposide: EC50 = 1125 ± 210 nM, Hill coefficient = 0.76 ± 0.12; doxorubicin: EC50 = 47.4 ± 5.99 nM, Hill coefficient = 1.13 ± 0.13; cytosine arabinodise: EC50 = 693 ± 119 nM, Hill coefficient = 0.70 ± 0.10; dexamethasone: EC50 = 62.6 ± 8.37 nM, Hill coefficient = 0.64 ± 0.05). Three-dimensional scatter plots show Loewe additivity zero-interaction response surfaces for each obatoclax–chemotherapy combination (gray spheres) derived from single-agent experiments. If actual experimental combination effects (black spheres) are above zero-interaction response surface, combination is synergistic; if below, antagonistic; and if on the response surface, additive. Note synergistic interactions between obatoclax and each chemotherapy agent tested. ADR, doxorubicin; Ara-C, cytosine arabinodise; DEX, dexamethasone; L-ASP, L-aspariginase; VCR, vincristine; VP16, etoposide.
Unique, mixed triple killing mechanism of obatoclax in MLL-AF4 ALL cell lines
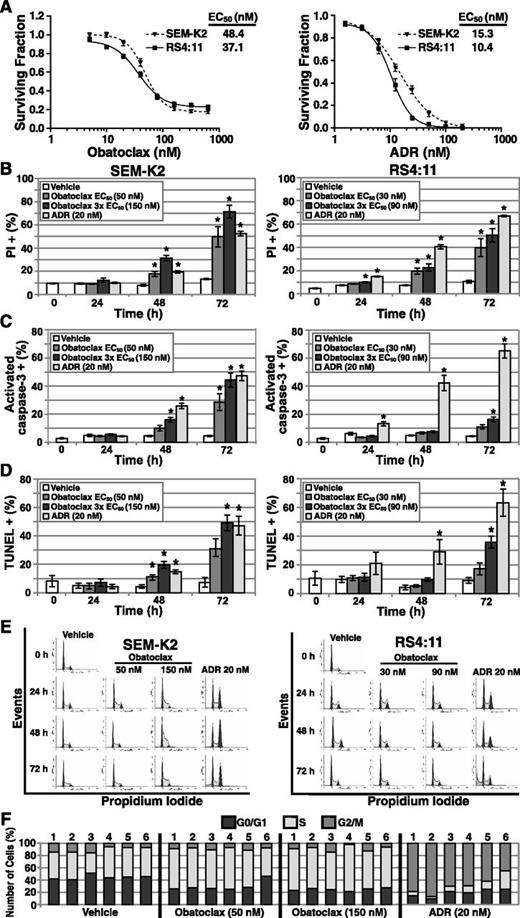
Obatoclax activity notwithstanding, its mechanism of killing infant ALL cells remained undefined. First the mechanism was investigated in the MLL-AF4 ALL cell lines SEM-K2 and RS4:11, which, similar to infant ALL,5,7 showed abundant antiapoptotic BCL-2 and MCL-1 expression (supplemental Figure 2). Furthermore, 72-hour single-agent obatoclax EC50s (SEM-K2, 48.4 nM; RS4:11, 37.1 nM) were similar to low nM EC50s in many primary infant cases (Figure 3A). Respective EC50s of ADR, the control for apoptosis,5 were 15.3 nM and 10.4 nM in SEM-K2 and RS4:11, respectively (Figure 3A). As confirmation of cell death, PI uptake increased significantly in a time- and dose-dependent manner in both cell lines treated with obatoclax at the 72-hour EC50 or 3 × EC50, similar to with high-dose ADR (Figure 3B). The potent obatoclax-induced cytotoxicity and death measured by MTT assays and PI uptake provided the basis to investigate the killing mechanism.
Flow cytometric assays showing induction of death, apoptosis, and cell cycle changes by single-agent obatoclax in MLL-AF4 ALL cell lines. (A) Surviving fraction plots of cell viability measured by MTT assays at 72 hours after obatoclax or ADR (positive control for apoptosis) exposure. Assays were performed at least 3 times (6 replicates/condition per experiment). Error bars indicate standard error. Results from MTT assays at 72 hours determined the low and high obatoclax (EC50; 3 × EC50) concentrations and the ADR concentration that were used in time-course flow cytometric assays (B-E). (B) Flow cytometric assays of cell death measured by PI fluorescence uptake in vehicle-, obatoclax-, or ADR-treated cell lines. Average percentage of dead (PI positive) cells from 5 independent experiments are plotted. (C) Flow cytometric assays of activated caspase 3 in vehicle-, obatoclax-, or ADR-treated cell lines. Activated caspase 3 positive cells were gated to determine percentages of apoptotic cells. Graphs represent average percentage of caspase 3 positive cells from 5 independent experiments. (D) FACS TUNEL assay of DNA fragmentation in vehicle-, obatoclax-, or ADR-treated cell lines. TUNEL-positive cells were gated to determine percentages. The average percentage of TUNEL positive cells from 4 independent experiments is plotted. Bars (B-D) show standard error. Asterisk indicates P ≤ .05 vs vehicle at individual points. (E) Representative cell cycle analyses in vehicle-, obatoclax-, or ADR-treated cell lines with cell cycle distribution determined using ModFit LT. (F) Bar graph plots of percentages of cells in G0/G1, S, and G2/M in vehicle-, obatoclax-, or ADR-treated SEM-K2 cells in 6 independent experiments.
Flow cytometric assays showing induction of death, apoptosis, and cell cycle changes by single-agent obatoclax in MLL-AF4 ALL cell lines. (A) Surviving fraction plots of cell viability measured by MTT assays at 72 hours after obatoclax or ADR (positive control for apoptosis) exposure. Assays were performed at least 3 times (6 replicates/condition per experiment). Error bars indicate standard error. Results from MTT assays at 72 hours determined the low and high obatoclax (EC50; 3 × EC50) concentrations and the ADR concentration that were used in time-course flow cytometric assays (B-E). (B) Flow cytometric assays of cell death measured by PI fluorescence uptake in vehicle-, obatoclax-, or ADR-treated cell lines. Average percentage of dead (PI positive) cells from 5 independent experiments are plotted. (C) Flow cytometric assays of activated caspase 3 in vehicle-, obatoclax-, or ADR-treated cell lines. Activated caspase 3 positive cells were gated to determine percentages of apoptotic cells. Graphs represent average percentage of caspase 3 positive cells from 5 independent experiments. (D) FACS TUNEL assay of DNA fragmentation in vehicle-, obatoclax-, or ADR-treated cell lines. TUNEL-positive cells were gated to determine percentages. The average percentage of TUNEL positive cells from 4 independent experiments is plotted. Bars (B-D) show standard error. Asterisk indicates P ≤ .05 vs vehicle at individual points. (E) Representative cell cycle analyses in vehicle-, obatoclax-, or ADR-treated cell lines with cell cycle distribution determined using ModFit LT. (F) Bar graph plots of percentages of cells in G0/G1, S, and G2/M in vehicle-, obatoclax-, or ADR-treated SEM-K2 cells in 6 independent experiments.
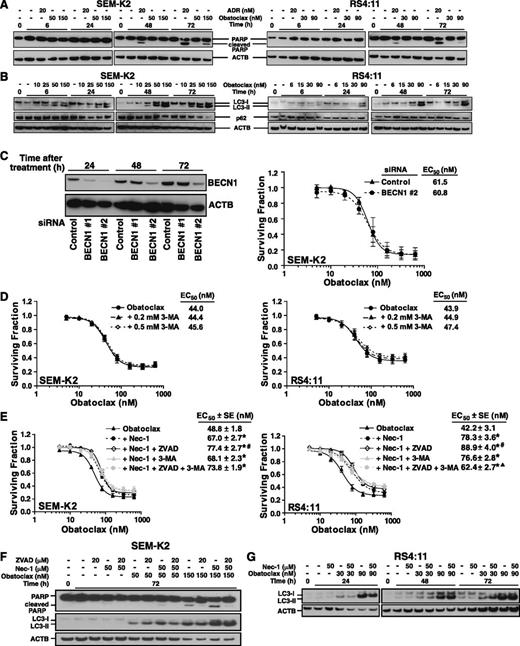
Obatoclax caused time- and dose-dependent increases in activated caspase 3 and TUNEL staining, which, respectively, indicate early- and late-stage apoptosis,22,23 although caspase 3 activation in RS4:11 cells was modest (Figure 3C-D; supplemental Figure 3). Obatoclax-treated SEM-K2 cells also showed time- and dose-dependent increases in S-phase cells (Figure 3E-F), whereas cell cycle was not appreciably altered by obatoclax in RS4:11 (Figure 3E). In contrast, ADR caused an increase in G2/M-phase cells, a decrease in S phase, and by 72 hours, a sub-G1 peak in both cell lines consistent with apoptosis (Figure 3E-F). Increased S-phase cells with obatoclax may promote synergy with cell cycle–dependent chemotherapy, as in Figure 2; however, the increased S-phase SEM-K2 cells were unlike the cell cycle changes with ADR and were not typical of apoptosis. In both cell lines, obatoclax caused time- and dose-dependent increases in PARP cleavage indicative of apoptosis, although the increases were smaller than with ADR (Figure 4A).
Western blot analyses and MTT assays of obatoclax effects on apoptosis, autophagy, and necroptosis by itself or combined with cell death pathway inhibition in MLL-AF4 ALL cell lines. (A) Time-course Western blot analysis of PARP cleavage using whole-cell lysates from SEM-K2 and RS4:11 cells after exposure to vehicle, or obatoclax at concentrations approximating 72-hour EC50 or 3 × EC50, or ADR (positive control for apoptosis) for indicated times. Increases in cleaved PARP by 72 hours with obatoclax treatment indicate apoptosis. (B) Time-course Western blot analyses of LC3-I to LC3-II conversion and p62 protein levels after exposure to vehicle or obatoclax at concentrations approximating 72-hour EC10, EC25, EC50, or 3 × EC50 for indicated times. Note time- and dose-dependent increases in LC3-I to LC3-II conversion and lack of p62 accumulation with obatoclax treatment. (C) Unchanged obatoclax-induced cytotoxicity with genetic autophagy inhibition in SEM-K2 cells. Representative Western blot depicting BECN1 protein knock down by BECN1 siRNAs #1 and #2 during obatoclax treatment compared with cells transfected with control siRNA. Cell lysates were prepared after obatoclax treatment of transfected cells for indicated times (left). Surviving fraction plots of cell viability measured by MTT assays performed after 72-hour obatoclax exposure (6 replicates/condition per experiment) in cells transfected with control siRNA or BECN1 siRNA 2 (right). The assay was performed 3 times. Bars indicate standard error. Transfection with BECN1 siRNA #1 gave the same result (not shown). (D) Unchanged obatoclax-induced cytotoxicity by chemical autophagy inhibition in SEM-K2 (left) and RS4:11 (right) cells. Surviving fraction plots show cell viability measured by MTT assays after 72-hour exposure to increasing obatoclax concentrations with indicated fixed 3-MA concentrations. Assays were repeated 4-7 times (6 replicates/condition per experiment). Bars indicate standard error. (E) Attenuation of obatoclax-induced cell death in SEM-K2 (left) and RS4:11 (right) cells by chemical necroptosis inhibition ± apoptosis and/or autophagy inhibition. Surviving fraction plots show cell viability measured by MTT assays 72 hours after treatment with increasing obatoclax concentrations ± 50 μM Nec-1 ± 20 μM zVAD-fmk and/or 0.5 mM 3-MA. MTT assays were performed 3-6 times (6 replicates/condition per experiment). Bars show standard error. * P ≤ .05 vs obatoclax alone; # P ≤ .05 vs obatoclax + Nec-1; ▲, P ≤ .05 vs obatoclax + Nec-1 + zVAD-fmk. Data in (D) and (E) were normalized to cells only treated with the relevant inhibitors alone or combined (supplemental Figure 1A). (F) Western blot analysis of SEM-K2 cells demonstrating dose-dependent increases in cleaved PARP indicative of more apoptosis with obatoclax treatment + Nec-1 for 72 hours compared with obatoclax alone, and reduction in obatoclax-induced PARP cleavage as well as attenuation of Nec-1 effect on PARP cleavage by zVAD-fmk (top). Also note the increase in LC3-I to LC3-II conversion induced by obatoclax in the presence of Nec-1 ± zVAD-fmk compared with obatoclax alone (bottom). Obatoclax concentrations are approximate 72-hour EC50 (50 nM) or 3 × EC50 (150 nM). Western blots are representative of 3 independent experiments. (G) Representative Western blot analyses of LC-I to LC-II conversion in RS4:11 cells treated with obatoclax at approximate 72-hour EC50 (30 nM) or 3 × EC50 (90 nM) ± Nec-1. Note increased (F) or unchanged (G) LC-I to LC-II conversion with Nec-1, indicating that obatoclax-induced autophagy was necroptosis-independent.
Western blot analyses and MTT assays of obatoclax effects on apoptosis, autophagy, and necroptosis by itself or combined with cell death pathway inhibition in MLL-AF4 ALL cell lines. (A) Time-course Western blot analysis of PARP cleavage using whole-cell lysates from SEM-K2 and RS4:11 cells after exposure to vehicle, or obatoclax at concentrations approximating 72-hour EC50 or 3 × EC50, or ADR (positive control for apoptosis) for indicated times. Increases in cleaved PARP by 72 hours with obatoclax treatment indicate apoptosis. (B) Time-course Western blot analyses of LC3-I to LC3-II conversion and p62 protein levels after exposure to vehicle or obatoclax at concentrations approximating 72-hour EC10, EC25, EC50, or 3 × EC50 for indicated times. Note time- and dose-dependent increases in LC3-I to LC3-II conversion and lack of p62 accumulation with obatoclax treatment. (C) Unchanged obatoclax-induced cytotoxicity with genetic autophagy inhibition in SEM-K2 cells. Representative Western blot depicting BECN1 protein knock down by BECN1 siRNAs #1 and #2 during obatoclax treatment compared with cells transfected with control siRNA. Cell lysates were prepared after obatoclax treatment of transfected cells for indicated times (left). Surviving fraction plots of cell viability measured by MTT assays performed after 72-hour obatoclax exposure (6 replicates/condition per experiment) in cells transfected with control siRNA or BECN1 siRNA 2 (right). The assay was performed 3 times. Bars indicate standard error. Transfection with BECN1 siRNA #1 gave the same result (not shown). (D) Unchanged obatoclax-induced cytotoxicity by chemical autophagy inhibition in SEM-K2 (left) and RS4:11 (right) cells. Surviving fraction plots show cell viability measured by MTT assays after 72-hour exposure to increasing obatoclax concentrations with indicated fixed 3-MA concentrations. Assays were repeated 4-7 times (6 replicates/condition per experiment). Bars indicate standard error. (E) Attenuation of obatoclax-induced cell death in SEM-K2 (left) and RS4:11 (right) cells by chemical necroptosis inhibition ± apoptosis and/or autophagy inhibition. Surviving fraction plots show cell viability measured by MTT assays 72 hours after treatment with increasing obatoclax concentrations ± 50 μM Nec-1 ± 20 μM zVAD-fmk and/or 0.5 mM 3-MA. MTT assays were performed 3-6 times (6 replicates/condition per experiment). Bars show standard error. * P ≤ .05 vs obatoclax alone; # P ≤ .05 vs obatoclax + Nec-1; ▲, P ≤ .05 vs obatoclax + Nec-1 + zVAD-fmk. Data in (D) and (E) were normalized to cells only treated with the relevant inhibitors alone or combined (supplemental Figure 1A). (F) Western blot analysis of SEM-K2 cells demonstrating dose-dependent increases in cleaved PARP indicative of more apoptosis with obatoclax treatment + Nec-1 for 72 hours compared with obatoclax alone, and reduction in obatoclax-induced PARP cleavage as well as attenuation of Nec-1 effect on PARP cleavage by zVAD-fmk (top). Also note the increase in LC3-I to LC3-II conversion induced by obatoclax in the presence of Nec-1 ± zVAD-fmk compared with obatoclax alone (bottom). Obatoclax concentrations are approximate 72-hour EC50 (50 nM) or 3 × EC50 (150 nM). Western blots are representative of 3 independent experiments. (G) Representative Western blot analyses of LC-I to LC-II conversion in RS4:11 cells treated with obatoclax at approximate 72-hour EC50 (30 nM) or 3 × EC50 (90 nM) ± Nec-1. Note increased (F) or unchanged (G) LC-I to LC-II conversion with Nec-1, indicating that obatoclax-induced autophagy was necroptosis-independent.
Obatoclax effects on the nonapoptotic cell death pathways autophagy and necroptosis24,25 also were examined. Autophagy was of interest because antiapoptotic BCL-2 proteins directly interact with and negatively regulate the key autophagy BH3-only protein BECN1.15,26-28 Death domain receptor engagement by ligand (tumor necrosis factor-α, CD95L [FASL], TNF-related apoptosis-inducing ligand [TRAIL; TNSF10]) activates RIP1 kinase–mediated necroptosis when intrinsic apoptosis is defective.29 Even though autophagy occurs independently of necroptosis, downstream death execution in necroptosis can occur through or without autophagy.30
In both cell lines, obatoclax caused time- and dose-dependent increases in LC3-I to LC3-II conversion indicative of LC3 lipidation during autophagy induction,24 and the lack of p62 accumulation suggested autophagic flux (Figure 4B).24 Thus, obatoclax effects on MLL-AF4 cell lines included potent cytotoxicity and some classical features of apoptosis and autophagy. Because autophagy can cause death or prolong cell survival,31 MTT experiments were performed that combined obatoclax with autophagy inhibition. Even though BECN1 siRNAs reduced BECN1 protein in SEM-K2 cells throughout obatoclax treatment, 72-hour obatoclax EC50s after BECN1 siRNA vs control siRNA transfection were similar (Figure 4C). In addition, the chemical autophagy inhibitor 3-MA (Figure 4D) and the pan-caspase inhibitor zVAD-fmk did not change the EC50 of obatoclax in either cell line. These results suggest that neither autophagy nor apoptosis was essential or rate-limiting to cell killing or that different death pathways were employed with their inhibition. The results also demonstrate that autophagy was not cytoprotective because obatoclax-induced death did not increase with autophagy inhibition.23
The RIP1 kinase inhibitor Nec-1 selectively prevents necroptosis, including autophagy, downstream of this pathway.25 In both cell lines, obatoclax-induced death was reduced significantly by Nec-1 alone or together with z-VAD-fmk and/or 3-MA and was decreased most by both RIP1 kinase and pan-caspase inhibition (Figure 4E).
There was more PARP cleavage consistent with more apoptosis in SEM-K2 cells, but not in RS4:11 cells treated with obatoclax in the presence of Nec-1 than with obatoclax by itself (Figure 4F). Additional Western blot analysis of SEM-K2 cells showed that, as expected, obatoclax-induced PARP cleavage was reduced significantly by z-VAD-fmk. Furthermore, exposure to obatoclax with Nec-1 and zVAD-fmk together blunted the increase in cleaved PARP associated with necroptosis inhibition (Figure 4F). There was some increased LC3-I to LC3-II conversion in SEM-K2 cells treated with obatoclax in the presence of Nec-1 compared with obatoclax alone, and a more pronounced increase indicative of more autophagy with both necroptosis and apoptosis inhibition (Figure 4F). In RS4:11 cells, obatoclax-induced LC3-I to LC3-II conversion was unaltered by necroptosis inhibition (Figure 4G). Because obatoclax-induced LC3-I to LC3-II conversion did not decrease in either cell line in the presence of Nec-1, the autophagy observed with obatoclax was not downstream of the necroptosis pathway.
Obatoclax induces apoptosis, autophagy, and necroptosis gene expression in MLL-AF4 ALL cell lines
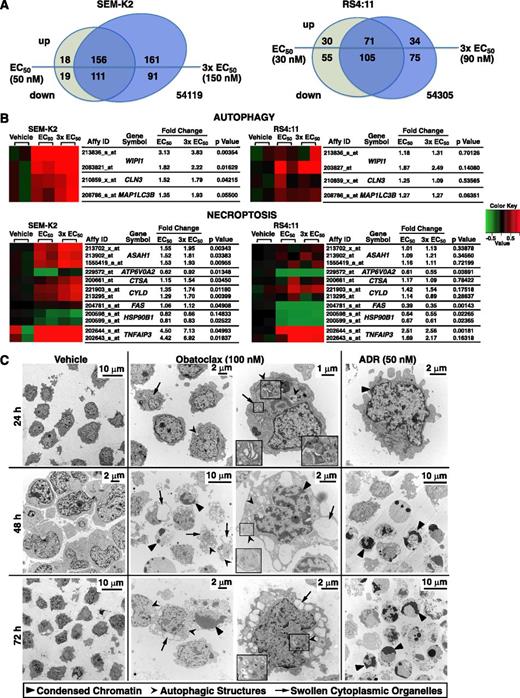
Obatoclax effects on gene expression were evaluated by microarray analysis to gain additional insights into this triple killing mechanism. Obatoclax treatment of 6 hours at the respective 72-hour EC50 or 3 × EC50 resulted in significant up- or downregulation of 891 probesets (GEO# GSE36149) in 1 or both cell lines (Figure 5A; supplemental Table 1). Changes in both cell lines were largely in the same direction (supplemental Figure 4).
Gene expression changes in SEM-K2 and RS4:11 cells and changes in cell morphology in SEM-K2 cells indicative of triple death mode killing by obatoclax. Cells were treated for 6 hours in duplicate with vehicle or obatoclax at approximate 72-hour EC50 or 3 × EC50 concentrations, and respective cDNAs were used for Affymetrix U133_Plus_2 microarray analysis. (A) Venn diagrams showing numbers of probesets up- or downregulated by obatoclax and their overlap between different obatoclax concentrations for each cell line. Significant changes were determined using differences between mean log2 expression levels in cells treated with obatoclax at EC50 or 3 × EC50 and mean log2 expression levels in vehicle-treated cells (analysis of variance, P ≤ .05 and >50% up-/downregulation in at least a single mean log2 expression level comparison of obatoclax at EC50 or 3 × EC50 vs vehicle in either cell line). For a complete list of significantly up- or downregulated probesets, see supplemental Table 1. (B) Mini heatmaps of autophagy- and necroptosis-related probesets showing upregulation (red) or downregulation (green) in expression at or near significance (P ≤ .05) with obatoclax treatment at EC50 or 3 × EC50 in either cell line. Expression levels for all 3 conditions (vehicle, obatoclax EC50, obatoclax 3 × EC50) and 2 biologic replicates per condition (6 samples) were drawn for each cell line; mean log2 expression levels of respective vehicle treatments were used as reference. (C) SEM-K2 cells treated with vehicle, obatoclax, or ADR (positive control for apoptosis) for indicated times were harvested, stained, and sectioned for electron microscopy. Indicated arrowheads mark condensed chromatin and/or nuclear fragmentation of apoptosis, autophagic structures, or swollen Golgi (obatoclax; 24 hours) or endoplasmic reticulum (obatoclax; 48 hours, 72 hours) of necroptosis. Magnified insets are in boxes. Note morphologic changes of all 3 modes of death in single cell (obatoclax, 48 hours; right middle panel).
Gene expression changes in SEM-K2 and RS4:11 cells and changes in cell morphology in SEM-K2 cells indicative of triple death mode killing by obatoclax. Cells were treated for 6 hours in duplicate with vehicle or obatoclax at approximate 72-hour EC50 or 3 × EC50 concentrations, and respective cDNAs were used for Affymetrix U133_Plus_2 microarray analysis. (A) Venn diagrams showing numbers of probesets up- or downregulated by obatoclax and their overlap between different obatoclax concentrations for each cell line. Significant changes were determined using differences between mean log2 expression levels in cells treated with obatoclax at EC50 or 3 × EC50 and mean log2 expression levels in vehicle-treated cells (analysis of variance, P ≤ .05 and >50% up-/downregulation in at least a single mean log2 expression level comparison of obatoclax at EC50 or 3 × EC50 vs vehicle in either cell line). For a complete list of significantly up- or downregulated probesets, see supplemental Table 1. (B) Mini heatmaps of autophagy- and necroptosis-related probesets showing upregulation (red) or downregulation (green) in expression at or near significance (P ≤ .05) with obatoclax treatment at EC50 or 3 × EC50 in either cell line. Expression levels for all 3 conditions (vehicle, obatoclax EC50, obatoclax 3 × EC50) and 2 biologic replicates per condition (6 samples) were drawn for each cell line; mean log2 expression levels of respective vehicle treatments were used as reference. (C) SEM-K2 cells treated with vehicle, obatoclax, or ADR (positive control for apoptosis) for indicated times were harvested, stained, and sectioned for electron microscopy. Indicated arrowheads mark condensed chromatin and/or nuclear fragmentation of apoptosis, autophagic structures, or swollen Golgi (obatoclax; 24 hours) or endoplasmic reticulum (obatoclax; 48 hours, 72 hours) of necroptosis. Magnified insets are in boxes. Note morphologic changes of all 3 modes of death in single cell (obatoclax, 48 hours; right middle panel).
As shown in the mini heatmaps (Figure 5B), analysis of autophagy-associated probesets suggested increased expression of WIPI1 (ATG18), MAP1LC3B (ATG8), and CLN3, gene products of which are autophagy mediators. Time-course treatment with obatoclax at the respective 72-hour EC50 or 3 × EC50 and quantitative reverse-transcriptase polymerase chain reaction confirmed WIPI1, MAP1LC3B, and CLN3 upregulation and showed increased BECN1 expression (supplemental Figure 5). MAP1LC3B (LC3-I) is converted to LC3-II through lipidation during autophagy induction.24 WIPI1 encodes the homolog of yeast Atg18, which forms distinct puncta at LC3-positive membrane structures.24 CLN3 aids in autophagosome/lysosome fusion.32
Analysis of published necroptosis genes19 indicated A20 (TNFAIP3), CYLD, ASAH1, and CTSA upregulation (Figure 5B). The acid ceramidase ASAH1 is important for necroptosis initiation.19 CYLD and A20 (TNFAIP3) are RIP1 deubiquitinases.19,29 CTSA is cathepsin and carboxyl pepsidase homologous to a Caenorhabditis elegans lysosomal protease family member released on lysosomal membrane permeabilization.19
In SEM-K2 cells treated at the 72-hour 3 × EC50 by annotation analysis, using the Database for Annotation, Visualization and Integrated Discovery (DAVID; National Institute of Allergy and Infectious Diseases, Bethesda, MD), the largest class of significantly upregulated genes (n = 145) encoded phosphoproteins (supplemental Table 2), many of which directly or indirectly are involved in cell death regulation. These genes included BCL-2L11 (BAM, BIM), CD44, ARHGEF2, SH3RF1, LYST, MAPK7, GADD34 (PPP1R15A), NEK6, NFKBIA, TGFbR1, TNFRSF14, AKT2, and VAV1; the autophagy genes SQSTM1 (p62) and CLN3; and the necroptosis gene A20 (TNFAIP3), further implicating activation/interplay of multiple pro- and antideath signals related to apoptosis, autophagy, or necroptosis (supplemental Table 3). Genes in vacuole (n = 15), lytic vacuole (n = 13), and lysosome (n = 13) classes including other autophagy and necroptosis genes also increased (supplemental Table 3). In RS4:11 cells, 19 upregulated phosphoprotein genes including the necroptosis gene A20 (TNFAIP3) were overlapping with those in SEM-K2 (supplemental Table 4). Thus, gene expression profiling gave further proof that obatoclax induces apoptosis, necroptosis, and autophagy in MLL-R ALL.
Morphologic evidence of apoptosis, necroptosis, and autophagy in obatoclax-treated SEM-K2 cells
Morphologic criteria were employed to define ultrastructural changes observed by electron microscopy over a course of obatoclax treatment in SEM-K2 cells. As early as 24 hours, obatoclax induced Golgi swelling indicative of necroptosis, as well as double membrane autophagic structures.23,24 By 48 and 72 hours, morphologic changes included chromatin condensation typical of apoptosis, autophagic structures (phagophores, autophagosomes, autophagolysosomes), and features of necroptosis such as prominent dilation and vesiculation of the endoplasmic reticulum compartment.23,24 Although features of one or another cell death mechanism sometimes were more prominent, features of 2 or all 3 modes of death occurred in single cells. In contrast, only prominent features of apoptosis including rounding-up of cells, chromatin condensation, and nuclear fragmentation occurred by 48 and 72 hours with ADR (Figure 5C). These findings further implicate multiple modes of killing by obatoclax, and the ADR control demonstrates intact apoptosis.
Triple killing mechanism of obatoclax in primary infant ALL cells
These results demonstrate that obatoclax is potently cytotoxic in MLL-AF4 ALL cell lines through apoptosis, necroptosis, and autophagy, rather than by simple disruption of BCL-2 family protein complexes.6 The next functional experiments investigated the mechanism of obatoclax in vitro killing in 6 primary diagnostic infant ALL samples (3 MLL-AF4, 1 MLL-ENL, 1 other MLL-R, and 1 MLL-G) included in Figure 1A-B. Single obatoclax concentrations likely to cause death were studied in flow cytometric (72-hour EC75) and Western blot (72-hour EC75 or EC50) analyses for sample conservation. Feasibility of certain flow cytometric assays in these primary samples also differed from in the cell lines. In 3 of 4 cases studied by flow cytometry (Figure 6A-D), the 72-hour EC75 was more than 150 nM; at these concentrations, spectral overlap of obatoclax fluorescence in multiple channels used for caspase 3 and TUNEL assays precluded signal specificity. Others previously used light side scatter (SSC) vs forward scatter (FSC) as an alternative to measure apoptosis in obatoclax-treated primary chronic lymphocytic leukemia cells because of the same phenomenon.33 Caspase 3 and TUNEL data on the cell lines nonetheless are sound because lower doses were employed and apoptosis was confirmed by other assays. Cell cycle analysis was not performed because of the inherently limited replication of primary cells in culture, unlike transformed cell lines.
Evidence for triple killing mechanism of obatoclax in primary infant ALL cells from flow cytometry, Western blot analyses, and MTT assays with cell death inhibitors. Molecular cytogenetic subtype and 72-hour obatoclax EC50s determined from surviving fraction plots in Figure 1 for each case are shown. In cases in (A-D), all 3 types of assays were performed; in 2 cases in (E), apoptosis and autophagy proteins were studied by Western blot. In the other MLL-R case in (C), AF4, AF9, and ENL partner genes were excluded.16 (A-D; left) show contour plots of FSC vs SSC in cells treated for indicated times at respective 72-hour obatoclax EC75 determined from Figure 1. Progressive decrease in FSC signal in all 4 cases indicates apoptosis. (A-D; middle) and (E) are Western blot analyses of apoptosis and autophagy. Note increases in cleaved PARP in all 6 cases in response to obatoclax consistent with apoptosis. Also note increases in LC3-II in all 6 cases in response to obatoclax (A-E); this occurs without increase in p62 in 4 cases in (A, D, E), indicating autophagy with autophagic flux. In MLL-AF4 case in (B), note increase in p62 at 24 hours but decrease by 48 hours after obatoclax treatment, suggesting that autophagy is not blocked. Also note increase in p62 at 48 hours in the other MLL-R case (C, middle panel), indicating either p62 induction or accumulation, the former of which is supported by gene expression profiling data in SEM-K2 (supplemental Table 2). (A-D; right) Surviving fraction plots for MTT assays 72 hours after obatoclax treatment at increasing concentrations by itself or with 50 μM Nec-1, 20 μM zVAD-fmk, or 0.5 mM 3-MA alone or altogether. Inhibitors were used at concentrations determined minimally toxic in cell lines as well as primary cases (supplemental Figure 1), rather than at case-by-case titrated concentrations, and were tested less extensively in obatoclax combinations. Data were normalized to respective primary samples only treated with relevant inhibitors alone or with each other (supplemental Figure 1B). Note inhibition of obatoclax-induced death by Nec-1 in both MLL-AF4 cases (A, B; right) and reduced obatoclax-induced death when all 3 pathways were inhibited (A; right).
Evidence for triple killing mechanism of obatoclax in primary infant ALL cells from flow cytometry, Western blot analyses, and MTT assays with cell death inhibitors. Molecular cytogenetic subtype and 72-hour obatoclax EC50s determined from surviving fraction plots in Figure 1 for each case are shown. In cases in (A-D), all 3 types of assays were performed; in 2 cases in (E), apoptosis and autophagy proteins were studied by Western blot. In the other MLL-R case in (C), AF4, AF9, and ENL partner genes were excluded.16 (A-D; left) show contour plots of FSC vs SSC in cells treated for indicated times at respective 72-hour obatoclax EC75 determined from Figure 1. Progressive decrease in FSC signal in all 4 cases indicates apoptosis. (A-D; middle) and (E) are Western blot analyses of apoptosis and autophagy. Note increases in cleaved PARP in all 6 cases in response to obatoclax consistent with apoptosis. Also note increases in LC3-II in all 6 cases in response to obatoclax (A-E); this occurs without increase in p62 in 4 cases in (A, D, E), indicating autophagy with autophagic flux. In MLL-AF4 case in (B), note increase in p62 at 24 hours but decrease by 48 hours after obatoclax treatment, suggesting that autophagy is not blocked. Also note increase in p62 at 48 hours in the other MLL-R case (C, middle panel), indicating either p62 induction or accumulation, the former of which is supported by gene expression profiling data in SEM-K2 (supplemental Table 2). (A-D; right) Surviving fraction plots for MTT assays 72 hours after obatoclax treatment at increasing concentrations by itself or with 50 μM Nec-1, 20 μM zVAD-fmk, or 0.5 mM 3-MA alone or altogether. Inhibitors were used at concentrations determined minimally toxic in cell lines as well as primary cases (supplemental Figure 1), rather than at case-by-case titrated concentrations, and were tested less extensively in obatoclax combinations. Data were normalized to respective primary samples only treated with relevant inhibitors alone or with each other (supplemental Figure 1B). Note inhibition of obatoclax-induced death by Nec-1 in both MLL-AF4 cases (A, B; right) and reduced obatoclax-induced death when all 3 pathways were inhibited (A; right).
Time-dependent decreases in the FSC signal suggested apoptotic death in response to obatoclax in both primary MLL-AF4 cases and the primary other MLL-R and MLL-G cases studied by flow cytometry (Figure 6A-D, left), which was confirmed by Western blot analysis. Obatoclax caused increases in PARP cleavage and LC3-I to LC3-II conversion, indicating apoptosis and autophagy, respectively, in each of these 4 cases (Figure 6A-D, middle) and in the cases with MLL-AF4 or MLL-ENL translocation in Figure 6E. Absence of p62 accumulation demonstrated that the autophagy resulting from obatoclax occurred without blockage in 2 of the MLL-AF4 cases (Figure 6A, middle, and 6E) and the MLL-ENL (Figure 6E) and MLL-G (Figure 6D, middle) cases. In the MLL-AF4 case in Figure 6B, increased p62 at 24 but not 48 hours suggested that whether the early increase was a result of induction of expression or accumulation, autophagy was brought to completion (Figure 6B, middle). Induction of p62 expression or accumulation also occurred with 48-hour obatoclax exposure in the other MLL-R case (Fig 6C, middle). Induction of p62 protein expression is plausible because SQSTM1 (p62) gene expression increased in obatoclax-treated SEM-K2 cells (supplemental Table 2).
Cell death inhibition by Nec-1 indicated necroptosis in both MLL-AF4 cases in which obatoclax was studied with inhibitors (Fig 6A-B, right). In the case in Figure 6A, obatoclax-induced death was unchanged by zVAD-fmk or 3-MA but decreased when all 3 pathways were inhibited. In the other MLL-R and MLL-G cases (Figure 6C-D, right), obatoclax-induced death was unchanged by any single inhibitor. Increased death with obatoclax and the 3 inhibitors together compared with obatoclax alone in the other MLL-R case (Figure 6C, right) may suggest untoward cytotoxicity, even though the data were normalized to cells treated with the triple inhibitor combination (supplemental Figure 1B). At concentrations employed singly or together, the inhibitors by themselves were minimally cytotoxic (supplemental Figure 1), but large cell number requirements precluded case-by-case titrations or further obatoclax-inhibitor combination testing, using primary samples.
Morphologic evidence of triple killing mechanism in obatoclax-treated primary infant ALL cells
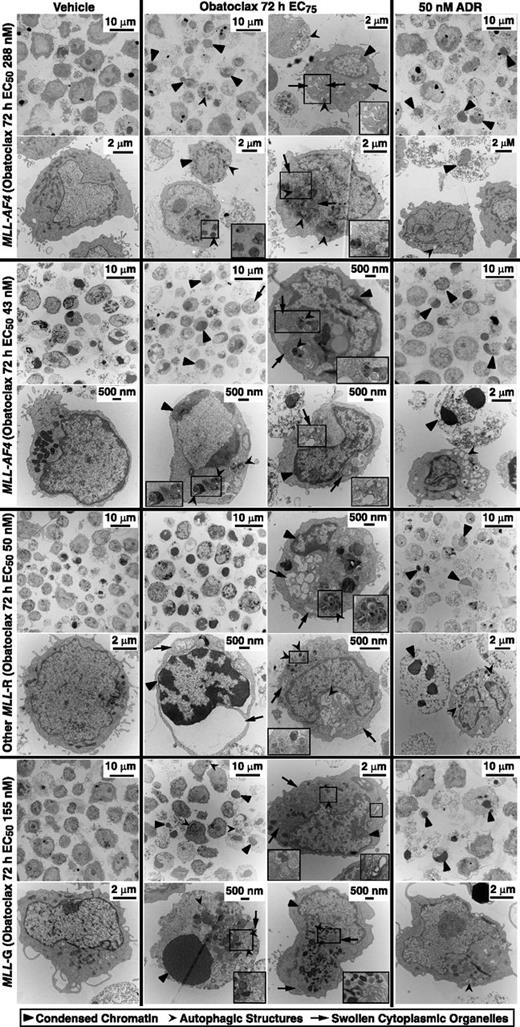
Electron microscopy was used to study the obatoclax response more directly and with fewer cells in 4 of the same primary patient samples (2 MLL-AF4, 1 other MLL-R, and 1 MLL-G). In each case, regardless of the subtype, obatoclax caused ultrastructural changes of apoptosis, necroptosis, and autophagy, including features of 2 or all 3 pathways in single cells (Figure 7). These findings directly implicate all 3 modes of killing by obatoclax across infant ALL. Prominent chromatin condensation as well as occasional autophagic structures in the ADR controls not only confirmed intact apoptosis, but also further indicated plasticity in death pathways able to be activated.
Electron microscopy evidence of apoptosis, autophagy, and necroptosis in obatoclax-treated primary infant ALL cells. Cases are the same as in Figure 6A-D. Molecular cytogenetic subtype and 72-hour obatoclax EC50s from surviving fraction plots in Figure 1 are at left. Cells were treated for 48 hours with vehicle, obatoclax at respective 72-hour obatoclax EC75 or 50 nM ADR, and harvested, stained, and sectioned for electron microscopy. Different arrowheads mark condensed chromatin of apoptosis, autophagic structures, or swollen organelles of necroptosis. Magnified insets are in boxes. Note morphologic findings indicating all 3 modes of death, including examples in single cells in all cases. Also note chromatin condensation as well as less prominent autophagic structures in ADR control cells in all cases.
Electron microscopy evidence of apoptosis, autophagy, and necroptosis in obatoclax-treated primary infant ALL cells. Cases are the same as in Figure 6A-D. Molecular cytogenetic subtype and 72-hour obatoclax EC50s from surviving fraction plots in Figure 1 are at left. Cells were treated for 48 hours with vehicle, obatoclax at respective 72-hour obatoclax EC75 or 50 nM ADR, and harvested, stained, and sectioned for electron microscopy. Different arrowheads mark condensed chromatin of apoptosis, autophagic structures, or swollen organelles of necroptosis. Magnified insets are in boxes. Note morphologic findings indicating all 3 modes of death, including examples in single cells in all cases. Also note chromatin condensation as well as less prominent autophagic structures in ADR control cells in all cases.
Discussion
In this study, obatoclax had potent single-agent in vitro activity in a large, particularly high-risk cohort of 54 primary infant ALL and bilineal acute leukemia cases. Furthermore, in primary pediatric MLL-AF4 ALL cells, obatoclax was synergistic with 6 antileukemia cytotoxic drugs used as essential components of infant ALL chemotherapy. Moreover, the mode of killing by this BH3 mimetic in MLL-AF4 ALL cell lines and primary MLL-R and MLL-G infant ALL cells occurred through a highly unique triple apoptosis/autophagy/necroptosis mechanism.
In contemporary trials, the prognosis of infant ALL with dose-intensive therapies remains dismal: The youngest infants with high WBCs and MLL-R disease experience the worst outcomes (Z.E.D. et al, manuscript submitted).1,2 Even applying a stringent definition, our data indicate that the preponderance of infant ALL is obatoclax-sensitive in vitro regardless of poor prognostic features, as evidenced by EC50s within clinically achievable concentrations. MLL-AF4 is the largest molecular cytogenetic subset within infant ALL.16 Although EC50s were higher than in other subsets, 19 (68%) of 28 MLL-AF4 cases were obatoclax-sensitive, and even greater sensitivity was found in MLL-ENL cases, a historically refractory infant ALL subset with significantly worse survival.1,16 In addition, most EC50s were well below that for normal PBMCs, suggesting that exposure requirements for target cell activity are lower than for normal blood cells.
Similar cell killing was observed in infant ALL diagnostic samples whether or not events occurred with COG P9407 treatment. Further study is required to determine whether obatoclax or a similarly acting compound will show activity regardless of traditional poor prognostic features and augment infant ALL cell killing by chemotherapy in vivo.
How obatoclax affects the cell cycle has implications for obatoclax–chemotherapy combinations because many chemotherapies are cell cycle–dependent and cell cycle determines chemosensitivity.34 Others implicated a S/G2 arrest in inhibition of proliferation by obatoclax in a non-MLL-R AML cell line.35
We observed increased cells in S phase in obatoclax-treated SEM-K2 cells but obatoclax did not notably alter the cell cycle in RS4:11 cells, indicating that obatoclax has variable effects on the cell cycle. The sub-G1 peak of apoptosis was seen with ADR but not obatoclax; however, the sub-G1 peak with PI staining reflecting DNA fragmentation is not always definitive for apoptosis.36 The extent of DNA degradation when cells are assayed, washing buffer pH, numbers of washes, and apoptotic body shedding can cause the sub-G1 peak to be undetected.36 Murine studies showing DNA laddering during G0/G1 and G2/M demonstrated that apoptotic cells are not constrained to sub-G1.37 Cells undergoing apoptosis in late S or G2/M result in a sub-G1 peak that is indistinguishable from G1/early S.36 Thus, we employed other assays alongside cell cycle analysis in MLL-AF4 ALL cell lines to quantify apoptosis.36
Our results on the mode of death induced by obatoclax from flow cytometric, protein, and electron microscopy assays and studies using cell death pathway inhibition in MLL-AF4 ALL cell lines and primary infant ALL cells, as well as gene expression profiling in the cell lines, are also highly novel. To our knowledge, infant ALL is the first disease in which single-agent obatoclax activity is mediated through triple death mode killing. This mechanism is important because multiple cell death pathway activation may help overcome chemotherapy resistance that results in relapse, which is a major cause of treatment failure in infant ALL. Because most anticancer drugs induce apoptosis, others have suggested that resistance may be surmountable by combining agents that are each able to induce a distinct class of death.38 Remarkably, in infant ALL cells, obatoclax induces all 3 classes as a single agent.
Obatoclax was reported to kill AML cell lines35 and primary CLL33 by apoptosis. Apoptosis and autophagy were observed when obatoclax was combined with histone deacetylase (HDAC) inhibition in HL-60 cells39 or with glucocorticoids in non-MLL-R ALL cell lines.40 However, the latter is controversial because others have reported that obatoclax–dexamethasone combination treatment in steroid-resistant ALL caused autophagy-dependent necroptosis.15 Nonetheless, our findings in infant ALL differ from these reports because obatoclax by itself activated 3 cell death pathways even within single cells. They also indicate that obatoclax killing mechanisms are heterogeneous and cell type–specific and reinforce that we uncovered a disease-specific mode of killing in infant ALL in which 3 death pathways are all activated.
In MLL-AF4 cell lines and primary MLL-R and MLL-G infant ALL cells, by MTT and flow cytometry assays, single-agent obatoclax was potently cytotoxic. In the cell lines, increased PI uptake gave evidence of death, and both increased caspase 3 activation, and PARP cleavage indicated apoptosis. Although insufficient evidence for apoptosis by itself, increased TUNEL positivity supported late-stage apoptosis.23 In the primary samples, light scatter analysis33 and increased PARP cleavage suggested apoptosis. Evidence for autophagy came from increased LC3-I to LC3-II conversion. In each cell line and 4 of 6 primary samples tested, there was no increase in p62, lack of accumulation of which indicates autophagic flux.24 Interestingly, one of the genes upregulated by obatoclax in SEM-K2 cells was SQSTM1 (p62).
In the second primary MLL-AF4 sample studied at a protein level, an early increase followed by a decrease in p62 also was consistent with autophagic flux, whether or not the early increase was a result of induced expression or accumulation. In the other MLL-R infant ALL sample in which increased p62 suggested either p62 induction or accumulation, the analyses of obatoclax effects on cell death proteins were performed at a single time-point after treatment. Autophagy also did not increase cell survival because cell death with obatoclax did not increase with autophagy inhibition, and it was RIP1 kinase–independent because LC3-I to LC3-II conversion was not decreased by Nec-1. The significantly reduced death with RIP1 kinase inhibition gave evidence of necroptosis as yet a third mode of killing by obatoclax in both MLL-AF4 ALL cell lines and 2 primary MLL-AF4 infant ALL cell samples. Interestingly, unlike with necroptosis inhibition, obatoclax-induced death was not critically affected by inhibiting apoptosis or autophagy alone. These observations further support obatoclax activation of multiple modes of death and may be consistent with a shift to killing by a different pathway or pathways with apoptosis or autophagy disruption. However, inhibitor concentrations were not optimized in the primary cases to the same degree as in the cell lines because of sample limitations.
In SEM-K2 cells, increased apoptosis with necroptosis inhibition and reversal of this effect, but increased autophagy with combined necroptosis and apoptosis inhibition, suggest plasticity in death modes able to be used and further implicate obatoclax induction of a mixed cell death mechanism. However, cell death was reduced significantly in MTT assays of obatoclax when 2 or all 3 death pathways were inhibited in either cell line or when all 3 death pathways were inhibited in the primary MLL-AF4 infant ALL.
Additional striking evidence for a triple obatoclax killing mechanism in both cell lines derives from upregulated gene expression pertinent to all 3 pathways. Although autophagy is not primarily regulated at the gene expression level,24 autophagy gene expression changes detected by microarray analysis were confirmed by quantitative reverse-transcriptase polymerase chain reaction. Further support for necroptosis came from upregulated genes for various necroptosis factors. The microarray analyses also implicate upregulated phosphoprotein, vacuole, lytic vacuole, and lysosome genes with products having roles in death as new obatoclax targets. The gene expression changes provide pharmacodynamic biomarkers of activity in the obatoclax response.
It recently was suggested that morphology remains the gold standard to define modes of death.36 Our electron microscopy findings indicative of simultaneous apoptosis, necroptosis, and autophagy in single SEM-K2 cells are unique, although 1 or 2 pathways predominated in some cells at a given time. Regardless, the severe morphologic changes and degree of PI uptake gave evidence that the cells were dead. In primary MLL-AF4, MLL-ENL, other MLL-R, and MLL-G infant ALL cell samples treated with obatoclax, there was flow cytometric and/or protein evidence of apoptosis, protein evidence of autophagy and, in 2 MLL-AF4 cases, there was evidence of necroptosis from MTT assays using necroptosis inhibition. However, cellular ultrastructure indicated even more directly that obatoclax kills primary infant ALL cells regardless of the subtype by the same unique, potent, triple apoptosis/autophagy/necroptosis mechanism.
Novel targeted strategies with potential to improve survival for patients with MLL-R leukemia have been of major interest.41 Because obatoclax activates multiple cell death pathways and augments leukemia cell killing, obatoclax-targeted agent combinations may also warrant testing. Obatoclax recently showed synergy with HDAC inhibition in a MLL-R AML cell line.39
We propose that obatoclax can overcome the intrinsic resistance of MLL-R infant ALL to die by inducing a highly novel, disease-specific form of killing that engages apoptosis, necoptosis, and autophagy. We show for the first time that despite abundant antiapoptotic factors5,7 and long-appreciated resistance to cell death,4 all 3 death pathways in MLL-R ALL are able to be activated. Furthermore, survival in infants with MLL-G ALL is inferior to that in children, and we show that MLL-G infant ALL cells are obatoclax-sensitive through similar triple death mode killing. With potent broad preclinical sensitivity in most cases of infant ALL and induction of prolonged remission by single-agent obatoclax in a case of adult MLL-R secondary AML,11 it is important to establish whether activity of a similarly acting compound against infant ALL and, possibly, MLL-R and MLL-G ALL in children occurs in the clinic.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank GeminX Pharmaceuticals for obatoclax.
This work was supported by grants from the Leukemia & Lymphoma Society Specialized Center of Research Program 7372-07 (L.-S.W., J.S.B., S.R.A., C.L.W., M.D., S.P.H., G.H.R., and C.A.F.), National Institutes of Health, National Cancer Institute (R25CA101871) Training Program in Cancer Pharmacology (A.Y.Z.E.); grants to COG, including the COG Chair’s grant (CA98543), U10 CA98413 (COG Statistical Center), U24 CA114766 (COG Specimen Banking); National Institutes of Health (K08HL084199) (M.K.); and American Lebanese Syrian Associated Charities (M.K.). S.P.H. is the Ergen Family Chair in Pediatric Cancer. C.A.F. is the Joshua Kahan Endowed Chair in Pediatric Leukemia.
Authorship
Contribution: A.Y.Z.E. and B.W.R. designed research, performed research, and collected, analyzed, and interpreted data. Q.-C.Y. analyzed and interpreted data and contributed to writing of the manuscript. A.H. performed research and collected, analyzed, and interpreted data. K.C., J.S.M., A.D.B., S.R.A., M.K., A.J.C., and N.A.H. analyzed and interpreted data. L.C. performed research and collected data. I.-M.L.C. performed research. C.L.W. designed research and analyzed and interpreted data. M.D. performed statistical analysis and collected, analyzed, and interpreted data. J.M.H. and Z.E.D. collected and interpreted data. S.P.H. designed research and collected and interpreted data. G.H.R. designed research, collected data, and contributed to writing of the manuscript. K.A.U. designed research; performed research; collected, analyzed, and interpreted data; and contributed to writing of the manuscript. L.-S.W., J.S.B., and C.A.F. designed research, analyzed and interpreted data, and contributed to writing of the manuscript.
Conflict-of-interest disclosure: C.A.F. owns the following patent: Methods and Kits for Analysis of Chromosomal Rearrangements Associated with Leukemia (US Patent #6 368 791, issued April 9, 2002).
Correspondence: Carolyn A. Felix, CTRB 4006, Children's Hospital of Philadelphia, 3501 Civic Center Blvd, Philadelphia, PA 19104; e-mail: felix@e-mail.chop.edu.