In this issue of Blood, Kim and colleagues demonstrate that the Fanconi anemia protein FANCP/SLX4 is multifunctional by identifying molecular domains that uniquely effect repair of DNA damage inflicted by either DNA cross-linkers or topoisomerase and PARP inhibitors.1
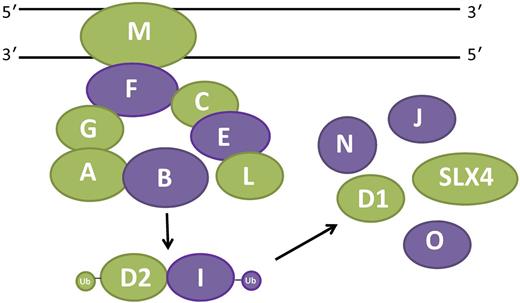
Fanconi anemia (FA) is characterized by bone marrow failure, clonal evolution to myelodysplastic syndrome (MDS) and acute myelogenous leukemia (AML), and epithelial malignancies and is caused by bi-allelic or X-linked inactivation of 1 of 15 different genes, including FANCP/SLX4.2 Eight of the gene products (FANCA, -B, -C, -E, -F, -G, -L, and -M) form a nuclear “core” complex that must be intact to mono-ubiquitinate FANCD2 and FANCI in response to DNA cross-links (see figure). Ubiquitinated FANCD2 and FANCI then directly facilitate repair of DNA cross-links in conjunction with SLX4 and other “downstream” FA proteins. If any 1 of the 8 core proteins is inactivated, FANCD2 and FANCI are not ubiquitinated and cross-linker–induced chromosomal instability results. The majorityof FA literature has focused on the roles of these proteins in cross-link repair and it has been widely speculated that each aspect of the Fanconi phenotype, including bone marrow failure, MDS, and AML, are direct consequences of DNA cross-links inflicted by either exogenous or endogenous chemicals.3 However, the work by Kim et al and other emerging evidence, as discussed below, demonstrates without ambiguity that FA protein function is not limited to interstrand cross-link repair (ICL).
Multifunctionality of the FA pathway. The 15 FA proteins shown here participate in a canonical pathway that repairs DNA cross-links. Eight proteins (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, and FANCM) form a nuclear core complex that facilitates mono-ubiquitination of the ID complex (FANCD2 and FANCI). The ID complex then mediates repair of cross-links in conjunction with FANCD1 (BRCA2), FANCJ (BRIP1), FANCN (PALB2), FANCO (RAD51C), and FANCP (SLX4). FA proteins with non-ICL repair functions that are referenced and discussed in the article are indicated in green.
Multifunctionality of the FA pathway. The 15 FA proteins shown here participate in a canonical pathway that repairs DNA cross-links. Eight proteins (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, and FANCM) form a nuclear core complex that facilitates mono-ubiquitination of the ID complex (FANCD2 and FANCI). The ID complex then mediates repair of cross-links in conjunction with FANCD1 (BRCA2), FANCJ (BRIP1), FANCN (PALB2), FANCO (RAD51C), and FANCP (SLX4). FA proteins with non-ICL repair functions that are referenced and discussed in the article are indicated in green.
Kim and colleagues ectopically expressed SLX4 deletion mutants in SLX4-null cells to delineate the molecular domains required for interaction with the nucleases XPF, MUS81, and SLX1 and to determine whether their roles in repair of 2 types of DNA damage, damage induced by either DNA cross-linking agents or topoisomerase inhibitors, could be uncoupled.1 The SLX4-XPF interaction was found to be required for cross-link repair, while the SLX4-MUS81 interaction was required for repair of damage induced by topoisomerase inhibitors and PARP inhibitors. The SLX4-SLX1 mutant resulted in partial resistance to both cross-linking agents and topoisomerase and PARP inhibitors. Expression of the SLX4-MUS81 mutant ameliorated the cross-linker hypersensitivity of SLX4-null patient cells. The genotype may also influence disease severity. For example, patients with deletions of only the UBZ domain that regulates cross-link repair had milder developmental defects than did SLX4 nullizygotes. While hematologic defects appear to be similar in both groups of patients, the numbers are too small to conclusively link mutation to phenotype.
The approach taken by these investigators should be exploited by those seeking to define pathways of bone marrow failure in this disease. There is a growing body of evidence supporting the notion that FA protein multifunctionality is not restricted to just SLX4. In fact, it is not unlikely that the majority of FA proteins participate in noncanonical pathways (see figure). Analogous to SLX4, other FA proteins possess DNA repair functions independent of core complex function and FANCD2 ubiquitination, including FANCM,4 FANCD1/BRCA2,1 and FANCG.5 FA proteins also protect cells against reactive oxygen species generation.6 Some of the FA proteins may influence hematopoietic cells in ways that are independent of DNA damage per se. For example, in normal hematopoietic progenitors and stem cells FANCA and FANCC suppress inhibitory responses to inflammatory cytokines and in macrophages suppress responses to toll-like receptor agonists.7 In addition, recent evidence suggests that FANCL promotes stem cell function by activating β-catenin.8
The work by Kim et al is a blueprint for the hematology community seeking to define the molecular pathogenesis of marrow failure in this disease and other inherited marrow failure syndromes. Because the development of strategies for prevention of marrow failure and clonal evolution in FA patients depends on a complete understanding of all potential functions of FA proteins, the research community must solve the problem of whether DNA damage in the stem cell pool is all that matters or whether there are other tractable targets. Aberrant molecular pathways induced by loss of noncanonical FA protein function may be inherently more druggable, as illustrated by the efficacies of antioxidants9 and p38 kinase inhibitors7,10 in preclinical FA models. Improvement in clinical diagnostics would also result from such research. Note that Kim and colleagues report that the SLX4-MUS81 deletion mutant ameliorates cross-linker hypersensitivity of SLX4-null patient cells. This raises the possibility that there may exist patients with SLX4 mutations that would test negative in conventional FA diagnostic tests. Mutations that abrogate noncanonical functions of SLX4 or other non-FA proteins, but do not confer cross-linker hypersensitivity, may hypothetically account for a subset of aplastic anemia patients who do not respond to immunosuppressive therapy.
Conflict-of-interest disclosure: The author declares no competing financial interests. ■

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal