Abstract
Human herpesvirus (HHV) 6 causes substantial morbidity and mortality in the immunocompromised host and has no approved therapy. Adoptive transfer of virus specific T cells has proven safe and apparently effective as prophylaxis and treatment of other virus infections in immunocompromised patients; however, extension to subjects with HHV6 has been hindered by the paucity of information on targets of cellular immunity. We now characterize the cellular immune response from 20 donors against 5 major HHV6B antigens predicted to be immunogenic and define a hierarchy of immunodominance of antigens based on the frequency of responding donors and the magnitude of the T-cell response. We identified specific epitopes within these antigens and expanded the HHV6 reactive T cells using a GMP-compliant protocol. The expanded population comprised both CD4+ and CD8+ T cells that were able to produce multiple effector cytokines and kill both peptide-loaded and HHV6B wild-type virus-infected target cells. Thus, we conclude that adoptive T-cell immunotherapy for HHV6 is a practical objective and that the peptide and epitope tools we describe will allow such cells to be prepared, administered, and monitored in human subjects.
Key Points
We have characterized, for the first time, the cellular immune response to HHV6 and defined a hierarchy of immunodominance.
We have developed a GMP-compliant approach to generate polyfunctional T-cell lines that effectively kill HHV6-infected cells and are suitable for clinical use.
Introduction
Adoptive T-cell immunotherapy can successfully prevent and treat otherwise fatal infections in the immunocompromised host. At present, these benefits have been restricted to adenovirus, CMV and EBV.1-9 However, it is increasingly apparent that a distinct herpesvirus, human herpesvirus 6 (HHV6) is also a significant cause of morbidity and mortality, particularly after hematopoietic stem cell transplantation (HSCT). To date, there have been no controlled studies on the use of antivirals as treatment for HHV6, but foscarnet, ganciclovir, and cidofovir have been used clinically, with variable results.10-14
HHV6 is a member of the β-herpesvirus subfamily and exists as 2 species, HHV6A and HHV6B, which share 75%-95% nucleotide sequence identity.15 Primary infection occurs in > 90% of individuals before the age of 2 years and is associated with clinical symptoms, including acute febrile illness and roseola infantum or exanthem subitum.16 The virus subsequently persists in latent form, in an analogous way to other herpesviruses. Although latency is usually asymptomatic, HHV6 reactivates in > 50% of allogeneic HSCT recipients and can produce clinically significant manifestations, including encephalitis, delayed engraftment, and an increased rate of graft-versus-host disease, substantially increasing mortality.10,11,14,17-19
Thus, while HHV6 might be a suitable candidate virus to benefit from adoptive immunotherapy, such treatments can only be developed if there is evidence that T cells recognizing the virus exist in (or can be generated from) the peripheral blood of healthy seropositive individuals. If such cells exist, then it is important to know the antigens and HLA-restricted epitopes they recognize, so that the most effective T cells can be identified and prepared.
We now demonstrate that HHV6-reactive CD4+ and CD8+ T cells do indeed exist in the peripheral blood of healthy subjects and HSCT recipients and that these cells can be expanded ex vivo. We have also analyzed the antigens recognized by these HHV6-specific T cells and identified CD8+ epitopes within those antigens that are most immunodominant. Finally, we show that HHV6 peptide-specific T cells can react to, and kill, virus-infected target cells. We anticipate such cytotoxic T lymphocytes (CTLs) will prove of value in the prevention and treatment of HHV6 infections in the immunocompromised.
Methods
Donors and cell lines
PBMCs from healthy volunteers and HSCT patients were obtained with informed consent per the Declaration of Helsinki on institutional review board–approved protocols. PBMCs were used to generate CTL lines and PHA blasts. PHA blasts were generated from PBMCs (2 × 106/mL) using PHA (5 μg/mL) and maintained in CTL media (RPMI 1640, 45% Click, Irvine Scientific; 2mM GlutaMAX TM-I, Invitrogen; and 5% Human AB Serum, Valley Biomedical) supplemented with IL-2 (100 U/mL, National Institutes of Health), which was replenished every 3 days.
CTL generation: peptide stimulation
Peptides/pepmixes.
For stimulation, we used either commercially available or custom-ordered pepmixes (15 mers overlapping by 11aa) spanning U54 (JPT Technologies), U90, U11, U14, and U71 (Genemed Synthesis). Control pepmixes spanning adenovirus-Hexon, CMV-pp65, and CMV-IE1 were also purchased from JPT. For epitope mapping, peptides were pooled into 25 (U14) and 33 (U90) mini-pools (see Figure 3A; supplemental Figure 2, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) such that each 15mer was represented in 2 mini-pools.20 To identify minimal epitopes, we generated a series of 9mers overlapping by 8 amino acids (aa) spanning immunogenic sequences of U14 aa597-610 VARRLTEMMNDARL, U90 aa53-71 CDVSFESLLFPELEAFDLF and aa29-51 RMQNYHPDPVVEESIKEILEESL, which were used as a stimulus in IFN-γ ELISPOT. Lyophilized peptides were reconstituted at 5 mg/mL in DMSO.
PBMC stimulation.
PBMCs were isolated from whole blood by Ficoll gradient, pelleted, and pulsed for 30-60 minutes at 37°C with peptides/pepmixes, individually or pooled (100 ng/peptide/15 × 106 PBMCs). After incubation, cells were resuspended in CTL media with IL-7 (10 ng/mL) + IL-4 (1000 U/mL; R&D Systems) and plated in a 24-well plate (2 × 106/well). Media and cytokines were replenished on day 5, and cultures split at a density > 3 × 106 cells/well. On days 9-11, CTLs were harvested, counted, and used for phenotypic/functional studies.
CTL generation: HHV6 Z29-infected PBMCs
HHV6B Z29 supernatant.
MOLT3 cells were maintained in cell culture media (RPMI 1640 + 10% FBS; Hyclone) and 2mM GlutaMAX. For infection, MOLT3 cells were resuspended in a 15-mL falcon tube at 5 × 106/mL in HHV6 Z29 supernatant and incubated for 90-120 minutes at 37°C. Subsequently, infected cells were resuspended at 0.8 × 106 cells/mL in cell culture media, cultured for 10-12 days, and split when > 80% confluence was reached. On days 10-12, when 90% of the cells showed cytopathic effects, cells were collected, pelleted (10 minutes/1000g), and virus-containing supernatant was harvested and frozen at −80C.
CTL generation using HHV6B Z29.
PBMCs were isolated, resuspended at 1 × 106 cells/mL in a 1:1 mix of viral supernatant and CTL media with IL-4 + IL-7, and cultured in a 24-well plate (2 × 106 cells/well). On days 5 or 6, cultures were replenished with media + cytokines and on days 9 or 10 harvested, counted, and used for phenotypic/functional analyses.
Immunohistochemistry.
Infected cells were resuspended in PBS, mounted onto glass slides (Shandon Cytospin 4 Cytocentrifuge; Thermo Scientific), and then placed in the steamer for 10 minutes in target retrieval solution (Dako North America). Slides were incubated in preblock/diluent for 30 minutes, followed by incubation with mouse-anti–human HHV6B mAb (monoclonal antibody MAB8535, 101-kDa protein, clone C3108-003; Chemicon) diluted 1:50 in diluent for 1 hour at room temperature. HRP/biotin-conjugated anti–mouse-Ig was used to detect positive cells, followed by enzymatic conversion using the chromogenic substrate 3,3 diaminobenzidine.
Mapping studies
Epitope mapping was performed using CD8-enriched T cell lines, produced by depletion of CD4/CD56/CD16 cells using MACS depletion (Miltenyi Biotec) as per the manufacturers' instructions. The purity of the CD8-enriched CTL fraction was evaluated using FACS; cells were then counted and used for functional studies.
Flow cytometry
Immunophenotyping.
CTLs were surface-stained with antibodies to: CD3, CD4, CD8, CD14, CD16, CD56, CD27, CD45RO, and CD62L (BD Biosciences). Cells were washed once with PBS + 2% FBS (Sigma-Aldrich), pelleted, and 10 μL antibody added. After 15 minutes at 4°C in the dark, cells were washed and analyzed. Approximately 20 000 live cells were analyzed. Samples were acquired on a FACSCalibur equipped with CellQuest Version 6 software (BD Biosciences).
Intracellular cytokine staining.
CTLs were harvested, resuspended at 5 × 106/mL in CTL media, plated at 200 μL/well in a 96-well U-bottom plate, and stimulated with 100 ng of test or control peptide/pepmix in brefeldin A (1 μg/mL), CD28 and CD49d (1 μg/mL; BD Biosciences) for 5-7 hours. Subsequently, CTLs were washed with PBS + 2% FBS, pelleted, and surface stained with CD3, CD8, and/or CD4 (10 μL/tube). After 15 minutes, cells were washed twice, pelleted, fixed, and permeabilized with cytofix/cytoperm (BD Biosciences) for 20 minutes at 4°C, then washed twice with PBS + 2% FBS + 0.1% saponin (Calbiochem, EMD Chemicals) and incubated with 20 μL IFN-γ and/or TNF-α antibodies (BD Biosciences) for 30 minutes at 4°C in the dark. Cells were then washed twice with cold PBS + 2% FBS + 0.1% saponin and at least 100 000 events analyzed.
FoxP3 staining.
FoxP3 staining was performed using the eBioscience FoxP3 kit. Briefly, CTLs were rested in CTL media for 48 hours; then 1 × 106 cells were resuspended in PBS + 2% FBS and surface stained for CD3/25/4. After washing cells were resuspended in 1 mL fixation/permeabilization solution, incubated for 1 hour (4°C), then washed, resuspended in permeabilization buffer, and incubated with 0.2 μL isotype or 10 μL FoxP3 antibody (Clone PCH101) for 30 minutes at 4°C, and then washed and acquired using a FACSCalibur.
Functional studies
Multiplex.
A total of 1 × 105 CTLs were stimulated using 100 ng/peptide HHV6B, pp65, or control pepmixes. After 16 hours, supernatant was collected and the cytokine profile assessed using the MILLIPLEX High Sensitivity Human Cytokine Panel (Millipore). Specifically, 50 μL supernatant was incubated overnight at 4°C with beads, then washed and incubated for 1 hour at room temperature with the biotinylated detection antibody. Finally, streptavidin-PE was added for 30 minutes at room temperature, and duplicate samples were washed and analyzed using the Luminex 200.
ELISPOT assay.
We used IFN-γ and Granzyme B ELISPOT to quantify antigen-specific T cells. Triplicate samples were serially diluted from 4 × 105 to 2.5 × 104 cells/well, and specificity measured after direct peptide/pepmix stimulation. Unstimulated or cells stimulated with an irrelevant pepmix served as controls. After 16-18 hours of incubation, plates were developed, dried, and sent to Zellnet Consulting for quantification. Spot-forming cells (SFCs) and input cell numbers were plotted, and a linear regression calculated after excluding plateau data points. The frequency of antigen-specific cells was expressed as SFCs/input cell number.
Chromium (Cr51) release assay.
Cytotoxicity was measured in a 4- to 6-hour Cr51 release assay, using effector/target ratios of 40:1, 20:1, 10:1, and 5:1. As targets, we used autologous HHV6B Z29-infected or peptide/pepmix-pulsed monocytes. Autologous mock-infected monocytes served as controls. To identify HLA restriction, we used autologous and partially HLA-matched peptide-pulsed PHA blasts as targets. Percentage specific lysis was calculated as specific lysis [(experimental release − spontaneous release)/(maximum release − spontaneous release)] × 100.
Coculture experiments.
CD14-isolated monocytes were infected by culture in a 1:1 mix of HHV6B Z29 viral supernatant and CellGenix media (CellGenix), 2mM L-GlutaMAX (1 × 106 cells/ 24-well) with 800 U/mL GM-CSF (Sargramostim Leukine, Immunex) and 1000 U/mL IL-4 for 24 hours. Subsequently, infected or mock-infected monocytes were mixed with HHV6B or control CTLs at a 1:2 monocyte/T-cell ratio in CTL media with IL-4 + IL-7 and after 24 hours were collected, counted, stained (CD3 and CD11c), and analyzed by FACS.
Results
Identification of immunogenic HHV6B antigens
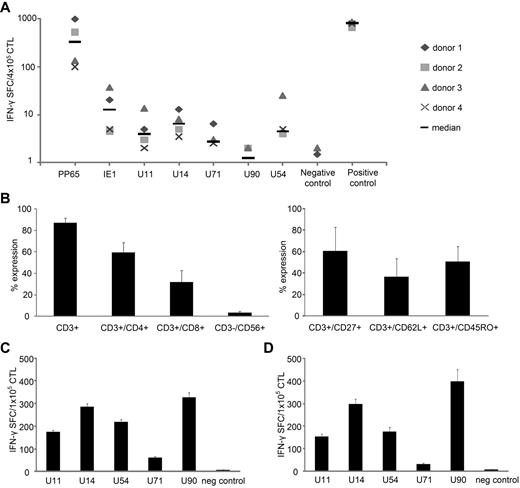
HHV6 shares approximately 70 conserved and colinear genes with CMV, in which the immediate early 1 (UL123) and tegument proteins (UL32:pp150, UL25:pp85, UL83:pp65, and UL99:pp28) are immunodominant T-cell targets. Because little is known about the T-cell immune response to HHV6B, we assessed whether the corresponding HHV6B genes (U90, U11, U14, U54, and U71, respectively; Table 1) had equivalent immunogenicity in seropositive individuals. We isolated PBMCs from healthy donors, stimulated them with pepmixes (peptide libraries of 15mers overlapping by 11 aa) spanning each antigen, and evaluated the frequency of antigen-specific T cells in peripheral blood. The frequency directed against our target antigens was low (median < 6.5 SFCs/4 × 105 PBMCs) as evaluated by IFN-γ ELISPOT (Figure 1A), in sharp contrast to responses against CMV, in which circulating T cells with reactivity against pp65 (HHV6 homolog = U54) and IE1 (HHV6 homolog = U90) may form 1%-10% of the total T-cell population21-25 (Figure 1A).
T-cell immunogenic CMV antigens, their HHV6B equivalent, and their gene function
| CMV gene . | HHV6B gene . | Gene function . |
|---|---|---|
| UL32 (pp150) | U11 | Major tegument phosphoprotein |
| UL25 (pp85) | U14 | Tegument phosphoprotein |
| UL83 (pp65) | U54 | Tegument phosphoprotein virion transactivator |
| UL99 (pp28) | U71 | Myristoylated virion protein |
| UL123 (IE1) | U90 | Immediately early transactivator |
| CMV gene . | HHV6B gene . | Gene function . |
|---|---|---|
| UL32 (pp150) | U11 | Major tegument phosphoprotein |
| UL25 (pp85) | U14 | Tegument phosphoprotein |
| UL83 (pp65) | U54 | Tegument phosphoprotein virion transactivator |
| UL99 (pp28) | U71 | Myristoylated virion protein |
| UL123 (IE1) | U90 | Immediately early transactivator |
HHV6B antigen-specific T cells can be expanded from peripheral blood. (A) The frequency of antigen-specific T cells in peripheral blood was determined by IFN-γ ELISPOT assay after overnight stimulation of PBMCs with CMV (pp65 and IE1) and HHV6 (U11, U14, U54, U71, and U90) antigen-spanning pepmixes in 4 donors. Stimulation with Staphylococcus aureus was used as positive control, and unstimulated PBMCs served as a negative control. Results are expressed as SFCs/4 × 105 input cells (median SFCs). (B) PBMCs were stimulated with the HHV6 pepmixes in the presence of IL-4 + IL-7. On day 9 after stimulation, the phenotype (mean expression ± SD) was assessed (n = 14). (C) PBMCs from 14 donors were stimulated with HHV6 pepmixes in the presence of IL-4 + IL-7, and specificity on day 9 was measured by IFN-γ ELISPOT. Results are expressed as SFCs/1 × 105 input cells. (D) Specificity of CTLs generated from 6 CMV-seronegative donors. IFN-γ production was assessed on day 9 after stimulation, and results are expressed as SFCs/1 × 105 input cells. The negative control was IFN-γ release detected in response to stimulation with an irrelevant pepmix.
HHV6B antigen-specific T cells can be expanded from peripheral blood. (A) The frequency of antigen-specific T cells in peripheral blood was determined by IFN-γ ELISPOT assay after overnight stimulation of PBMCs with CMV (pp65 and IE1) and HHV6 (U11, U14, U54, U71, and U90) antigen-spanning pepmixes in 4 donors. Stimulation with Staphylococcus aureus was used as positive control, and unstimulated PBMCs served as a negative control. Results are expressed as SFCs/4 × 105 input cells (median SFCs). (B) PBMCs were stimulated with the HHV6 pepmixes in the presence of IL-4 + IL-7. On day 9 after stimulation, the phenotype (mean expression ± SD) was assessed (n = 14). (C) PBMCs from 14 donors were stimulated with HHV6 pepmixes in the presence of IL-4 + IL-7, and specificity on day 9 was measured by IFN-γ ELISPOT. Results are expressed as SFCs/1 × 105 input cells. (D) Specificity of CTLs generated from 6 CMV-seronegative donors. IFN-γ production was assessed on day 9 after stimulation, and results are expressed as SFCs/1 × 105 input cells. The negative control was IFN-γ release detected in response to stimulation with an irrelevant pepmix.
To determine whether the apparent paucity of HHV6-reactive T cells in peripheral blood reflected a frequency below the limit of detection of the ELISPOT, we stimulated PBMCs with each of the HHV6B pepmixes and cultured the cells for 9-10 days in vitro in the presence of IL-4 + IL-7 to support the expansion of antigen-specific CTLs.26 When we analyzed the specificity of the expanded lines by IFN-γ ELISPOT, we now readily detected immunity to all stimulating antigens. These expanded HHV6-reactive cultures comprised CD3+ T cells (mean, 86.9% ± 0.3%), with a majority of helper (CD4+) T cells (59.6% ± 0.6%) and a minor CD8+ T-cell component (31.8% ± 0.8%; n = 14). A subset of the expanded CTLs expressed the memory markers CD62L (36.5% ± 17%), CD27 (60.7% ± 22%), and CD45RO (50.6% ± 14%; n = 4; Figure 1B). These data imply that the expanded HHV6 response was derived from a memory population. There was no evidence of regulatory T-cell outgrowth, as assessed by CD4/CD25/FoxP3+ staining (supplemental Figure 1).
Based on the frequency of IFN-γ–secreting cells and the proportion of responding donors, we identified a hierarchy of immunodominance. Thus, all 14 donors (100%) responded to U90, U14, U54, and U11 (mean, 327.8 ± 20.6, 285.4 ± 14, 218.2 ± 11, and 174.3 ± 4.8 SFCs/1 × 105, respectively), whereas only 8 of 14 (57%) responded to U71 (mean 61.1 ± 5.4 SFCs/1 × 105; Figure 1C).
To confirm that the expanded, cytokine-producing cells truly reflected expanded HHV6B-specific cells and not cross-reactive CMV-specific T cells expanded by HHV6-derived peptides, we evaluated whether T cells reactive for our chosen HHV6B antigens could be expanded from CMV-seronegative donors. Results from 6 donors in Figure 1D show a profile similar to lines generated from CMV+/HHV6+ donors, with activity against U90, U14, U54, and U11 (mean, 397.8 ± 53.5, 297.5 ± 23.2, 176.3 ± 19.5, and 154.5 ± 11.5 SFCs/1 × 105) detected in all (100%), whereas only 3 of 6 (50%) lines reacted to U71 (mean, 31.3 ± 3.8 SFCs/1 × 105).
Characterization of HHV6B-specific T cells
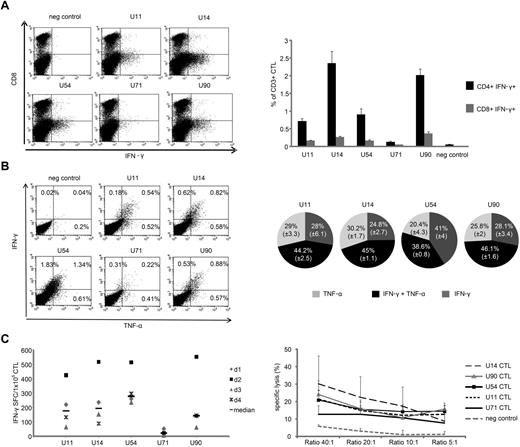
To evaluate whether HHV6B specificity was mediated predominantly by the major population of CD4+ or the minor CD8+ T-cell component, we used ICS and gated on IFN-γ–producing CD3/CD8+ or CD3/CD8− (CD4+) T cells. For all antigens in 5 donors studied, specificity was detected predominantly in the CD4+ compartment, with a minor CD8+ component. Figure 2A shows a representative FACS plot while summary data from all 5 donors is shown in Figure 2A (right panel).
HHV6B antigen-specific CTLs are polyfunctional. (A) IFN-γ cytokine production, as evaluated by ICS, from CD3+/CD8− (CD4 helper; bottom quadrant) and CD3+/CD8+ (cytotoxic; top quadrant) CTLs on day 9 after stimulation in 1 representative donor (dot plots shown were gated on CD3+ cells; left panel). Right panel: Summary results from 5 donors (mean ± SEM). (B) Polycytokine production by CTLs as assessed using ICS in 1 representative donor (left panel) and the summary results from 5 donors showing double-cytokine (IFN-γ and TNF-α) ■ or single cytokine-producing T cells IFN-γ only  or TNF-α only
or TNF-α only  with specificity for U11, U14, U54, and U90 (right panel). U71 was omitted from the analysis because of the low frequency of reactive T cells detected. (C) HHV6-specific T-cell lines generated from 4 donors (IFN-γ ELISPOT; top panel) evaluated 9 days after pepmix stimulation were also cytotoxic (bottom panel), as evaluated by standard 4-6 hours Cr51 release assay using pepmix-pulsed monocytes as targets. Results are presented as percentage specific lysis (mean ± SD, n = 4).
with specificity for U11, U14, U54, and U90 (right panel). U71 was omitted from the analysis because of the low frequency of reactive T cells detected. (C) HHV6-specific T-cell lines generated from 4 donors (IFN-γ ELISPOT; top panel) evaluated 9 days after pepmix stimulation were also cytotoxic (bottom panel), as evaluated by standard 4-6 hours Cr51 release assay using pepmix-pulsed monocytes as targets. Results are presented as percentage specific lysis (mean ± SD, n = 4).
HHV6B antigen-specific CTLs are polyfunctional. (A) IFN-γ cytokine production, as evaluated by ICS, from CD3+/CD8− (CD4 helper; bottom quadrant) and CD3+/CD8+ (cytotoxic; top quadrant) CTLs on day 9 after stimulation in 1 representative donor (dot plots shown were gated on CD3+ cells; left panel). Right panel: Summary results from 5 donors (mean ± SEM). (B) Polycytokine production by CTLs as assessed using ICS in 1 representative donor (left panel) and the summary results from 5 donors showing double-cytokine (IFN-γ and TNF-α) ■ or single cytokine-producing T cells IFN-γ only  or TNF-α only
or TNF-α only  with specificity for U11, U14, U54, and U90 (right panel). U71 was omitted from the analysis because of the low frequency of reactive T cells detected. (C) HHV6-specific T-cell lines generated from 4 donors (IFN-γ ELISPOT; top panel) evaluated 9 days after pepmix stimulation were also cytotoxic (bottom panel), as evaluated by standard 4-6 hours Cr51 release assay using pepmix-pulsed monocytes as targets. Results are presented as percentage specific lysis (mean ± SD, n = 4).
with specificity for U11, U14, U54, and U90 (right panel). U71 was omitted from the analysis because of the low frequency of reactive T cells detected. (C) HHV6-specific T-cell lines generated from 4 donors (IFN-γ ELISPOT; top panel) evaluated 9 days after pepmix stimulation were also cytotoxic (bottom panel), as evaluated by standard 4-6 hours Cr51 release assay using pepmix-pulsed monocytes as targets. Results are presented as percentage specific lysis (mean ± SD, n = 4).
Production of multiple proinflammatory cytokines may correlate with improved cytolytic function and contribute to in vivo activity.27-29 Therefore, we assessed whether the reactivated CTL produced more than 1 cytokine in response to stimulation. For all but U54, we found that the majority of IFN-γ–producing cells also produced TNF-α (detailed results from 1 donor; Figure 2B; summary of 5; Figure 2B right panel). Supplemental Figure 1 shows that, in addition to IFN-γ and TNF-α, the lines produced GM-CSF as well as granzyme B (supplemental Figure 2) with production of Th2 cytokines, including IL-5, IL-10, and IL-13 (supplemental Figure 1), detected at levels similar to pp65-specific CTLs. Thus, our expansion of HHV6-reactive cells induced polyclonal, Th1-polarized cells that produce a similar spectrum of effector cytokines as CMVpp65-specific CTLs.
Finally, to determine whether the polyclonal, HHV6B-directed CTL were cytolytic, we incubated specific T cells (confirmed by ELISPOT; Figure 2C) with autologous CD14-selected monocytes alone (control) or pulsed with U14, U90, U54, U11, and U71 pepmixes, and measured Cr51 release after 6 hours. At an effector/target ratio of 40:1, HHV6-specific CTLs killed U14-, U90-, U54-, and U11-pulsed targets (30% ± 16%, 24% ± 8%, 21% ± 6%, and 21% ± 6%, respectively). In contrast, lysis mediated by U71-specific CTLs was weak (13% ± 8%; 40:1), probably because of the low frequency of reactivated cells (median, 22.3; range, 8-53.5 IFN-γ SFCs/1 × 105CTL). Control target cells were not recognized (6% ± 1%; Figure 2C).
Identification of T-cell epitopes
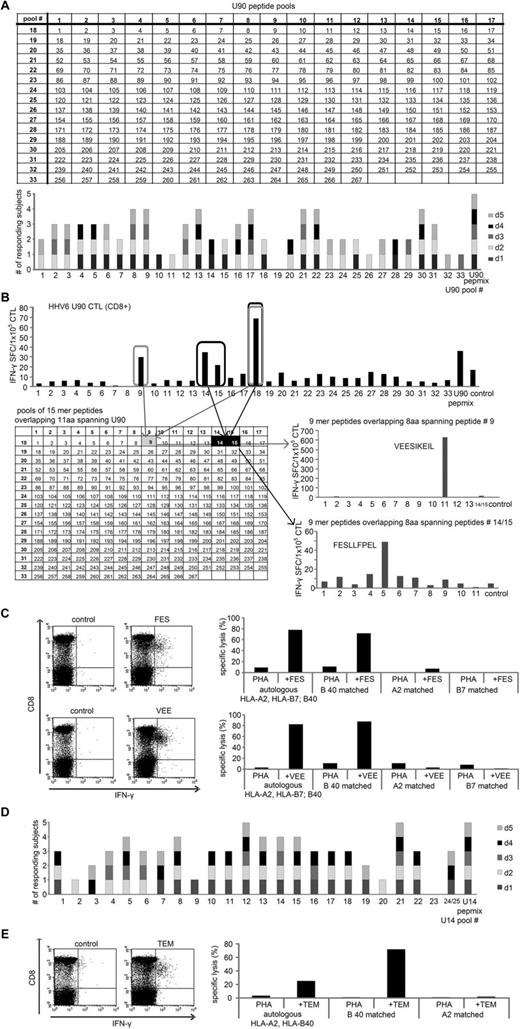
We assessed the breadth of epitope specificity within U90 and U14, the 2 most immunogenic antigens. We measured responses to 267 and 150 15mers (overlapping by 11 amino acids) spanning U90 and U14, respectively, and arranged these into 33 and 25 pools such that each peptide was represented in 2 pools (Figure 3A; supplemental Figure 3). CTL lines with specificity for these individual antigens were exposed to the relevant mini-pools, and positive responses were detected by IFN-γ ELISPOT. Figure 3A (bottom panel) shows the results from U90-specific CTLs generated from 5 donors with disparate HLA types. Each line recognized multiple mini-pools [minimum 9 (donor 3), maximum 27 (donor 2)]. All mini-pools, except #19, were recognized by at least 1 donor, demonstrating that the CTL lines were polyclonal and that immunogenic T-cell epitopes were present throughout the antigen.
Identification of U90 and U14 HLA-restricted CD8+ T-cell epitopes. The breadth of T-cell reactivity in pepmix-activated U90-specific CTLs generated from 5 donors was evaluated by IFN-γ ELISPOT on day 9 using a total of 33 mini-peptide pools, each containing 15-17 peptides, representing all U90 peptides. Mini-pools were arranged such that each peptide was represented in 2 pools (A top panel). Each mini-pool that induced a specific response by IFN-γ ELISPOT was scored as 1, and the summary data from 5 lines screened are shown in panel A (bottom panel). To specifically identify CD8+ T-cell epitopes, we depleted CD4 and NK cells from the CTL lines and rescreened using the U90 mini-pools. (B) Results from 1 donor screened who demonstrated specific activity against the mini-pools 9, 14, 15, and 18, which intersected at the 15mer peptides 9, 14, and 15 of U90 (B top panel). To identify the minimal epitope, CTLs were rescreened against a panel of 9mers overlapping by 8 amino acids spanning the sequence of the stimulatory 15mer peptides. The CD8+ epitope within peptide 9 corresponded to a single 9mer peptide, VEE (aa 39-47), whereas the stimulatory peptide contained within peptides 14 and 15 corresponded to the 9mer peptide FES (aa 57-65). Results of the ELISPOT screening are presented as SFCs/1 × 105 CTL. (C left panel) Confirmation that these peptides indeed reflect stimulatory CD8+ epitopes, as assessed by ICS after CTL stimulation with the 9mer peptides. HLA restriction was evaluated by standard 4-hour Cr51 release assay using autologous and partially HLA-matched PHA blasts alone or pulsed with the VEE and FES peptides as targets (C right panel). The breadth of T-cell reactivity directed against U14-specific CTLs was also evaluated by IFN-γ ELISPOT after exposure to 25 mini-peptide pools representing all U14 peptides (D). (E) Left panel: Confirmation by ICS that the 9mer peptide TEM (aa 602-610) is recognized by CD8+ T cells. Right panel: peptide-pulsed HLA-B40 matched targets are recognized in a 4-hour Cr51 assay.
Identification of U90 and U14 HLA-restricted CD8+ T-cell epitopes. The breadth of T-cell reactivity in pepmix-activated U90-specific CTLs generated from 5 donors was evaluated by IFN-γ ELISPOT on day 9 using a total of 33 mini-peptide pools, each containing 15-17 peptides, representing all U90 peptides. Mini-pools were arranged such that each peptide was represented in 2 pools (A top panel). Each mini-pool that induced a specific response by IFN-γ ELISPOT was scored as 1, and the summary data from 5 lines screened are shown in panel A (bottom panel). To specifically identify CD8+ T-cell epitopes, we depleted CD4 and NK cells from the CTL lines and rescreened using the U90 mini-pools. (B) Results from 1 donor screened who demonstrated specific activity against the mini-pools 9, 14, 15, and 18, which intersected at the 15mer peptides 9, 14, and 15 of U90 (B top panel). To identify the minimal epitope, CTLs were rescreened against a panel of 9mers overlapping by 8 amino acids spanning the sequence of the stimulatory 15mer peptides. The CD8+ epitope within peptide 9 corresponded to a single 9mer peptide, VEE (aa 39-47), whereas the stimulatory peptide contained within peptides 14 and 15 corresponded to the 9mer peptide FES (aa 57-65). Results of the ELISPOT screening are presented as SFCs/1 × 105 CTL. (C left panel) Confirmation that these peptides indeed reflect stimulatory CD8+ epitopes, as assessed by ICS after CTL stimulation with the 9mer peptides. HLA restriction was evaluated by standard 4-hour Cr51 release assay using autologous and partially HLA-matched PHA blasts alone or pulsed with the VEE and FES peptides as targets (C right panel). The breadth of T-cell reactivity directed against U14-specific CTLs was also evaluated by IFN-γ ELISPOT after exposure to 25 mini-peptide pools representing all U14 peptides (D). (E) Left panel: Confirmation by ICS that the 9mer peptide TEM (aa 602-610) is recognized by CD8+ T cells. Right panel: peptide-pulsed HLA-B40 matched targets are recognized in a 4-hour Cr51 assay.
We next defined the minimal CD8+ epitopes and HLA class I-restricting alleles associated with CD8+ responses. We depleted CTL lines of CD4/CD56/CD16+ cells using magnetic beads, achieving a purity of > 85% CD8+ T cells, and then reexposed these CD8-enriched cells to the mini-pools. Results for a representative donor (donor 1: HLA-A2; B7; B40) are shown in Figure 3B. We mapped CD8+ T-cell reactivity to 4 mini-pools (9, 14, 15, and 18), which intersected to identify 2 stimulatory 15mers (#9: aa33-47 YHPDPVVEESIKEIL; and #14: aa53-67 CDVSFESLLFPELEA). To map the minimal epitopes, we synthesized a panel of 9mer peptides (overlapping by 8 amino acids) starting from aa29 (RMQNYHPDP) to aa51 (IKEILEESL) to encompass all potential CD8+ epitopes within the 15mer #9. Similarly, we generated a panel of 9mers spanning aa53-71 CDVSFESLLFPELEAFDLF (corresponding to 15mers #14 and #15). The minimal epitope recognized in peptide #9 corresponded to aa39-47 VEESIKEIL (VEE), whereas reactivity against peptides #14 and 15 reflected recognition of aa57-65 FESLLFPEL (FES). Recognition of these peptides by CD8+ T cells was confirmed by ICS after peptide stimulation (Figure 3C left panel).
To determine the HLA restricting alleles of VEE and FES, we used autologous (HLA-A2; B7; B40) and partially HLA-matched peptide-pulsed PHA blasts as targets. Detailed results are shown in Figure 3C. T cells reactive against FES and VEE recognized autologous and allogeneic targets matched at HLA-B40 with no significant reactivity detected against targets matched at other alleles (Figure 3C right panel). We were subsequently able to raise epitope-specific T cells from at least 2 other donors sharing the restricting HLA allele (not shown), confirming our mapping results.
Using the same approach, we analyzed the breadth of specificity to U14. Figure 3D shows results from 5 donors, with recognition of 7-17 of the 25 mini-pools. All pools, except for #23, were recognized by at least 1 donor. By rescreening CD8-enriched T cells and epitope mapping, we identified 1 novel HLA B40-restricted epitope peptide, aa602-610 TEMMNDARL (TEM), by ICS and a cytotoxicity assay using autologous and partially HLA-matched peptide-pulsed PHA blasts (Figure 3E). Table 2 shows a summary of the 3 novel CD8+ epitope peptides identified in U90 and U14 with amino acid coordinates and identified restricting HLA allele.
Peptide sequences and amino acid coordinates of novel HLA-B40–restricted CD8+ T-cell epitopes identified in U90 and U14
| HHV6B antigen . | Peptide sequence . | HLA restriction . |
|---|---|---|
| U90 | VEESIKEIL (39-47) | B40(60) |
| FESLLFPEL (57-65) | B40(60) | |
| U14 | TEMMNDARL (602-610) | B40(60) |
| HHV6B antigen . | Peptide sequence . | HLA restriction . |
|---|---|---|
| U90 | VEESIKEIL (39-47) | B40(60) |
| FESLLFPEL (57-65) | B40(60) | |
| U14 | TEMMNDARL (602-610) | B40(60) |
Multiantigen-targeted CTLs kill virus-infected targets
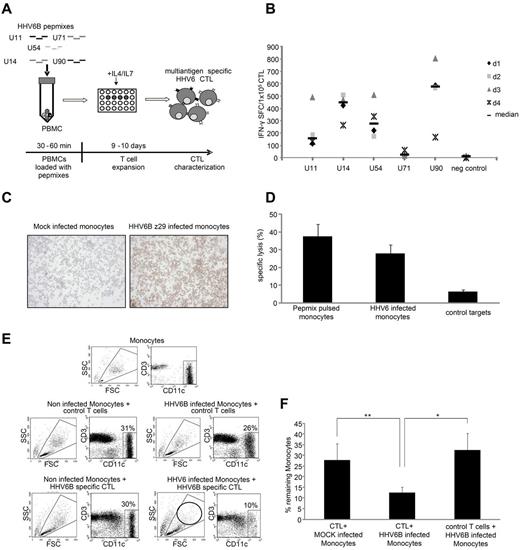
Because adoptive immunotherapy should target multiple viral antigens to prevent immune escape,30 we sought to generate CTL lines reactive against multiple HHV6B antigens simultaneously. We pulsed PBMCs with a mastermix of pepmixes spanning U90, U11, U14, U54, and U71 and then expanded the cells for 9-10 days in IL-4 + IL-7 (Figure 4A schematic). The resulting CTLs were specific for all 5 stimulating antigens (Figure 4B) at frequencies similar to those achieved using a single antigen stimulus (Figure 2C). Next, we evaluated the cytolytic ability of these multiantigen-specific CTLs using, as targets (1) HHV6B pepmix-pulsed monocytes and (2) HHV6 wild-type virus-infected monocytes. Eighty percent of monocytes were HHV6-infected, as assessed by IHC (Figure 4C), and confirmed by analyzing cells for the presence of HHV6 transcripts associated with active infection (not shown).31 In a 6-hour Cr51 release assay, multiantigen-specific CTLs specifically lysed pepmix-pulsed (37% ± 7%, 40:1) and virus-infected targets (28% ± 5%, 40:1), with no recognition of mock-infected targets (6% ± 1%, 40:1) (n = 5; Figure 4D). Cytolytic ability was confirmed in a 24-hour coculture assay of virus- or mock-infected monocytes with HHV6B-specific or control T cells. Surface staining for CD11c and CD3 was used to discriminate monocytes from T cells. At initiation, T cells and monocytes were cocultured at a 2:1 ratio. This T cell/monocyte ratio remained stable in all 3 control groups: (1) noninfected monocytes + control T cells (31%:69%), (2) HHV6B-infected monocytes + control T cells (26%:74%), and (3) mock-infected monocytes + HHV6B-specific CTLs (30%:70%). However, coculture of HHV6B-specific CTLs with infected monocytes for 24 hours eliminated 66% of monocytes (with the 10% residual cells probably representing noninfected targets; Figure 4E). Figure 4F summarizes these results from 4 donors in whom HHV6B-specific CTLs killed infected (12.5% ± 2.5% residual monocytes) more effectively than mock-infected targets (27.8% ± 7.7%, P = .01) and than coculture of control T cells with virus-infected monocytes (32.6% ± 7.7%, P = .05).
HHV6B multiantigen-specific CTLs kill HHV6B wild-type virus-infected targets. (A) Schematic of our multiantigen-specific HHV6 CTL generation process. (B) By IFN-γ ELISPOT, these multiantigen-specific lines were specific for the stimulating pepmixes. Results are expressed as SFCs/1 × 105 input cells. (C) IHC analysis of CD14-selected monocytes infected with the HHV6B wild-type virus strain Z29 or the mock-infected controls. The IHC pictures were taken using 10× magnification at an exposure of 6 ms on an Olympus BX41 microscope, and images were captured using Q-capture software (Q-imaging). Autologous monocytes alone, or pepmix-pulsed or HHV6 wild-type virus-infected cells, were used as targets in a standard 4-6 hour Cr51 release assay (D). Data are mean ± SEM (% specific lysis at an effector/target ratio of 40:1; n = 4). (E) Representative FACS results of a 24-hour coculture assay where either uninfected or HHV6 Z29-infected autologous monocytes (CD11c+) were cultured with nonspecific or HHV6 multiantigen-specific CTLs (CD3+). (F) Summary results from 4 donors where the percentage of residual monocytes after treatment, as quantified by FACS analysis, were plotted (mean ± SD).
HHV6B multiantigen-specific CTLs kill HHV6B wild-type virus-infected targets. (A) Schematic of our multiantigen-specific HHV6 CTL generation process. (B) By IFN-γ ELISPOT, these multiantigen-specific lines were specific for the stimulating pepmixes. Results are expressed as SFCs/1 × 105 input cells. (C) IHC analysis of CD14-selected monocytes infected with the HHV6B wild-type virus strain Z29 or the mock-infected controls. The IHC pictures were taken using 10× magnification at an exposure of 6 ms on an Olympus BX41 microscope, and images were captured using Q-capture software (Q-imaging). Autologous monocytes alone, or pepmix-pulsed or HHV6 wild-type virus-infected cells, were used as targets in a standard 4-6 hour Cr51 release assay (D). Data are mean ± SEM (% specific lysis at an effector/target ratio of 40:1; n = 4). (E) Representative FACS results of a 24-hour coculture assay where either uninfected or HHV6 Z29-infected autologous monocytes (CD11c+) were cultured with nonspecific or HHV6 multiantigen-specific CTLs (CD3+). (F) Summary results from 4 donors where the percentage of residual monocytes after treatment, as quantified by FACS analysis, were plotted (mean ± SD).
In vitro reactivation of HHV6B-specific T cells using virus-infected APCs
To assess the frequency of T cells specific for our predicted antigens in an in vitro model of HHV6 reactivation, we expanded cells directed against our target antigens using virus-infected, rather than pepmix-pulsed, APCs. We infected healthy donor PBMCs with the HHV6B Z29 wild-type strain, which has tropism for monocytes, dendritic cells, and CD4+ T cells within PBMCs.32 To expand antigen-activated T cells, we cultured the infected PBMCs in IL-4 + IL-7 for 9-10 days and subsequently evaluated the phenotype and specificity of the lines.
Flow cytometry demonstrated that, similar to pepmix-stimulated T cells, these lines were almost exclusively CD3+ T cells (93.3% ± 1.6%; Figure 5A) and dominated by the CD4+ subset (54.9% ± 6.9%); CD8+ T cells formed 40.2% ± 6.5% of the total (Figure 1D). To evaluate the specificity of these CTLs, we stimulated them with pepmixes spanning U11, U14, U54, U71, and U90 and measured responses by IFN-γ ELISPOT. We confirmed reactivity against all antigens previously identified as immunogenic [U11 (56.5 ± 3.7 SFCs/1 × 105cells), U14 (67.4 ± 8.3 SFCs/1 × 105cells), U54 (116.6 ± 17.9 SFCs/1 × 105cells), and U90 (73.2 ± 5.1 SFCs/1 × 105cells)]. No or minimal activity was detected after stimulation with U71 pepmix (4.8 ± 1.1 SFCs/1 × 105cells). In contrast, mock-infected PBMCs showed no evidence of activity for these antigens (Figure 5B). Thus, HHV6B virus-infected APCs induce T cells with reactivity for the antigens U11, U14, U54, and U90, confirming that our chosen antigens are both immunologically and physiologically relevant in considering T-cell targeting against HHV6-infected cells.
HHV6B-specific CTLs induced using wild-type virus-infected PBMCs. (A) The phenotype of CTL lines generated using HHV6B Z29-infected PBMCs as stimulators (mean expression ± SD; n = 6). (B) In the same lines, specificity was assessed by IFN-γ ELISPOT. Results of HHV6B Z29-stimulated CTLs (HHV6+) and MOCK-stimulated CTLs (HHV6−) are expressed as SFCs/1 × 105 input cells.
HHV6B-specific CTLs induced using wild-type virus-infected PBMCs. (A) The phenotype of CTL lines generated using HHV6B Z29-infected PBMCs as stimulators (mean expression ± SD; n = 6). (B) In the same lines, specificity was assessed by IFN-γ ELISPOT. Results of HHV6B Z29-stimulated CTLs (HHV6+) and MOCK-stimulated CTLs (HHV6−) are expressed as SFCs/1 × 105 input cells.
Detection of HHV6B-specific T cells after in vivo viral reactivation
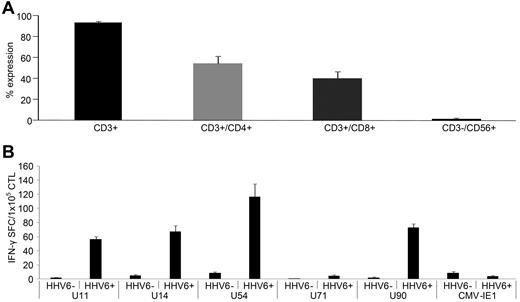
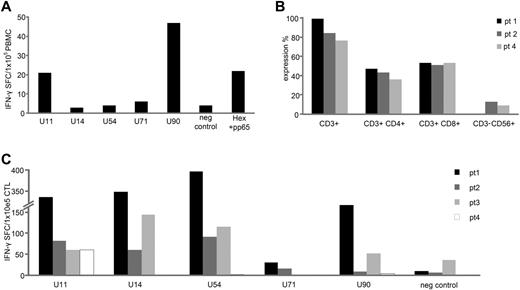
To assess the in vivo relevance of HHV6B antigen-specific T cells for viral elimination, we investigated whether allogeneic HSCT recipients had evidence of circulating T cells directed against our identified immunodominant target antigens after HHV6B reactivation. First, we measured the frequency of HHV6-reactive T cells in the peripheral blood of patients. In 1 patient who presented with an active viral HHV6B reactivation (day 29 after transplantation), we were able to measure HHV6B-specific T cells directed against U11 (21 SFCs/1 × 105 cells) and U90 (47 SFCs/1 × 105 cells; Figure 6A), even without the ex vivo expansion steps invariably required for normal donors. To ensure that the cytokine-producing T cells detected in our patients' PBMCs were specifically induced by HHV6B, we also confirmed lack of expansion of T cells reactive against other viral antigens, including CMV-pp65 and adenovirus-Hexon. Four additional patients who recovered from HHV6B reactivation (ranging from days 25-82 after transplantation) had reactive CD4+ and CD8+ T cells after ex vivo expansion (Figure 6B), which were specific for at least 1 of our chosen antigens (Figure 6C), further confirming the in vivo relevance of U11, U14, U54, and U90 specific T cells for viral elimination.
HHV6B-specific T cells are detected in the peripheral blood of HSCT recipients. PBMCs isolated from a HSCT recipient at the time of acute HHV6 infection were tested for specificity against our panel of HHV6 antigens, using IFN-γ ELISPOT as a readout. Unstimulated PBMCs were used as a negative control, whereas stimulation with a cocktail of CMV-pp65 and Adv-Hexon pepmixes was used to assess the relative frequency of circulating T cells directed against other ubiquitous viruses. Results are expressed as SFCs/1 × 105 input PBMCs (A). PBMCs from 4 other HSCT recipients with recently controlled HHV6 reactivations were stimulated with HHV6B antigen-spanning pepmixes and expanded for 9-10 days in presence of IL-4 + IL-7. (B) Phenotype of the expanded cells assessed by flow cytometry (mean ± SD expression). (C) Specificity by IFN-γ ELISPOT assay; results are expressed as SFCs/1 × 105 input cells.
HHV6B-specific T cells are detected in the peripheral blood of HSCT recipients. PBMCs isolated from a HSCT recipient at the time of acute HHV6 infection were tested for specificity against our panel of HHV6 antigens, using IFN-γ ELISPOT as a readout. Unstimulated PBMCs were used as a negative control, whereas stimulation with a cocktail of CMV-pp65 and Adv-Hexon pepmixes was used to assess the relative frequency of circulating T cells directed against other ubiquitous viruses. Results are expressed as SFCs/1 × 105 input PBMCs (A). PBMCs from 4 other HSCT recipients with recently controlled HHV6 reactivations were stimulated with HHV6B antigen-spanning pepmixes and expanded for 9-10 days in presence of IL-4 + IL-7. (B) Phenotype of the expanded cells assessed by flow cytometry (mean ± SD expression). (C) Specificity by IFN-γ ELISPOT assay; results are expressed as SFCs/1 × 105 input cells.
Discussion
In this study, we characterized the cellular immune response to 5 HHV6B antigens predicted to be immunogenic and defined a hierarchy of immunodominance with respect to both the frequency of responding donors and the magnitude of reactive T cells. U90, U14, U54, and U11 were recognized by all 20 donors screened with progressively smaller frequencies of reactive T cells, whereas U71 induced the lowest frequency of specific cells and was immunogenic in just more than half of the donors. This profile of antiviral activity was subsequently confirmed in patients with active/recent HHV6 reactivations, demonstrating the in vivo relevance of our chosen antigens. We show that T cells reactive against these antigens, which can be expanded ex vivo using either peptides or virus-infected monocytes, were able to produce multiple effector cytokines and kill both peptide-pulsed and HHV6B virus-infected targets. Finally, we were able to successfully map 3 novel CD8+ epitope peptides in U90 and U14, the first identified in HHV6, and define their HLA class I–restricting allele.
HHV6 is a lymphotropic virus (genus Roseolovirus) of the β-herpesvirus family, which also contains CMV and HHV7.33 There are 2 species of HHV6: HHV6A and HHV6B.15 Although the HHV6 genome is highly conserved between the species, significant differences are observed between their biology, immunology, and epidemiology.15 Primary infection with HHV6B presents with roseola infantum or exanthem subitum and occurs early in most individuals, resulting in a prevalence of nearly 100%.16 By contrast, the epidemiology and clinical symptoms of primary HHV6A infection remain unclear.13 Although not usually associated with disease in the immunocompetent, in immunocompromised individuals, such as solid-organ or HSCT recipients, reactivation of HHV6 can produce CNS disease (including encephalitis and meningitis), pneumonitis, transplant rejection, or delayed engraftment.10-12,17-19,34,35 Reactivation has also been associated with seizures,36 myocarditis, and pityriasis rosea. Beyond the immunocompromised host, HHV6 has been proposed to have a pathogenic role in multiple sclerosis, chronic fatigue syndrome, and drug-induced hypersensitivity syndrome, but these associations remain controversial.37-40 Although ganciclovir, cidofovir and foscarnet have been used to treat HHV6 infection and severe disease, these agents are associated with substantial toxicities and have produced variable results. Given the success of adoptive immunotherapy for other herpesvirus diseases in the immunocompromised, we set out to identify appropriate antigens against which an effector T-cell response might be effectively generated and we focused our efforts specifically on HHV6B, which is responsible for > 95% of reactivations detected in transplant patients.41
A comprehensive understanding of virus immune control in healthy individuals and the identification of immunogenic/protective viral antigens are prerequisites for developing any immunotherapeutic strategy. To date, there have been few analyses of the specificity of the immune response to HHV6, a complex virus expressing > 100 genes, any or all of which could be potential T-cell targets. Given that HHV6B shares ∼ 70 conserved and collinear genes with the immunologically well characterized CMV virus, we focused on the HHV6B equivalents of 5 CMV antigens known to be important in viral recognition/protection by T cells, namely, IE1 (UL123) and tegument proteins (UL25-pp85, UL99-pp28, UL32-pp150, and UL83-pp65). In HHV6, these antigens are expressed at immediate early (U90), early (U71 and U14), and late (U11 and U54) stages of the lytic cycle.33 Unlike CMV, in which the majority of seropositive individuals have circulating reactive T cells at frequencies readily detected in peripheral blood, the frequency of T cells reactive to our chosen HHV6B antigens was low/undetectable when assessed directly ex vivo (Figure 1). This result is consistent with data from others who used whole virus (including cell lysate and purified/disrupted virus) as the antigen source.37,42-44 Thus, we find it unlikely that our failure to find measurable reactivity was the result of inappropriate selection of targets but instead reflects the low frequency in peripheral blood of T cells reactive to this persistent herpesvirus. Notwithstanding this lack of measurable numbers of HHV6-directed T cells, we were able to expand specific populations by amplifying the scanty population of reactive cells with a single in vitro stimulation using overlapping peptide libraries spanning our target antigens or with virus-infected APCs.
In CMV-seropositive donors, there is a clear hierarchy of immunodominance with pp65 (analog of HHV6 U54) being the most frequently detected T-cell population, followed by IE1 (analog of U90), UL99 (analog of U71), and UL32 (analog of U11). Finally, UL25 (analog of U14) is immunogenic in a minority of donors and is recognized predominantly by CD4+ T cells, in contrast to the CD8+ responses favored by other viral antigens.25,45 HHV6B is different. All donors tested had detectable activity against 4 of the 5 antigens tested (U90, U14, U54, and U11), but the magnitude of the response induced was strongest against U90 (CMV analog = IE1), followed by U14 (UL25 analog) and U54 (pp65 analog). We were concerned that the homology, albeit limited, between our 5 HHV6 antigens and their CMV counterparts might lead to a specious hierarchy profile if CMV-reactive T cells were cross-reactive. However, neither the distinct pattern of target recognition nor the magnitude of the responses induced was influenced by whether HHV6-specific CTL lines were generated from CMV-seropositive (n = 14) or seronegative donors (n = 6; Figure 1C-D). However, the situation may be different with HHV6A, which shares 75%-95% homology with HHV6B.15 Indeed, recent work by Nastke et al reported that T cells raised against HHV6A cross-reactively recognized HHV6B and vice versa.42
We next addressed whether recognition of HHV6B, like CMV, was mediated predominantly by CD8+ T cells.24,29 We found that all our target antigens, except for U14, induced a predominantly CD4+ effector population with a minor CD8+ component. It is possible that the profile we detect is an artifact related to our use of just 5 antigens as others, not examined in this study, might induce a predominantly CD8-mediated response. However, this explanation is unlikely because T-cell lines generated with virus-infected monocytes as stimulators produced similarly CD4-polarized products, a finding that is in agreement with reports from others.37,43,44,46 The bias to CD4-reactive T cells might instead be influenced by HHV6-derived immune-modulating genes. Such immune modulation is a common feature of herpesviruses. For example, CMV expresses 6 different viral genes (pp65, US2, US11, US3, US6, and US10) dedicated to inhibiting T-cell recognition by enhancing HLA class I and II internalization, inducing HLA degradation or preventing peptide loading.47 Although less well studied, it is likely that equivalent genes may alter HLA class I-associated antigen processing/presentation in HHV6. For example, in HHV6B, the lytic glycoprotein U21 has been shown to divert HLA class I to the lysosomal compartment for degradation as a means of evading CD8-mediated elimination.48 Despite this bias to a CD4 response, we found that both the predominant CD4+ population as well as the minor CD8+ fraction produced Th1-polarized effector molecules, including IFN-γ, TNF-α, and granzyme B, and both subsets effectively killed both peptide-pulsed and virus-infected targets, suggesting a potential future clinical utility.49,50
A more extensive and detailed characterization of HHV6 epitopes greatly enhances our ability to produce effective vaccines and to evaluate the in vivo effects of adoptively transferred CTL lines. Such studies are difficult with epitopes presented on HLA class II and recognized by CD4+ T cells as they are often promiscuously presented in the context of multiple HLA alleles.51 We therefore depleted helper and NK cells from our lines and limited our mapping to the epitope specificity of CD8+ T cells recognizing U90 and U14. Given both the prevalence of the HLA-A2 allele in whites and the fact that it is an immunodominant allele for CMV epitopes,22,23 we initially focused on HLA-A2+ donors and analyzed CTL lines generated from 5 donors, 3 of whom were HLA-A2 homozygous and 2 heterozygous. In contrast to similar studies of CMV minimal epitopes, we were unable to identify a single HLA-A2-restricted CD8+ T-cell epitope in either U90 or U14. Instead, the 3 CD8+ epitopes we identified (the first described for HHV6) were all found to be presented in the context of HLA-B40(60).
To confirm that our chosen target antigens were clinically relevant and induced effective T cells, we used wild-type virus-infected cells to activate bulk HHV6-reactive T cells from healthy donors and demonstrated that the expanded populations had activity against our chosen antigens at comparable levels to those detected using peptide libraries as a stimulus. Next, in HSCT recipients with either active disease or evidence of recent HHV6 reactivation that was controlled, thus excluding the possibility that the patients had chromosomally integrated HHV6,52 we detected HHV6-specific T cells for our chosen antigens. Finally, we generated single CTL lines specific for U90, U11, U14, U54, and U71 using pepmixes as a stimulus and showed that such lines were able to recognize and kill HHV6B Z29-infected monocytes but not control targets. Taken together, this evidence suggests that HHV6-specific CTLs with specificity for U90, U11, U14, U54, and U71 will support viral control in vivo. Given these data and that our process for generating T-cell lines targeting multiple HHV6-specific antigens is compatible with current Good Manufacturing Practices, we conclude that adoptive T-cell immunotherapy for HHV6B is a practical objective and our newly identified CD8+ T cell epitopes will provide valuable tools to better understand the behavior of the virus and transferred immunity in vivo.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
A.M.L. was supported by the National Marrow Donor Program through funding from the Amy Strelzer Manasevit Research Program. U.G. was supported by a Leukemia & Lymphoma Society Special Fellow in Clinical Research Award, an ASBMT Young Investigator Award, and an HHV-6 Foundation pilot grant. H.E.H. was supported by a Dan L. Duncan Chair. M.K.B. was supported by a Fayez Sarofim Chair. The design and execution of this study were supervised at the EHA-ASH Translational Research Training in Hematology.
Authorship
Contribution: U.G. and A.M.L. developed and designed the study and analyzed data; U.G., A.M.L., and M.K.B. wrote the manuscript; U.G. developed the method for generating HHV6-specific CTLs using peptides and virus-infected targets; U.G., L.K., J.M.K., U.L.K., and C.T.Q.N. grew the CTL lines and performed phenotypic and functional studies; A.P.d.P. provided critical reagents and technical expertise on expanding specific cells with virus-infected targets; C.A.R., A.K.-N., and S.M.G. provided patient samples; H.E.H. and C.M.R. provided expertise in CTL generation; and all authors critically reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ann M. Leen, Center for Cell and Gene Therapy, Baylor College of Medicine, 1102 Bates St, Suite 1770, Houston, TX 77030; e-mail: amleen@txch.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal