Abstract
Human cancers display substantial intratumoral genetic heterogeneity, which facilitates tumor survival under changing microenvironmental conditions. Tumor substructure and its effect on disease progression and relapse are incompletely understood. In the present study, a high-throughput method that uses neutral somatic mutations accumulated in individual cells to reconstruct cell lineage trees was applied to hundreds of cells of human acute leukemia harvested from multiple patients at diagnosis and at relapse. The reconstructed cell lineage trees of patients with acute myeloid leukemia showed that leukemia cells at relapse were shallow (divide rarely) compared with cells at diagnosis and were closely related to their stem cell subpopulation, implying that in these instances relapse might have originated from rarely dividing stem cells. In contrast, among patients with acute lymphoid leukemia, no differences in cell depth were observed between diagnosis and relapse. In one case of chronic myeloid leukemia, at blast crisis, most of the cells at relapse were mismatch-repair deficient. In almost all leukemia cases, > 1 lineage was observed at relapse, indicating that diverse mechanisms can promote relapse in the same patient. In conclusion, diverse relapse mechanisms can be observed by systematic reconstruction of cell lineage trees of patients with leukemia.
Introduction
Even with the current standard of treatment for acute leukemia, aggressive chemotherapy and stem cell transplantation, close to one-half of young patients with leukemia succumb to the disease within 5 years, and survival prospects for elderly patients are worse still, mainly because of relapse, but also because of induction therapy failure and treatment-related mortality.1 Several known mechanisms dominate treatment failure and can be broadly separated into 3 main categories, the first being population level effects, including tumor burden and tumor kinetics.2 One of the main mechanisms associated with tumor kinetics is related to dormancy and quiescence of some tumor subpopulations, such as leukemia stem cells (LSCs). A second, closely related, but distinct mechanism is the single cancer cell effect, which relates to preexisting or de novo (during the course of treatment) genetic or epigenetic alterations, with consequent functional heterogeneity contributing to single cell resistance to chemotherapy. In this regard, cells can acquire genetic instability traits (such as microsatellite instability [MSI]) that enable a higher mutation rate and, therefore, increased population diversity and better fitness in the face of environmental stress, including chemotherapy,2 or specific mutations conferring chemotherapy resistance, such as ATP-dependent drug efflux.3 A third category relates to tumor environment-related chemotherapy resistance mechanisms.4 Current cancer treatment by conventional chemotherapy does not take into account all of the various chemotherapy resistance mechanisms and therefore, often, only reduces the genetic diversity of the cancer as a result of dramatic reduction in the number of cancer cells, followed by relapse of the surviving cells. Although the existence of intratumoral phenotypic and genotypic heterogeneity has been recognized from the early days of experimental cancer research, the relative contribution of different cellular phenotypes and a multiplicity of subpopulations to chemotherapy resistance are still not clear.5 Therefore, it has been suggested that a population genetic-based approach can advance the understanding of mechanisms wherein subpopulations of leukemic cells lead to relapse and treatment resistance.6
Despite some efforts to uncover tumor population substructure and evolution among human cancers,7-10 the paucity in single-cell technology and lack of appropriate models that incorporate the bidirectional effects of genes and environment on phenotype over time have hampered comprehensive population genetic analysis of clinical tumors in affected patients. Recent studies have shown that acute lymphoblastic leukemia (ALL) comprises diverse subpopulations that had distinct functional growth properties in xenografts.11 However it was not possible to identify relapse mechanisms, because the study was not designed for longitudinal sample analysis. In the present study we hypothesized that reconstruction of lineage trees from individual sorted leukemia cells at both diagnosis and relapse might shed light on relapse mechanisms. To reconstruct lineage trees we used a method developed in our laboratory that uses somatic MS mutations (predominantly mono- and di-nucleotide repeats) for reconstructing cell lineage trees12-16 (Figures 1–2A). This retrospective method, which has also been applied by others,17-20 is based on the concept that somatic mutations accumulated during cell divisions endow each cell with a genomic signature that is unique with high probability.12 The distances between the genomic signatures of different cells, as measured with the use of standard phylogenetic methods,21 can then be used to reconstruct a cell lineage tree relevant to an organ, organism, or tumor.
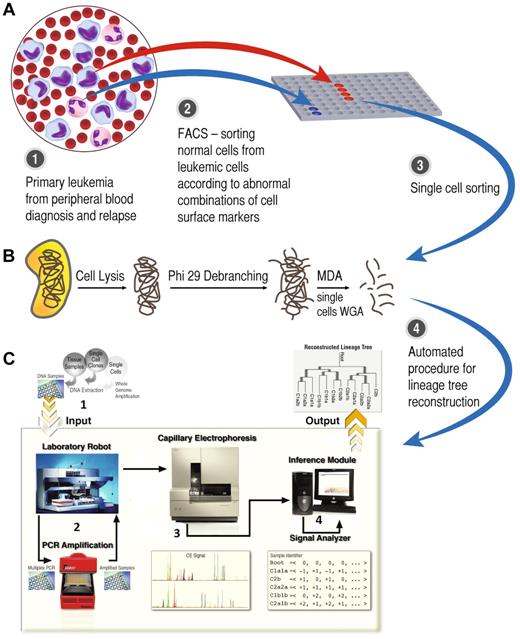
Flowchart of the study method. (A) Paired diagnosis and relapse bulk samples were sorted by FACS, yielding a pure leukemic cell population apart from the normal fraction. (B) Single cells were sorted and whole genome amplification (WGA) was taken by a multiple displacement loop (MDA) procedure. (C1) The product of single-cell WGA entered the automatic lineage tree reconstruction module. (C2) Microsatellites (MSs; n = 120) were amplified by the laboratory robot for each single cell. (C3) Fluorescently labeled PCR products were analyzed by capillary electrophoresis. (C4) Automated analysis of capillary signals was translated to the number of MS repeat units and to reconstruct a lineage tree by phylogenetic methods (panel C was adapted from Frumkin et al12 ).
Flowchart of the study method. (A) Paired diagnosis and relapse bulk samples were sorted by FACS, yielding a pure leukemic cell population apart from the normal fraction. (B) Single cells were sorted and whole genome amplification (WGA) was taken by a multiple displacement loop (MDA) procedure. (C1) The product of single-cell WGA entered the automatic lineage tree reconstruction module. (C2) Microsatellites (MSs; n = 120) were amplified by the laboratory robot for each single cell. (C3) Fluorescently labeled PCR products were analyzed by capillary electrophoresis. (C4) Automated analysis of capillary signals was translated to the number of MS repeat units and to reconstruct a lineage tree by phylogenetic methods (panel C was adapted from Frumkin et al12 ).
Our method was validated with the use of ex vivo cell lineage trees12 and was applied to the lineage analysis of cells from a mouse tumor,15 the estimation of depth of different cell populations14 (Figure 2A), the study of the development of muscle stem cells,22 and the study of preservation and development of female germline.23 Most recently, we reported the reliability of this method for the detection of stem cells and tissue dynamics in the colon.16
In light of the recent advances in single-cell genotyping,24,25 it is important to emphasize the merit of using neutral mutations in noncoding regions of the genome (such as the MS loci used in this study) to reconstruct cell lineage trees, rather than mutations that affect cell phenotype, for example, cancer driver mutations that are mainly located in the exome. MS mutations have been widely used as a molecular clock to approximate relative numbers of cell replication events26 and have already been used effectively for cell lineage reconstruction in mice as reported earlier. Although further calibration is needed to determine the actual human cell depth in various settings (eg, phylogenetic coalescence calculations, somatic replication events) on the basis of MS mutations, computed depth provides valuable information about the relative actual depth of cells (Figure 2A). By contrast, a driver mutation in an oncogene provides a selective advantage and hence cannot be used to infer depth, because cells that lose this mutation will grow more slowly and be flushed out of the population. Hence, although the number of divisions of such tumor cells could be large, no diversity in the driver-mutation region will be apparent; therefore, variability in this specific region cannot be used to infer depth.
The other main advantage of cell lineage reconstruction with the use of MS stems from the high mutation rate of MSs is that examining a rather limited subset of the cell genome can show useful lineage information. Cell lineage trees thus reconstructed show groups of cells that are genetically close and, therefore, are clustered together on the reconstructed cell lineage tree and will be termed lineages in this study. Some of these lineages correspond to distinct functional cell groups, as retrieved and analyzed from patients, for example, cancer cells at diagnosis, at relapse, T cells, LSCs, and so forth. Of course, it is gratifying and illuminating when such functionally distinct cells have characteristic features in the reconstructed cell lineage tree, such as distinct depth, or membership in distinct clusters.
In the present study the reconstructed cell lineage trees show that, for at least some patients with acute myeloid leukemia (AML), relapse after chemotherapy was initiated by cells that divided rarely before relapse. Furthermore, at least in 1 case the cells that initiated relapse are close to LSC-enriched populations (LSCEPs) sampled at diagnosis. In contrast, this phenomenon could not be observed in the reconstructed cell lineage trees of patients with ALL.
Methods
Research cohort
Peripheral blood (PB) and BM biopsy samples paired at diagnosis and relapse were collected from 2 patients with AML, 2 patients with ALL, and 1 patient with chronic myeloid leukemia (CML) blast crisis admitted to the Rambam Health Care Campus in Haifa, Israel, and at The Ohio State University Comprehensive Cancer Center. Another patient with AML was analyzed at diagnosis only because no relapse occurred. Patients with AML and ALL were randomly selected, and the only inclusion criteria were male sex (to facilitate single-cell phylogenetic analysis as described below) and having paired samples from diagnosis and relapse. The clinical, genetic, and cytogenetic characteristics of the patients are described in supplemental Table 2 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All patients provided signed informed consent in accordance with the Declaration of Helsinki, and the study was approved by the Rambam and The Ohio State University Institutional Review Boards (approval no. 028009).
The method of the present study is shown in Figure 1. The first step was cell preparation (supplemental Methods).
Immunofluorescence and sorting of leukemic cells
After relapse, PB leukemia cells harvested from both diagnosis and relapse were thawed, and leukemic cells were then sorted by FACS (FACSAria II; BD Biosciences) according to each patient-specific leukemia immunophenotype (LIP; supplemental Figure 1; supplemental Tables 3-4). Sorted T cells were used as control cells for the patients with AML.
LSCEP separation
For the separation of LSCEPs of patient L2, only the BM cells from diagnosis were thawed and lineage-negative (LIN−) cells were separated by the lineage cell depletion kit (Miltenyi Biotec). LIN− cells were further sorted by FACS to purify CD34+CD38−CD90+ cells (supplemental Figure 1E). This method does not separate only LSCs and does not separate all LSCs.27 However, the cell population it separates is enriched with LSCs and therefore will be designated as an LSCEP.
WGA
After single cell isolation (supplemental Methods) whole genome amplification (WGA) was performed with the Illustra GenomiPhi V2 DNA Amplification kit (GE Healthcare Life Sciences) according to the manufacturer's instructions28 (see supplemental Methods for more details).
Tree and depth reconstruction
MSs were amplified by a multiplex PCR (details in supplemental Methods; supplemental Table 1). Only cells in which > 25 alleles were amplified were included in the analysis (supplemental Tables 5-10). The number of cells was determined to be ∼ 30 cells from each group analyzed, for practical considerations, including cost of analysis. To achieve coverage of 25 amplified alleles, ∼ 50 cells were first amplified by the WGA reaction, and cells with successful amplification were chosen. The exact number of cells for each sample is detailed in supplemental Table 11.
Capillary signals were analyzed with an automatic signal analysis program developed in our laboratory.13,14 Lineage trees were reconstructed with the distance-based neighbor joining (NJ) algorithm29 that used mean normalized absolute difference in repeat size between each pair of cells as the genetic distance (for details see supplemental Methods).The number of MS mutations was calculated by comparing the signal in 120 MSs of each cell with a putative root (root signature was taken as the median of the allele size values of all cells from both diagnosis and relapse14 ). This genetic distance from the root correlates well with the number of cell divisions and is defined as cell depth (Figure 2A).
Statistical analysis
Depth distributions cannot be assumed to be normal; hence, the nonparametric test, Kolmogorov-Smirnov, was used to calculate P values for the difference in depth distributions (PKS). To avoid outliers we only used data between the 25th and 75th percentiles. The Wilcoxon rank sum test was also applied for the depth differences P values to validate the results (PRS). Hypergeometric tests were performed for each internal branch to assess whether subtree leaves are clustered for a cell population. P values declared as significant are corrected for multiple hypothesis testing with the use of a false discovery rate of .2.
Reliability of reconstructed lineage trees
Bootstrap analysis was used to evaluate the accuracy of tree topology, the clustering on the tree, and the depth separation according to cell type (100 iterations were used). We also validated the results by randomly removing leaves in the tree, and measured the robustness of the clustering and depth separation. Details are provided in supplemental Table 12.
Quality controls
We took precautions to address potential experimental errors. One source of error could be misclassification of leukemia cells at diagnosis or relapse. For example, incorrect labeling of normal hematopoietic stem cells (expected to be shallow cells because of low replication rate) as leukemia cells at relapse might have caused the observed difference in depth between diagnosis and relapse. To preempt such error we used several approaches. In general the aim was to sort the same LIP from both diagnosis and relapse. The sorting scheme was designed according to the following principles: (1) sample the largest leukemic cell population possible, to include as many subpopulations as possible; (2) optimize the definition of leukemic cells to exclude normal cells. This aim was achieved by the following techniques: (1) Sort cells from PB which were in the blast window and expressed markers of early differentiation (CD34 or CD117; patient L2 in supplemental Figure 1C-D, patient L3 in supplemental Figure 1F-G, and patient L5 in supplemental Figure 1J-K). These kinds of cells are usually rare in the normal PB; therefore, when found at high frequencies among leukemic patients, they were considered appropriate surrogates for leukemic cells. (2) When the main leukemic population did not express markers of early differentiation, a combination of abnormal cell surface markers were used to define the main leukemic population. The explicit separation scheme for each of the patients is presented in supplemental Figure 1 and follows the above-mentioned principles.
FLT-3/internal tandem duplication
PCR mutation analysis was performed on the WGA DNA product of patient L2 only according to previous specifications30 (supplemental Methods).
PCR repeats
MSs are unstable and are prone to replication errors during multiplex PCR. To validate that our signal is not a result of PCR errors, we divided the DNA of several cells in each experiment into 2 portions, performed PCR independently on each portion, and included both results in the cell lineage tree, as if they came from different cells. The distances between PCR repeats in the reconstructed cell lineage tree provide a good indication of the contribution of PCR errors to the signal.
MSI
MSI was defined as instability at 40% of analyzed markers.31 To calculate the frequency of loci in which MSs have been mutated, an “equal (E)” or “not equal (NE)” genetic distance was used to reconstruct lineage trees and calculate the distance matrices. As a proof of concept mutation analysis was performed on patients L2 and L6 to localize the cause for the MSI. Mutational analysis of the hMLH1, hMSH2, and hMSH6 (mismatch repair, MMR, genes) was performed by denaturing high-performance liquid chromatography, followed by sequencing analysis as previously described.30 The deepest cells that were designated as MSI cells were excluded from the depth analysis and are not included in the lineage figures.
Results
AML lineage analysis
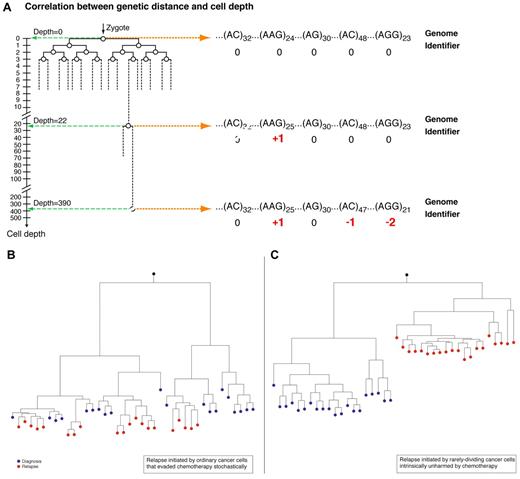
Considering the lineage relations between leukemia cells at diagnosis and at relapse, one can envision ≥ 2 possible mechanisms (Figure 2B-C) that correspond to distinct biologic scenarios. One (Figure 2B) is that relapse is initiated by rapidly dividing leukemia cells that evade chemotherapy by expansion of cells that have pharmacologic resistance to chemotherapeutic agents. Another scenario is that relapse is initiated by rarely dividing cells (RDCs; Figure 2C) that have undergone relatively few cell divisions before initiating relapse and hence would appear in a predicted shallow position in the overall leukemia cell lineage tree according to depth calculation.
Cell depth and the role of replication rate heterogeneity in relapse. (A) As cells replicate they accumulate random microsatellite (MS) alterations which create a unique genetic identifiers. The genetic distance of each cell from the root is defined as cell depth. The accumulation of MS mutations can be translated to the number of replications (deduced from MS mutation rate). Therefore, deep cells, namely cells that are far away from the root, had undergone more replications and the opposite is true for shallow cells (adapted from Wasserstrom et al14 ). (B) Relapse is initiated by cells that escape chemotherapy stochastically. Cancer cells at relapse are descendants of these cells at diagnosis. Y-axis represents depth in arbitrary units. (C) Relapse is initiated by quiescent cells unharmed by chemotherapy. The cells that initiated relapse are shallow because they divided rarely before relapse. Cells at relapse are descendants of these cells.
Cell depth and the role of replication rate heterogeneity in relapse. (A) As cells replicate they accumulate random microsatellite (MS) alterations which create a unique genetic identifiers. The genetic distance of each cell from the root is defined as cell depth. The accumulation of MS mutations can be translated to the number of replications (deduced from MS mutation rate). Therefore, deep cells, namely cells that are far away from the root, had undergone more replications and the opposite is true for shallow cells (adapted from Wasserstrom et al14 ). (B) Relapse is initiated by cells that escape chemotherapy stochastically. Cancer cells at relapse are descendants of these cells at diagnosis. Y-axis represents depth in arbitrary units. (C) Relapse is initiated by quiescent cells unharmed by chemotherapy. The cells that initiated relapse are shallow because they divided rarely before relapse. Cells at relapse are descendants of these cells.
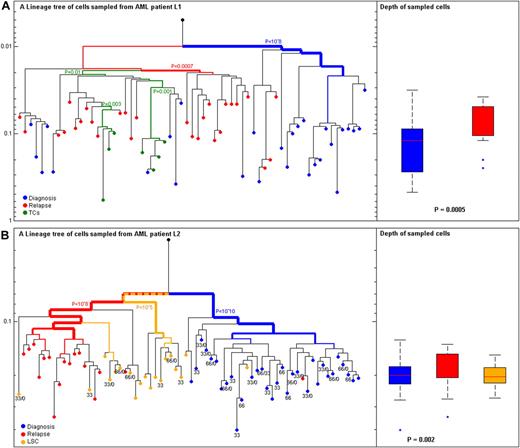
In the present study we reconstructed lineage trees of individual cells from 2 patients with AML (L1 and L2) sampled longitudinally during the course of their disease (relapse occurred 4 and 5 months after diagnosis, respectively). The clinical and LIP characteristics of the patients are described in supplemental Tables 2 and 3. For both patients with AML the depth of cells at diagnosis (L1: n = 28 cells, depth = 0.1; L2: n = 32 cells, depth = 0.082) was significantly higher than cells at relapse (L1: n = 28 cell, depth = 0.05; L2: n = 17cells, depth = 0.054), L1 PKS-test = .0005, L2 PKS-test = .002 (Figure 3). We further studied LSCEPs at diagnosis, which were available for analysis for patient L2 (n = 19). Although LSCEPs are regarded quiescent and slowly replicating and, therefore, predicted to be shallow on a lineage tree, no differences in depth between LSCEPs at diagnosis and other leukemic cells from diagnosis were found. However, most LSCEPs clustered with leukemic cells from relapse and not with leukemic cell from diagnosis (Figure 3B). T cells from patient L1 (n = 7) were found to cluster to a separate lineage from leukemia lineages (Figure 3A; hypergeometric P = .01). The validation of all results is presented in supplemental Table 12.
Leukemia cells at diagnosis and relapse from 2 patients with AML. (A) Patient L1 with acute myeloid leukemia (AML). (Left) Reconstructed lineage tree of CD33+CD4+CD15− blasts from peripheral blood (PB) individual leukemia cells at diagnosis (blue) and relapse (red). T cells (TCs) from PB both at diagnosis and relapse were CD3+CD33−CD4− (green); root (black) was calculated from the median signal of all cells. (Right) Comparison of depth between cells at diagnosis (blue) and relapse (red; P < .0005). (B) Patient L2 with AML (Left) Reconstructed lineage tree of CD117+ blasts from PB individual leukemia blast cells at diagnosis (blue) and relapse (red). CD34+CD38−CD90+ leukemia stem cell (LSC)–enriched population from BM at diagnosis (orange). Root (black) was calculated from the median signal of all cells. (Right) Comparison of depth between cells at diagnosis (blue) and relapse (red; P = .002). The most significant P value for clustering is indicated. The width of the colored branches represents the level of clustering significance (wider = lower P value). Y-axis represents depth in arbitrary units. The numbers below the cells represent the FLT3/internal tandem duplication genotype of each cell only if duplication was present; wild types are blanks.
Leukemia cells at diagnosis and relapse from 2 patients with AML. (A) Patient L1 with acute myeloid leukemia (AML). (Left) Reconstructed lineage tree of CD33+CD4+CD15− blasts from peripheral blood (PB) individual leukemia cells at diagnosis (blue) and relapse (red). T cells (TCs) from PB both at diagnosis and relapse were CD3+CD33−CD4− (green); root (black) was calculated from the median signal of all cells. (Right) Comparison of depth between cells at diagnosis (blue) and relapse (red; P < .0005). (B) Patient L2 with AML (Left) Reconstructed lineage tree of CD117+ blasts from PB individual leukemia blast cells at diagnosis (blue) and relapse (red). CD34+CD38−CD90+ leukemia stem cell (LSC)–enriched population from BM at diagnosis (orange). Root (black) was calculated from the median signal of all cells. (Right) Comparison of depth between cells at diagnosis (blue) and relapse (red; P = .002). The most significant P value for clustering is indicated. The width of the colored branches represents the level of clustering significance (wider = lower P value). Y-axis represents depth in arbitrary units. The numbers below the cells represent the FLT3/internal tandem duplication genotype of each cell only if duplication was present; wild types are blanks.
The only driver mutation analyzed in the present study was of FLT-3/internal tandem duplication (ITD) of patient L2 who was known to be positive for the mutation from routine clinical analysis at diagnosis. The results showed that the majority of the leukemia cells at diagnosis (65%) were positive for the FLT-3/ITD mutation. After TA cloning and sequencing of ≥ 10 different clones from each cell, the 5 different alleles were categorized as either heterozygous for a 33/WT (wild-type) or 66/WT base pair duplication or hemizygous for these duplications (−/33 and −/66; Figure 3B), the fifth allele was WT/WT. The only sample that had a WT/33/66 genotype was the bulk cell sample (Figure 3B). A similar phenomenon of deletion of the WT allele and the presence of several different duplications in different variants in the same patient has been described before.32 The 8 leukemia cells at diagnosis negative for the mutation were not clustered with any subpopulation. All T cells were negative for the mutation. Note that a lower proportion of LSCs at diagnosis were FLT-3/ITD positive (42%), and none of the cells at relapse were positive.
PCR repeats
The analysis of signal-to-noise ratio indicated that most PCR repeats were genetically closer to each other than to all other cells (supplemental Figure 2), confirming the relative accuracy of the method. Although some inaccuracies were observed (the signal-to-noise ratio was estimated at 10:1, namely) for each 10 MS mutations identified, one was a false-positive mutation that differed between the PCR repeats. The analysis of leukemic cells from BM versus PB showed that BM and PB leukemic cells have the same depth and are not enriched on different sublineages (supplemental Figure 3).The raw data with fragment length analysis are displayed in supplemental Tables 5 through 10.
ALL lineage analysis
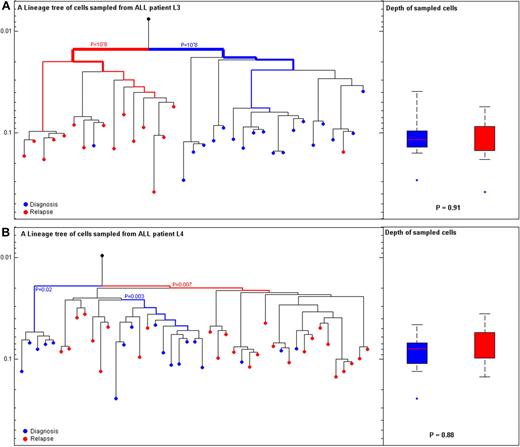
In contrast to the differences in depth observed between patients with AML, no depth differences were observed between cells at diagnosis and relapse for patients with ALL, L3 (PKS-test = .91) and L4 (PKS-test = .88; Figure 4). Relapse occurred 6 and 9 months after diagnosis, respectively, among patients with ALL. Both patients with ALL showed clonal selection of a major lineage at relapse which was different from lineages at diagnosis (Hypergeometric P L3 < .001 and L4 = .007; Figure 4).
ALL cells at diagnosis and relapse cluster to different lineages. (A) Patient L3 with B-cell acute lymphoblastic leukemia (B-ALL): (Left) Reconstructed lineage tree of CD34 blasts from peripheral blood (PB) individual leukemia cells at diagnosis (blue) and relapse (red) for patient L3. Root (black) was calculated from the median signal of all cells. (Right) Comparison of depth between cells at diagnosis (blue) and relapse (red; P|=|.91). (B) Patient L4 with B-ALL: (Left) Reconstructed lineage tree of CD2CD4CD3 blasts from PB individual leukemia blast cells at diagnosis (blue) and relapse (red) for patient L4. (Right) Comparison of depth between cells at diagnosis (blue) and relapse (red; P|=|.88). The most significant P value for clustering is indicated. The width of the colored branches represents the level of clustering significance (wider lower P value). Y-axis represents depth in arbitrary units.
ALL cells at diagnosis and relapse cluster to different lineages. (A) Patient L3 with B-cell acute lymphoblastic leukemia (B-ALL): (Left) Reconstructed lineage tree of CD34 blasts from peripheral blood (PB) individual leukemia cells at diagnosis (blue) and relapse (red) for patient L3. Root (black) was calculated from the median signal of all cells. (Right) Comparison of depth between cells at diagnosis (blue) and relapse (red; P|=|.91). (B) Patient L4 with B-ALL: (Left) Reconstructed lineage tree of CD2CD4CD3 blasts from PB individual leukemia blast cells at diagnosis (blue) and relapse (red) for patient L4. (Right) Comparison of depth between cells at diagnosis (blue) and relapse (red; P|=|.88). The most significant P value for clustering is indicated. The width of the colored branches represents the level of clustering significance (wider lower P value). Y-axis represents depth in arbitrary units.
MSI and CML blast crisis relapse
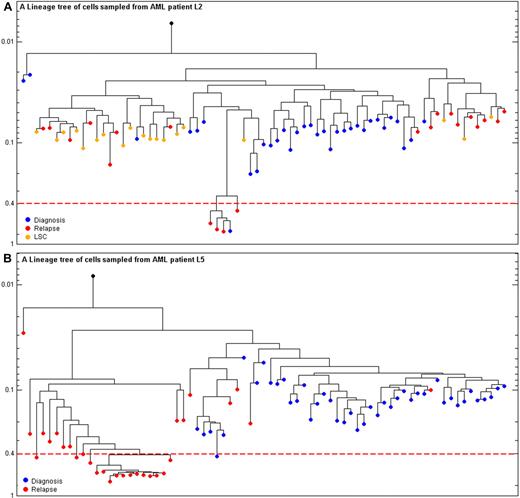
The longitudinal analysis of individual cells enabled the monitoring of minor lineages and their evolution during tumor progression. One of these minor lineages was a MS-unstable lineage whose detection was enabled by inspecting MSs. As described in “Methods,” MSI was defined according to accepted guidelines. Plotting the MSI cells on the same tree as MS-stable cells shows the relation of mutation rates variability attributable to MSI (supplemental Figure 4). Of the 6 patients analyzed, 5 patients developed an MSI lineage at some stage of leukemia, and for 3 patients (L2, L3, and L5) the MSI lineage was enriched at relapse (Table 1; Figure 5). Remarkably, for patient L5 (CML blast crisis) a substantial portion of cells at relapse, which occurred 9 months after blast crisis, were MSI (16 of 30, 53%) in comparison with the rare frequency of MSI cells at diagnosis (1 of 45, 2.2%; Figure 5). For the cause of MSI among our patients, a novel missense mutation in MSH6 (I1054F) was identified in 2 L6 leukemia cells (supplemental Figure 4). For patient L2 no mutations in MMR genes were found. The mutational status of MMR genes was not determined for any of the other patients.
MSI among the cohort patients
| Pt . | Disease . | Status . | Bulk DNA MSI . | MSI + leukemia (%)* . |
|---|---|---|---|---|
| L1 | AML | Diagnosis | Negative | (2.7) 1/38 |
| L1 | AML | Relapse | Negative | (3.7) 1/27 |
| L2 | AML | Diagnosis | Negative | (2.8) 1/35 |
| L2 | AML | Relapse | Negative | (10.5) 4/38 |
| L3 | ALL | Diagnosis | Negative | (0) 0/29 |
| L3 | ALL | Relapse | Negative | (9.7) 3/31 |
| L4 | ALL | Diagnosis | Negative | (0) 0/18 |
| L4 | ALL | Relapse | Negative | (0) 0/27 |
| L5 | CML blast crisis | Diagnosis | Negative | (2.2) 1/45 |
| L5 | CML blast crisis | Relapse | Negative | (55) 18/33 |
| L6 | AML | Diagnosis | Negative | (6.9) 2/29 |
| Pt . | Disease . | Status . | Bulk DNA MSI . | MSI + leukemia (%)* . |
|---|---|---|---|---|
| L1 | AML | Diagnosis | Negative | (2.7) 1/38 |
| L1 | AML | Relapse | Negative | (3.7) 1/27 |
| L2 | AML | Diagnosis | Negative | (2.8) 1/35 |
| L2 | AML | Relapse | Negative | (10.5) 4/38 |
| L3 | ALL | Diagnosis | Negative | (0) 0/29 |
| L3 | ALL | Relapse | Negative | (9.7) 3/31 |
| L4 | ALL | Diagnosis | Negative | (0) 0/18 |
| L4 | ALL | Relapse | Negative | (0) 0/27 |
| L5 | CML blast crisis | Diagnosis | Negative | (2.2) 1/45 |
| L5 | CML blast crisis | Relapse | Negative | (55) 18/33 |
| L6 | AML | Diagnosis | Negative | (6.9) 2/29 |
MSI indictes microsatellite instability; Pt, patient; AML, acute myeloid leukemia; ALL, acute lymphoblastic leukemia; and CML, chronic myeloid leukemia.
The total number of cells used for the analysis of the percentage of MSI cells was the actual number of cells which underwent successful whole genome amplification (> 25% amplified alleles). Cells that were excluded from the lineage reconstruction were not excluded from the MSI analysis.
Enrichment of MSI cells at relapse. (A) Patient L2: reconstructed lineage tree of CD117+, PB individual leukemia blasts cells at diagnosis (blue) and relapse (red) and CD34+ CD38− CD90+ leukemia stem cells from bone marrow at diagnosis (orange) for patient L2. The dashed red line is the threshold above which cells are designated as MSI. Y-axis represents depth. The scale is the fraction of MSs with mutated allele divided by total number of MSs amplified for each cell. (B) CML blast crisis patient L5: reconstructed lineage tree of CD34+ CD117+ CD33+, from PB, individual leukemia blast cells at diagnosis (blue) and relapse (red) for patient L5. The dashed red line is the threshold above which cells are designated as MSI. Y-axis represents depth. The scale is the fraction of MSs with mutated allele divided by total number of MSs amplified for each cell.
Enrichment of MSI cells at relapse. (A) Patient L2: reconstructed lineage tree of CD117+, PB individual leukemia blasts cells at diagnosis (blue) and relapse (red) and CD34+ CD38− CD90+ leukemia stem cells from bone marrow at diagnosis (orange) for patient L2. The dashed red line is the threshold above which cells are designated as MSI. Y-axis represents depth. The scale is the fraction of MSs with mutated allele divided by total number of MSs amplified for each cell. (B) CML blast crisis patient L5: reconstructed lineage tree of CD34+ CD117+ CD33+, from PB, individual leukemia blast cells at diagnosis (blue) and relapse (red) for patient L5. The dashed red line is the threshold above which cells are designated as MSI. Y-axis represents depth. The scale is the fraction of MSs with mutated allele divided by total number of MSs amplified for each cell.
A summary of the number of cells analyzed for each patient and the exclusion criteria are presented in supplemental Table 11.
Reliability of reconstructed lineage trees
Bootstrapping showed that the robustness of any particular branch in the tree is low (supplemental Figure 5), but the robustness of our results on clustering and depth separation according to cell type is high. Greater than 95% of the trees show the same significant clustering of cells from diagnosis, relapse, and LSCs (L1 and L2). In addition, 90% of the trees show significant depth separation between cells from diagnosis and cells from relapse (L1; supplemental Table 12).
The results were also validated by randomly removing one leaf in the tree. Significant clustering and depth separation according to cell type were shown in almost 100% of the reconstructed trees (for L1 and L2). In fact, the majority of the trees (> 70%) show significant clustering even after the random removal of ∼ 25% of the leaves and depth separation after the random removal of > 50% of the leaves (supplemental Table 12). Significant clustering and depth differences between diagnosis and relapse among patients with AML were shown with the use of both maximum likelihood and absolute distance (supplemental Figures 6-7; supplemental Table 12). Euclidian distance and mean squared distance, yielded inconclusive results (ie, the clustering and the depth differences) were not always significant, and the bootstrap values were low (supplemental Figures 8-9; supplemental Table 12).
Discussion
In the present study cells of patients with various leukemia subtypes were sampled at 2 different time points (diagnosis and relapse), somatic mutations in MS were analyzed in the sampled cells and were used to reconstruct cell lineage trees for each patient. Diversity among leukemic cells is not merely the result of random mutations that accumulate during the large number of replications that create the tumor. Rather, diversity is shaped by various mechanisms that may exist even within seemingly homogeneous environments.33
Among one of the patients with AML in the present study (Figures 3B and 4A, patient L2), ≥ 3 different genetic lineages were either enriched or involved during the course of clinical relapse. Remarkably, all of the 3 different lineages at relapse were FLT-3/ITD negative, although the major lineage at diagnosis was positive for the duplication. This phenomenon of disappearance of FLT-3/ITD during relapse has been described in the past and supports the notion that the duplication is a secondary event and not necessarily needed for relapse.34
The first lineage of patient L2 at relapse includes a cell which is most probably a direct descendent of the cells at diagnosis (the single red [relapse] cell in the group of blue cells [diagnosis]; Figures 3B and 4A). This phenomenon probably represents clonal evolution, as was recently reported.35
The second lineage is comprised of cells that are not presented in Figure 3 but are shown in Figure 4A and are MSI cells that were enriched during relapse. Enrichment of MSI cells was observed in other patients too and will be further discussed.
The third lineage includes cells at relapse that cluster together with LSCEPs from diagnosis, a sublineage that is related to cells from diagnosis but distinct from the main clone at diagnosis. This is the major lineage at relapse which is composed of mainly RCDs that are shallow on the lineage tree and have a strikingly lower history of mitotic divisions compared with the sampled leukemia cells at diagnosis. Such a lineage was also enriched during relapse of the other patient with AML analyzed (L1 in Figure 3A). The presence of the RDC lineage at relapse that clustered with LSCEPs harvested at diagnosis is consistent with the enrichment of RDCs after chemotherapy among patients with AML. Heterogeneity in leukemia cell replication rate has been known for a long time.36 The role of quiescent cells in relapse has been shown in chronic leukemia37,38 but never proven for an acute leukemia where cancer cells divide frequently.39 The results of the present study might explain at least some of the failure of chemotherapy to eradicate AML and indicate that, to prevent relapse, leukemia therapy must also target RDCs and other subpopulations. The results of the present study cannot shed light on the magnitude of the RDC relapse mechanism among patients with AML or about the polyclonality of the relapse process because it is based on only 2 patients with AML and lacks functional analysis. However, the existence of such a mechanism is supported by the large number of individual cells sampled. Applying cell lineage analysis to additional patients may enable determination of the relative contribution of this mechanism and hence the design of improved therapy for leukemia and other types of cancer with similar relapse mechanisms.
In contrast to the patients with AML, no RDC enrichment was observed during the relapse process of ALL (Figure 4). However, both patients with ALL showed > 1 lineage at relapse. The major lineage at relapse was clustered separately from the major lineage at diagnosis, a new distinct lineage. Other lineages at relapse were clustered with cells at diagnosis; these cells most probably represent a simple clonal evolution. Previous studies of patients with ALL have shown that LSCs were found to be prevalent, implying that not all cancer stem cells replicate slowly,40 and therefore does not exclude the role of LSCs in the progression of ALL.
One common relapse mechanism observed among patients with AML and patients with ALL and specifically in the patient with CML blast crisis was the enrichment of MSI cells during the relapse process (Figure 5). MSI is known to increase mutation rates in MS sites.41 We designated a cell in our cohort as MSI when > 40% of its analyzed MS sites were mutated.31 MSI cells were found in variable proportions in 83% of the patients (Table 1). MSI has been previously described in patients with AML by bulk cell analysis42,43 and was found to be related to disease progression among patients with CML.44 A more recent study45 presented a novel mechanism whereby somatic deletions of genes regulating hMSH2 (one of the MMR genes) degradation result in undetectable levels of hMSH2 protein in leukemia cells, DNA MMR deficiency, and drug resistance. Accordingly, the finding of MSI among subpopulations of cells and sometimes enriched in relapse is consistent with current findings of the importance of this mechanism in different types of leukemia relapse. In the current analysis an hMSH2 missense mutation was diagnosed. However, not only mutations in MMR genes are relevant but also other genes involved in MMR gene degradation or other mechanism might result in MSI. Specifically in CML it was found that the BCR/ABL mutant inhibits the MMR genes44 ; therefore, no attempt to find MMR genes mutation was performed for patient L5. We suggest that the discovery of minor but clinically relevant lineages with MSI might require single-cell analysis, although it might be captured by a specifically designed deep sequencing assay of bulk cells.
As can be observed in the various cell lineage trees in the present study, some cells from various subpopulations (diagnosis, relapse LSCEP, and T cells) can be located not in their major lineage. For example, with regard to patient L2, not all RDCs clustered with the LSCEP (Figure 3B). Several explanations are possible: inaccurate assignment of the presumed population because of misclassifications of cells during the sorting process. The sorting of leukemic cells from the bulk population is complicated because an abnormal LIP could not be assigned in all cases. Sorting the blast population only might also introduce mistakes because normal early progenitors might also be included in this sorted population. Our sorting scheme aimed at excluding this kind of bias. Another possible explanation for the malposition of RDCs is that some RDCs do not originate from the LSCEP analyzed in the present study (LIN−CD34+CD38−CD90+), but rather from other LSC subpopulations, or not from LSCs at all. Inaccurate lineage tree reconstruction could also be because of several limitations of the tree reconstruction methods and the limited number of genetic markers used in the present study. As was shown by the bootstrap analysis, low bootstrap scores were calculated for any specific branch of the tree, most probably because of the low number of mutations. However, because we do not place biologic significance on any particular branch in the tree, only on its overall shape, as measured by clustering and depth separation according to cell type, this analysis indicates that our data provide robust support for our claims. To accurately reconstruct a tumor lineage tree more genetic variants should be genotyped in the maximum number of cells. Recent studies have found that deep whole genome sequencing of diagnosis and relapse–paired bulk DNA samples35 can shed some light on the organization of leukemia subpopulations and uncover functionally important variants. However, one cannot accurately infer the relation between different genetic variants because they are all pooled together in the bulk DNA, whereas in single-cell analyses each cell has a unique genetic signature that can be compared with other cells.
It has been suggested in the past that highly polymorphic genetic markers (such as MS) can lead to underestimation of divergence between populations and, accordingly, to inaccurate tree topologies.42,46 However, it should be noted that despite these drawbacks MS-based NJ was used by our group in the past and validated on various organisms (mouse and Arabidopsis), both in vivo and also in an ex vivo model of lineage trees.12,15 Furthermore, NJ reconstruction algorithm, with absolute distance measure, was consistently better comparing with several other algorithms, and using the normalized absolute distance measure gives a slightly more precise results than comparing with other distance measures (unpublished data). Furthermore, the results of the maximum likelihood–based reconstructed lineage trees (supplemental Figure 6) correlated well with the normalized absolute distance NJ trees and showed the same depth differences between diagnosis and relapse among patients with AML. Other distance measures (such as Euclidian distance measure)were also used and showed similar clustering and depth differences between diagnosis and relapse among patients with AML (supplemental Figures 6-9).47
The distinct clustering of presumably normal T cells from leukemia cells (Figure 3A) validates the classification and sorting process in the present study. The appearance of leukemia cells in the T-cell lineage may be the result of imprecision in cell lineage tree reconstruction because of the reasons mentioned earlier, or because of cell fusion between a cancer cell and a normal cell closer to the T-cell lineage.48,49
To conclude, the present study correlates replication rate heterogeneity with relapse and possibly to some degree LSCEP among patients with AML and sheds light on the role of MSI in disease progression. Despite the limitations of the current methods to precisely reconstruct leukemia cell lineage tree, the main results on clustering and depth differences among cells sampled at diagnosis and relapse, the clustering of LCSEP and leukemic cells at relapse, and the presence of MSI lineages are robust regardless of the precise tree topology. The enrichment of RCDs and MSI cells after chemotherapy cannot be explained by a stochastic process and, therefore, is more likely to reflect a selective advantage because of the pressure exerted by chemotherapy. The application of cell lineage analysis enabled the uncovering of these resistance mechanisms and the multiplicity of lineages contributing to the relapse process. Replication rate heterogeneity and MSI are most probably only the tip of the iceberg. Other kinds of heterogeneity probably also exist10 and should be further explored in a larger cohort and other types of cancer. Understanding the whole spectrum of tumor heterogeneity at different stages of the disease may enable the development of a repertoire of therapies directed at specific cancer cell subpopulations. This is consistent with the approach of viewing cancer therapy in terms of turning a rapidly lethal disease to a chronic manageable disease, in which the individual patient with cancer and the individual cancer cells comprising the malignancy for that patient are followed and treated over time.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Shalev Itzkovitz for helpful discussions and the European Union FP7-COSTEuGESMA action.
This work was supported by The ISF Converging Technologies (grant 1694/07), The European Union (grant FP7-ERC-AdG), Miel de Botton Aynsley Foundation, Paul Sparr Foundation, the Arthur and Rosalinde Gilbert Foundation (K.S.), the Soref Foundation (American Technion Society; K.S.), Slava Smolakovski Fund (Rambam Medical Center; K.S.), the Etai Sharon Atidim grant program of Rambam Medical Center (L.I.S.), and the Israel Cancer Association (grant 20110054-B 2011; L.I.S.). E.S. is the incumbent of The Harry Weinrebe Professorial Chair of Computer Science and Biology. K.S. is the incumbent of the Annie Chutick Chair in Medicine at the Technion–Israel Institute of Technology. S.I. would like to thank the Israeli Cancer Foundation.
Authorship
Contribution: L.I.S., J.M.R., S.I., T.Z., K.S., and E.S. designed the research; L.I.S., N.C.-I., R.A., N.P., R.S., D.B., S.I., G.M., C.D.B., M.T., and T.Z. and performed research; L.I.S., N.C.-I., Y.M., A.S., S.I., and E.S. analyzed and interpreted data; and L.I.S., T.Z., K.S., and E.S. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Ehud Shapiro, Department of Computer Science and Applied Math and Department of Biological Chemistry, Weizmann Institute of Science, Rehovot 76100, Israel; e-mail: ehud.shapiro@weizmann.ac.il.
References
Author notes
L.I.S., N.C.-I., and R.A. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal