Abstract
During the past decade it was recognized that homeobox gene families such as the clustered Hox genes play pivotal roles both in normal and malignant hematopoiesis. More recently, similar roles have also become apparent for members of the ParaHox gene cluster, evolutionarily closely related to the Hox gene cluster. This is in particular found for the caudal-type homeobox genes (Cdx) genes, known to act as upstream regulators of Hox genes. The CDX gene family member CDX2 belongs to the most frequent aberrantly expressed proto-oncogenes in human acute leukemias and is highly leukemogenic in experimental models. Correlative studies indicate that CDX2 functions as master regulator of perturbed HOX gene expression in human acute myeloid leukemia, locating this ParaHox gene at a central position for initiating and maintaining HOX gene dysregulation as a driving leukemogenic force. There are still few data about potential upstream regulators initiating aberrant CDX2 expression in human leukemias or about critical downstream targets of CDX2 in leukemic cells. Characterizing this network will hopefully open the way to therapeutic approaches that target deregulated ParaHox genes in human leukemia.
Introduction
The last decade has seen large advances in our understanding of the regulatory network in prenatal and adult hematopoiesis. As one pivotal class of regulatory factors, the highly conserved family of homeobox genes have been identified as playing major roles not only in normal hematopoiesis but also in leukemogenesis.1 The homeobox was first identified in the 1980s as a sequence motif shared among Drosophila homeotic genes (the HOM-C complex) that play key roles in embryonic differentiation along the anterior-posterior axis. The existence of homeotic genes was already proposed in the early 1900s, based on observations of Drosophila mutants with changes in the body structure.2 The homeobox is now known to be present in many genes in virtually all eukaryotic species.3 It is estimated that the human genome contains ≥ 200 homeobox genes.4 Unlike the HOM-C/HOX genes, which are organized in gene clusters, most homeobox genes are dispersed throughout the genome.5,6 Homeobox-containing genes constitute a gene family characterized by a highly conserved 183-nucleotide sequence encoding a 61-aa domain, the homeodomain (HD). These HDs are structurally related to the helix-turn-helix motif of prokaryotic DNA-binding proteins and have sequence-specific DNA binding activity.7 Homeobox genes are divided into 2 classes. Class I includes clustered genes (HOX) that comprises 39 members, and the class II divergent homeobox genes are dispersed through the genome and include smaller families such as the MSX, ParaHox (CDX), PAX, DLX, and Engrailed (EN) groups. Although individual members of homeobox families often share little sequence similarity other than the homeobox, some families have additional conserved sequence motifs that contribute to their distinct functional properties.5,8
A key role of homeobox genes for normal and malignant stem and progenitor cell behavior was first shown for Hox genes. Initial hints that these genes are essential for regulating HSCs and progenitor cells came from data showing that Hox genes of the A and B cluster are highly expressed in normal murine and human HSCs and progenitor cells, but silenced in their more differentiated progeny.9,10 Studies in experimental models have convincingly proven the functional relevance of Hox genes for normal hematopoietic behavior in mouse and man; for example, Hoxa9−/− BM cells manifest a severe defect at the level of HSCs and committed progenitors11,12 ; however, aberrant expression of Hox/HOX genes such as Hoxa9 or Hoxa10 can induce acute myeloid leukemia (AML) in mice, whereas constitutive expression of Hoxb4 increases the self-renewal capacity of murine HSCs without initiating leukemic transformation.13-15 Similar data were obtained for the human system with the use of retroviral gene transfer techniques and the NOD/SCID xenograft model.16,17 Early data indicated aberrant expression of HOX genes in patients with acute leukemia,18 and this was confirmed later by microarray analyses in large cohorts of patients with AML, thereby associating aberrant expression of HOX genes such as Hoxa9 with clinical outcome.19-22 The rich body of data on the role of Hox genes in normal and malignant hematopoiesis has been reviewed in detail before.1,7,8,23 In the past years, however, it was shown that other homeobox genes, not belonging to the clustered Hox genes, have a crucial role in regulating early hematopoietic cells and facilitating leukemic transformation. One example is the so-called “three-amino-acid loop extension” superfamily, forming an “atypical class” of homeobox genes that show low sequence identity with other classes of homeobox genes.24 Most prominent members of this family are Meis1 and Pbx1. Both can directly interact with Hox genes and with each other, thereby forming trimeric complexes, which alter Hox–DNA binding properties and in the case of Meis1 accelerate Hox-induced leukemogenesis.25 This review focuses on another homeobox gene family, the ParaHox cluster, which was shown to be involved in normal as well as malignant hematopoiesis.6,24
The ParaHox cluster genes: emerging key players in normal and malignant hematopoiesis
Genomic organization and molecular characteristics
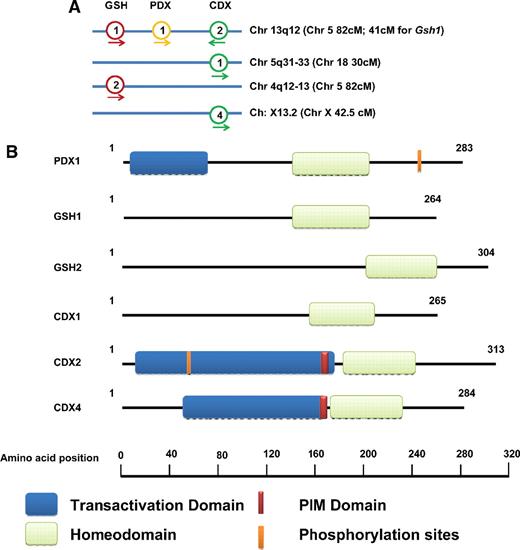
The ParaHox gene cluster was discovered in 1998 by Brooke et al, who reported that Gsx (genomic screened homeobox), Xlox (Xenopus laevis homeobox), and Cdx (Caudal-type homeobox) genes form their own cluster in the cephalochordate amphioxus.26 This cluster was termed the ParaHox cluster because it showed homology to the different paralogue groups of the Hox cluster. In mice and man homologous genes cluster on chromosome 5 and 13q12, respectively; these comprise the genes Gsh1/GSH1, Xlox/XLOX (also known as Pdx1/PDX1), and CDX2.27,28 In contrast to chordates, mammals have additional ParaHox genes, namely Gsh2/GSH2, Cdx1/CDX1, and Cdx4/CDX4, which are spread over 3 different chromosomes in the human genome (Table 1; Figure 1A).
Characteristics of ParaHox genes
| Human gene . | Protein size, kDa . | Homology between man and mouse, %* . | Chromosomal translocation . | Aberrant expression in leukemia . | Transforming potential . | Phenotype of knockout mice . |
|---|---|---|---|---|---|---|
| GSH1 | 27 | 98.01 | — | Unknown | Not tested | Phenotypically normal at birth, but 75% of mice die at 4 wks of age; marked growth retardation, infertile, and low WBC count29 |
| GSH2 | 32 | 89.14 | CHIC2-ETV630 | AML (case reports)30 | Transforms NIH 3T3 cells in vitro30 | Born healthy, but death within 24 hours after birth; homozygous mutants have apnea and severe brain alterations31 |
| PDX1 | 30 | 87.99 | — | Unknown | Not tested | Death at 1 dpp; apancreatic32 |
| CDX1 | 28 | 85.28 | — | Not expressed33 | Not tested | Viable and fertile, anterior homeotic transformations; posterior shifts of Hox gene expression in the somitic mesoderm34 |
| CDX2 | 33 | 93.89 | ETV6-CDX235 | AML, ALL, CML in blast crises33,35,36 | Causes AML in mice37 | Peri-implantation embryonic lethality; heterozygous mutants show growth retardation and anterior transformation38 |
| CDX4 | 30 | 82.98 | — | AML39 | Overexpression induces AML in mice39,40 | Viable, only a mild anterior transformation with low penetrance41 |
| Human gene . | Protein size, kDa . | Homology between man and mouse, %* . | Chromosomal translocation . | Aberrant expression in leukemia . | Transforming potential . | Phenotype of knockout mice . |
|---|---|---|---|---|---|---|
| GSH1 | 27 | 98.01 | — | Unknown | Not tested | Phenotypically normal at birth, but 75% of mice die at 4 wks of age; marked growth retardation, infertile, and low WBC count29 |
| GSH2 | 32 | 89.14 | CHIC2-ETV630 | AML (case reports)30 | Transforms NIH 3T3 cells in vitro30 | Born healthy, but death within 24 hours after birth; homozygous mutants have apnea and severe brain alterations31 |
| PDX1 | 30 | 87.99 | — | Unknown | Not tested | Death at 1 dpp; apancreatic32 |
| CDX1 | 28 | 85.28 | — | Not expressed33 | Not tested | Viable and fertile, anterior homeotic transformations; posterior shifts of Hox gene expression in the somitic mesoderm34 |
| CDX2 | 33 | 93.89 | ETV6-CDX235 | AML, ALL, CML in blast crises33,35,36 | Causes AML in mice37 | Peri-implantation embryonic lethality; heterozygous mutants show growth retardation and anterior transformation38 |
| CDX4 | 30 | 82.98 | — | AML39 | Overexpression induces AML in mice39,40 | Viable, only a mild anterior transformation with low penetrance41 |
WBC indicates white blood cell; CML, chronic myeloid leukemia; and —, not described.
At amino acid level.
Characteristics of ParaHox genes. (A) Chromosomal location of human ParaHox genes. Chromosomal locations of murine ParaHox genes are given in brackets. The direction of transcription is indicated by an arrow. (B) Protein structure of ParaHox genes; shown is the protein structure depicting the N-terminal transactivation domain (blue), the Pbx-interacting motif (PIM; brown), the HD (green), and validated phosphorylation sites (orange).
Characteristics of ParaHox genes. (A) Chromosomal location of human ParaHox genes. Chromosomal locations of murine ParaHox genes are given in brackets. The direction of transcription is indicated by an arrow. (B) Protein structure of ParaHox genes; shown is the protein structure depicting the N-terminal transactivation domain (blue), the Pbx-interacting motif (PIM; brown), the HD (green), and validated phosphorylation sites (orange).
The ParaHox and Hox clusters are thought to originate from an ancient hypothetical ProtoHox cluster by duplication before the split of protostomes and deuterostomes, characterizing the ParaHox cluster as one of the evolutionary early homeobox gene clusters.6,42,43 This also explains the homologies of the ParaHox genes to paralogue groups of the Hox cluster. The paralogue groups of the Hox cluster subdivide the Hox genes into 4 groups: the anterior group (paralogue group [PGs] 1-2), group 3, central group (PGs 4-8), and posterior group (PGs 9-13).6,43 The Gsx gene is more closely related to Hox anterior group genes than to other ParaHox genes; Xlox is more similar to Hox group 3, and Cdx is more similar to the Hox posterior group genes.
Structure–functional analyses have characterized the molecular characteristics of ParaHox proteins in detail (Table 1; Figure 1B). As for homeobox genes, all the ParaHox genes are characterized by a DNA-binding HD and in part by a N-terminal transactivation domain. However, homology between the different family members of the ParaHox genes is relatively low and proteins display individual features. Thus, the ParaHox gene PDX1 contains an antennapedia-type hexapeptide motif (PFPWMK), which preferentially binds the DNA motif 5′-[CT]TAAT[TG]-3′. PDX1 is phosphorylated by HIPK2 on Ser-268 on glucose accumulation, which mediates shifting of the protein from nuclear periphery to nucleoplasm.44 GSH1 contains an additional Snail/Gfi domain essential for transcriptional repression. On the basis of bioinformatics analysis it is postulated that GSH genes bind to DNA as monomers or as homodimers and/or heterodimers, in a sequence-specific manner. CDX1, in contrast to other CDX members, does not contain the highly conserved pentapeptide domain for binding of the three-amino-acid loop extension cofactor Pbx1.
Lessons from knockout mice: initial clues to function of ParaHox genes
Knockout mice have been an important tool for our understanding of ParaHox gene function and their essential role in body development (Table 1). Gsh1 homozygous mutant mice are phenotypically normal at birth but fail to grow properly after 7-10 days. Homozygous mutants exhibit extreme dwarfism, sexual infantilism, and significant perinatal mortality with total white blood cell count 5-fold decreased in mutant mice. Both lymphoid and myeloid cells were similarly reduced, whereas macrophages were not affected.29 Gsh2 knockout mice display severe structural alterations in the brain, accompanied by apnea and reduced blood oxygen levels, leading to death within the first day after birth.31 Pdx-1 was shown to be crucial for the development of the pancreas and anterior duodenum.32 Cdx1-null mutant mice are viable and fertile, but they display anterior homeotic transformations of vertebrae accompanied by posterior shifts of Hox gene expression in the somatic mesoderm.34 Cdx2-null mutant embryos die between 3.5 and 5.5 days after coitum, and heterozygotes have tail abnormalities and exhibit anterior homeotic transformations that involve the cervical and upper thoracic vertebrae, ribs, and midgut endoderm, underlining the crucial role of Cdx genes for anterior-posterior regional identity. Ninety percent of Cdx2 heterozygote mutant mice develop multiple intestinal adenomatous polyps, particularly in the proximal colon, suggesting that Cdx2 mutations might be the primary event in the genesis of some intestinal tumors.38,45,46 In contrast to Cdx2, Cdx4-null murine embryos are born healthy and do not show any major structural or histologic abnormalities.41
Cdx2 and Cdx4 are key regulators of embryonic hematopoiesis
Over the past several years it has become evident that ParaHox Cdx family members are critical for ordered proliferation and differentiation of embryonic hematopoietic cells.47 This was particularly found for the Cdx genes Cdx4 and Cdx2. Evidence for a key role of Cdx genes in embryonic hematopoiesis initially came from studies in zebrafish; homozygous mutations in cdx4 caused a severe defect in embryonic hematopoiesis with a profound reduction of hemoglobin-positive erythroid cells that resulted in severe anemia.48 These blood defects were accompanied by deregulated or reduced expression of hoxb4, hoxb5a, hoxb6b, hoxb7a, hoxb8a, hoxb8b, and hoxa9a. Of note, the blood deficiency in zebrafish embryos could be rescued by overexpression of hoxb7a or hoxa9a but not by scl, indicating that the hematopoietic defect is at least partly caused by perturbation of distinct Hox genes.48 The same group found that morpholino-mediated knockdown of cdx1a in a cdx4-mutant background caused a severe perturbation of Hox gene expression and a complete failure to specify blood in zebrafish. Furthermore, these Cdx-deficient embryos displayed a significant reduction in expression of key hematopoietic genes such as scl, runx1, and gata1.49 This hematopoietic defect could be efficiently rescued by hoxa9 overexpression, but not by hoxb7.49
In mouse, ectopic expression of Cdx4 enhances hematopoietic mesoderm formation and further promotes blood progenitor specification and hematopoietic engraftment of murine embryonic stem cells (ESCs) in adult mice.50 In addition, Cdx4 overexpression rescues defective blood progenitor formation in mouse ESCs deficient in Mll, a Hox regulator involved in definitive hematopoiesis.51 Surprisingly, Cdx4 knockout mice do not show any major hematopoietic defects.40 Indeed, in both germline and conditional Cdx4 knockout models, there is no apparent defect in HSCs and progenitor cells as well as mature blood cells. This indicates that Cdx4 as a single factor is not essential for normal adult hematopoietic stem cell functions in mice and that Cdx4 function can be compensated at least partly by other Cdx genes.40 One candidate for this is Cdx2; Cdx2-deficient ESCs show impaired production of multipotential blood progenitor colonies in vitro consistent with an essential role of Cdx2 in early embryonic hematopoiesis.52 By injecting Cdx2−/− or Cdx2+/+ ESCs into normal lacZ+ blastocysts, Wang et al could show an intrinsic requirement for Cdx2 during primitive embryonic hematopoiesis in vivo.52 According to genome-wide expression analysis they reported that Cdx2 deficiency dramatically perturbs expression profiles of Hox genes and genes involved in signal transduction, cell growth and proliferation, and hematopoiesis. Expression of several components of the canonical signal transduction pathways such as the Jak/Stat and sonic hedgehog pathways were altered in Cdx2−/− embryonic bodies (EBs). Expression of several hematopoiesis-specific genes, including Scl, Gata1, Runx1, and β-H1 globin, were markedly decreased in Cdx2-defecient EBs,52 supporting the notion that Cdx2 is a key player during hematopoietic development.
In contrast to Cdx4 and Cdx2, loss of Cdx1 gene function does not affect hematopoietic development as shown by homozygous Cdx1 mutant mice,34 but also by studies in zebrafish, whereby cdx1a had no effect on rostral blood island hematopoiesis and the intermediate cell mass development.49 The distinct effects of the different members of the Cdx gene family was elegantly shown in a study,52,53 which ectopically expressed Cdx1, Cdx2, and Cdx4 in murine ESCs; whereas Cdx1 and Cdx4 both promoted the hematopoietic potential of the EB-derived CD41+c-kit+ population, Cdx2 grossly reduced EB-derived hematopoietic potential. Expression of all the different Cdx genes increased Hox gene expression with the notable difference that Cdx2 induced changes in both the anterior and posterior HoxA and HoxB genes, whereas Cdx1 and Cdx4 only altered posterior Hox gene expression.53
Taken together, these data indicate that Cdx genes play essential and both distinct and overlapping roles in embryonic hematopoiesis.
ParaHox genes in malignant hematopoiesis
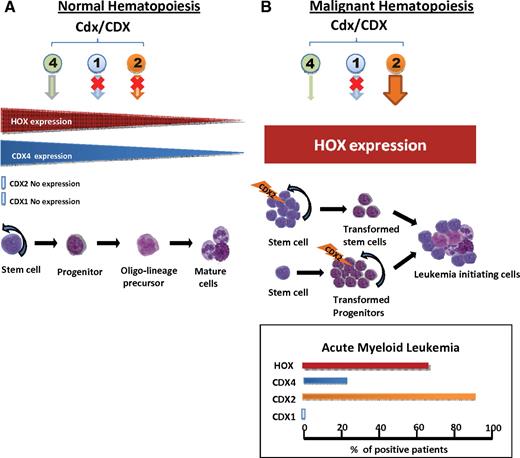
ParaHox genes have emerged as pivotal players in leukemogenesis with a rich body of evidence to link aberrant expression of the ParaHox gene Cdx2 and other ParaHox genes with acute leukemia (Table 1; Figure 2).
Cdx expression in normal and malignant hematopoiesis. (A) Normal hematopoiesis.Cdx4 and HOX A and B cluster genes are expressed in normal stem and progenitor cells, whereas Cdx2 and Cdx1 remain silenced. (B) Malignant hematopoiesis. Ectopic expression of CDX2 at the level of stem or progenitor cells induces increased expression of Hox A and B cluster genes and leukemic transformation of hematopoietic cells. The proportion of patients with AML, which ectopically express CDX2 and overexpress HOX genes, is indicated. CDX4 is expressed only in a minority of patients, and CDX1 expression is not detectable in patients with leukemia.
Cdx expression in normal and malignant hematopoiesis. (A) Normal hematopoiesis.Cdx4 and HOX A and B cluster genes are expressed in normal stem and progenitor cells, whereas Cdx2 and Cdx1 remain silenced. (B) Malignant hematopoiesis. Ectopic expression of CDX2 at the level of stem or progenitor cells induces increased expression of Hox A and B cluster genes and leukemic transformation of hematopoietic cells. The proportion of patients with AML, which ectopically express CDX2 and overexpress HOX genes, is indicated. CDX4 is expressed only in a minority of patients, and CDX1 expression is not detectable in patients with leukemia.
GSH2
The Gsh2 gene was previously characterized as a brain-specific ParaHox gene important in forebrain and hindbrain formation of the mouse. Peter Marynen's group (Cools et al30 ) has shown that GSH2 is not expressed in adult human normal hematopoietic cells. However, the group has reported ectopic expression of GSH2 in 4 patients with AML carrying the translocation t(4;12)(q11-q12;p13), which itself generated no detectable fusion gene. GSH2 is located at 4q11-q12 and thus located closely to the respective breakpoint, which implies that the translocation induces ectopic expression of GSH2. Indeed the researchers could demonstrate that aberrant expression of GSH2 caused focus formation in the NIH3T3 assay as a surrogate marker for the oncogenic potential of this ParaHox gene.30
Cdx1
In normal adult hematopoietic tissue, expression of this homeobox gene was not reported so far, and there are no data to suggest involvement of Cdx1 in malignant hematopoiesis. However, in the adult Cdx1 is known to be involved in the intestinal differentiation and the development of intestinal metaplasias.54-56
CDX2
Aberrant expression of CDX2 is a frequent event in patients with AML.
Several studies have reported that CDX2 is normally not expressed in adult murine and human normal hematopoiesis (Figure 2). Ectopic expression of CDX2 in AML was reported by Chase et al in 1999 in a patient with AML M1 carrying the chromosomal translocation t(12;13)(p13;q12), creating a novel fusion of ETV6 and CDX2.35 In addition, aberrant expression of CDX2 was found in a case of blast crisis of chronic myeloid leukemia (CML-BC) with no detectable chromosomal abnormality.35 The fusion partner ETV6 is an important regulator of HSC survival and frequently affected by translocations.57,58 Later, we and others have reported that CDX2 is aberrantly expressed in > 80% of human AML patients, characterizing CDX2 as one of the most frequent aberrantly expressed proto-oncogenes in human leukemia (Figure 2). Aberrant expression of CDX2 was found in all types of cytogenetic subgroups. Scholl et al reported the highest expression of CDX2 in patients with t(9;11) followed by normal karyotype AML (CN-AML) and t(15;17)–positive patients,36 similar to our results with highest expression of CDX2 in CN-AML followed by t(9;11)– and t(15;17)–positive cases.33 Aberrant expression of CDX2 was not only more common in patients with CN-AML than in patients with abnormal karyotype (89% vs 64%, respectively), but the expression level of CDX2 was significantly and > 15-fold higher in patients with CN-AML as well. Furthermore, the level of CDX2 expression correlated with aberrant HOX gene expression in human AML.33 The analyses did not show any correlation of the NPM1 mutational status with the frequency and level of aberrant CDX2 expression. Aberrant CDX2 expression is also detectable in most of the human AML cell lines. The mechanisms responsible for disruption of the normal CDX2 silencing in human AML are unknown and are not because of promoter mutation, gene amplification, or abnormal promoter methylation.36 In contrast to CDX2, all CN-AML cases were negative for expression of the other members of the CDX gene family CDX1 and CDX4.33
CDX2 is aberrantly expressed in most patients with acute lymphoblastic leukemia (ALL).
Ectopic expression of CDX2 is not restricted to patients with AML; we showed that CDX2 is aberrantly expressed in most adult patients with ALL, with 81% overall positivity and a higher overall expression level than detectable in patients with CN-AML. The expression of CDX2 showed high variation between the different ALL subgroups (classified according to the criteria of the European Group for the Immunologic Characterization of Leukemias59 ) with highest median expression in pre-T-ALL, followed by c-ALL and pro-B-ALL. In contrast, patients with B-ALL/Burkitt lymphoma and thymic T-ALL exhibited only low CDX2 expression. In addition, the number of patients aberrantly expressing CDX2 differed considerably between ALL subtypes, with 100% positivity in patients with pro-B-ALL, c-ALL, and Ph+ ALL versus only 40% positivity in B-ALL/Burkitt lymphoma and ∼ 70% in thymic and pre-T-ALL.60 The expression level of CDX2 in patients with ALL significantly correlated with treatment outcome (high expression levels were associated with inferior overall survival) of the analyzed patients. Although that study was limited by a low number of patients, CDX2 remained an independent risk factor even after adjusting for the risk factors of age and presence of molecular markers by bivariate Cox regression.60 These data were confirmed for pediatric ALL whereby most patients also showed aberrant expression of CDX2. Also in this study, aberrant expression of CDX2 significantly correlated to an inferior disease outcome.61 Another recent independent study reported CDX2 expression in 61% of patients with de novo adult ALL and decrease of CDX2 expression in patients with complete remission and increase in relapse.62 Similar to AML the mechanism of aberrant CDX2 expression in ALL is not understood. We have not identified any difference in the methylation level of the CDX2 promoter in patients with CDX2-positive and -negative ALL.60 In contrast to AML, whereby aberrant CDX2 expression correlated with HOX gene deregulation, there was only a low correlation between the expression of HOX genes with the expression of CDX2, in line with reports that HOX gene deregulation is less common in ALL than in AML.63,64 Taken together, these data link aberrant expression of CDX2 to myeloid as well as lymphoid leukemia. So far it is unknown whether the stage of differentiation of the cell, which is initially hit by aberrant CDX2 expression, determines the phenotype of the leukemia. Experiments testing whether aberrant expression of CDX2 in lymphoid progenitor cells generates ALL in mice are lacking so far.
Cdx2 is highly leukemogenic in experimental models.
In a murine BM transplantation model it was shown that ectopic expression of the ParaHox gene CDX2, but not expression of the fusion gene ETV6/CDX2, which was described in a patient with translocation t(12;13)(p13;q12)–positive AML,35 was highly leukemogenic.37 This was the first direct evidence that the ParaHox gene Cdx2 is highly leukemogenic and induces aggressive AML when aberrantly expressed in hematopoietic progenitor cells. Retrovirally enforced expression of Cdx2 in murine hematopoietic progenitors resulted in increased expression of leukemogenic Hox genes such as Hoxa5, Hoxa7, Hoxa9, Hoxa10, as well as Hoxb3, Hoxb6, and Hoxb8.33 These data are in agreement with previous findings that Cdx2 can deregulate Hox gene expression in embryonic as well as adult nonhematopoietic tissues.45,65 The ability to up-regulate Hox gene expression and to induce AML clearly depended the N-terminal transactivation domain of Cdx2, known to harbor serine phosphorylation sites, which affect the transcriptional activity of the gene.33,66 Interestingly, the ETV6-CDX2 fusion gene also lacks the N-terminal transactivation domain of CDX2, which would explain the obvious discrepancy of the oncogenic potential between Cdx2 and the ETV6-CDX2 fusion.
Aberrant Cdx2 expression also led to an altered expression of a number of genes involved in lymphopoiesis such as Lef1, Tcf3, or Id3. These genes are frequently deregulated in lymphoid leukemia60,67,68 and are able in the case of Lef1 to induce ALL in mice that received a transplant.33
Most data on the leukemogenicity of Cdx2 come from murine models. However, at least at the level of human leukemia cell lines it could be shown that leukemic growth depends on aberrant CDX2 expression; thus, shRNA-mediated depletion of CDX2 in the human pre-B cell line Nalm-6 and several AML cell lines significantly impaired leukemic cell proliferation and clonogenicity.56,60
CDX4 is less leukemogenic than Cdx2.
It has been shown that overexpression of Cdx4 confers enhanced replating and proliferative potential to transduced murine hematopoietic cells. Mice transplanted with Cdx4-overexpressing BM developed a myeloproliferative disorder, which progressed to AML over time.39 In this model aberrant expression of Cdx4 had to collaborate with the homeobox gene Meis1 to induce AML in all experimental animals,39 thus clearly differing from Cdx2, which caused rapid disease in all mice that received a transplant.37 In line with this, the long latency of leukemia development in primary animals that received a transplant suggested that aberrant expression of Cdx4 alone is not sufficient to induce leukemia but rather induces a preleukemic stage. Loss of Cdx4 significantly prolonged the latency of disease onset in a mouse model of mixed-lineage leukemia-AF9–induced AML and altered the phenotype of the resultant disease with increased expression of B- and T-lymphoid markers.40 The mechanism of Cdx4-induced leukemogenesis is not completely understood. However, recently it has been shown that CDX4 activates transcription of the Hox gene HOXA10 in myeloid cells and binds to a cis element of its promoter. Vice versa HOXA10 is able to activate CDX4 expression, pointing to a positive feedback loop between CDX4 and a HOX gene, which itself is linked to myeloid leukemogenesis.17,69,70 It was also found that Cdx4 collaborates with Menin to activate Hoxa9 expression by binding with Menin at the same regulatory region at the Hoxa9 locus.71 In summary, these data categorize CDX4 as an important collaborative partner in Hox-associated leukemogenesis.
In patients with AML CDX4 is expressed aberrantly in ∼ 25% of all cases (Figure 2). CDX4 expression, however, did not correlate to a certain leukemia subtype, and its expression did not correlate to HOX gene deregulation.39 This finding suggests that the ability of CDX4 to deregulate HOX gene expression is of minor relevance for its role in human leukemia.
Mechanisms of ParaHox-induced leukemogenesis: speculations and future directions
Gene expression profiling and functional data clearly support a critical involvement of CDX2 in human leukemogenesis. However, molecular mechanisms of Cdx2-induced leukemogenesis are poorly understood. One obvious possibility is that CDX2 exerts its leukemogenic potential by deregulating expression of leukemogenic genes of the HOXA and HOXB cluster, known to be dysregulated in 65% of all AML cases, in particular in normal karyotype (CN-AML) and mixed-lineage leukemia–rearranged AML33,36 (Figure 2). This would classify the CDX-HOX axis as one of the main driving forces in human AML. However, it could be clearly shown that aberrant Cdx2 expression, besides Hox deregulation, also led to an altered expression of a number of genes involved in embryonic development, lymphopoiesis, Wnt pathway, and genes involved in stem cell function. These data support the hypothesis that Cdx2 might deregulate multiple pathways in leukemia, which would also explain the differences between Cdx2 and Cdx4 in their leukemogenic potential.
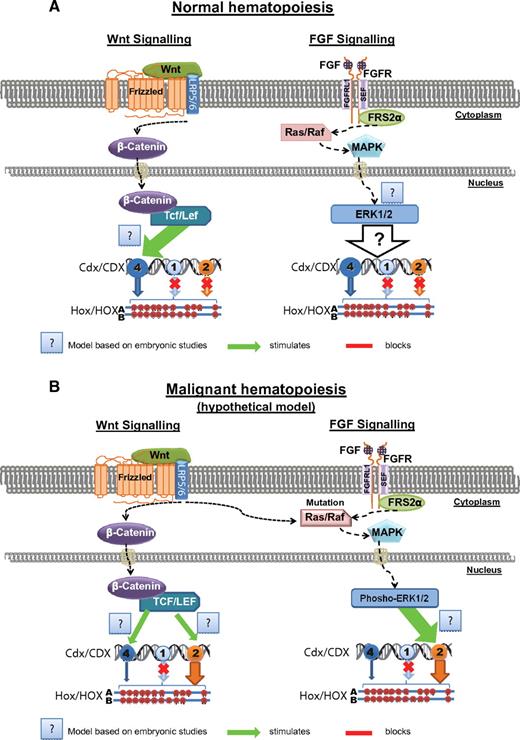
So far strategies to target Cdx2-induced transformation remain elusive. One possibility would be to identify signaling pathways that are able to be affected by drugs on which Cdx2 activity is dependent. Interestingly, it was shown in a colon cancer cell line that the transcriptional activity of Cdx2 depends on phosphorylation of the N-terminal S60 site by ERK kinases. We have shown that the N-terminal activation domain of Cdx2, at which the S60 site is localized, is essential for Cdx2-induced leukemogenesis and perturbation of Hox gene expression.33 Recent studies documented that Erk1/2 and their upstream effectors Mek1/2 are constitutively activated in primary human AML cells.72-74 Elevated p-ERK levels were found in 83.3% of the patients with AML.75 Because aberrant expression of CDX2 is detected in 80% of all AML cases, regulation of Cdx2 could be indirectly or directly linked with MEK/ERK signaling72,76 (Figure 3A-B). Therefore, it will be interesting to test whether post-transcriptional modification of CDX2 is crucial for its leukemogenic properties and whether MAPK inhibitors are able to block Cdx2-induced leukemogenesis.
Hypothetical model of Cdx gene regulation in normal and malignant hematopoiesis. (A) Normal hematopoiesis, based on results from studies in embryogenesis, the canonical Wnt pathway regulates the expression of CDX4 by the Lef1/Tcf complex in hematopoiesis. The FGF-MAPK pathway is not constitutively active in stem and progenitors cells. (B) Malignant hematopoiesis, either deregulation of the WNT pathway or FGF signaling induces aberrant CDX2 expression in human leukemia.
Hypothetical model of Cdx gene regulation in normal and malignant hematopoiesis. (A) Normal hematopoiesis, based on results from studies in embryogenesis, the canonical Wnt pathway regulates the expression of CDX4 by the Lef1/Tcf complex in hematopoiesis. The FGF-MAPK pathway is not constitutively active in stem and progenitors cells. (B) Malignant hematopoiesis, either deregulation of the WNT pathway or FGF signaling induces aberrant CDX2 expression in human leukemia.
Another possibility to block Cdx2-induced leukemogenesis would be to inhibit mechanisms that induce and maintain aberrant Cdx2 expression. However, mechanisms that lead to aberrant expression of CDX2 in leukemogenesis are still a mystery. As reported by Scholl et al, the methylation levels of the promoter region of CDX2 in patients with AML did not correspond to its expression levels.36 In addition, treatment of CDX2-negative cell lines with demethylating agents did not lead to an induction of CDX2 expression,77 suggesting that CDX2 expression in leukemia is independent of promoter hypomethylation as well as of the methylation level of cis-regulatory elements. Amplification of the CDX2 locus was detected only in a small percentage of patients with complex karyotype, however, not in AML patients with normal karyotype, who are characterized by high CDX2 expression levels.36,78 Another possibility, leading to aberrant transcription of CDX2 could be deregulation of trans-acting pathways, which target cis-regulatory elements in the 5′ region of the CDX2 gene as suggested by Hinoi et al.77 However, it was reported that CDX2 expression is monoallelic in most of the analyzed patients with leukemia,36 which indicates that the altered CDX2 expression is not primarily because of differences in the expression of trans-acting factors but rather because of altered cis-regulatory elements, which affect only one allele. Mutations were not detected in the promoter region of CDX2. However, this does not exclude still unknown mutations in cis-regulatory elements of the CDX2 gene, which cause aberrant expression. It would be interesting, therefore, to expand the sequence analysis of the CDX2 gene beyond its coding and promoter region and to conduct a more comprehensive sequence analysis. One target of genetic alterations, for example, could be negative regulators, which Wang and Shashikant postulated to be present in an 11.4-kb sequence flanking Cdx2.79 Disruption of these elements in hematopoietic cells could lead to an aberrant expression of the normally silenced CDX2 as well as mutations within 2 enhancer regions in the first intron of CDX2, described to induce ectopic expression of Cdx2 in the murine embryo.79 Interestingly, several binding sites for fibroblast growth factor (FGF) and Wnt signaling were identified in these regions. Mutations therefore could possibly affect the binding of factors such as FGF4, which previously was shown to target Cdx280 (Figure 3). Furthermore, it is probable that Cdx2 not only regulates but also is targeted by Wnt signaling as already demonstrated for other ParaHox genes such as Cdx1 and Cdx4.81,82 Another pathway that was shown to regulate Cdx2 expression is Ras/MAPK signaling,76 a pathway that links extracellular stimuli among others to proliferation, differentiation, and cell survival. Because both the Wnt and Ras/MAPK pathways are frequently deregulated in leukemia, this deregulation in combination with genetic alterations in the cis-regulatory elements of Cdx2 could possibly affect the transcription of the gene (Figure 3).
Conclusion
In summary, there is a great body of evidence that ParaHox genes play a pivotal role in normal hematopoiesis but also in leukemogenesis. These data add another homeobox gene family to the growing number of homeobox genes known to be critically involved in orchestrating normal hematopoiesis and promoting leukemogenesis. Thus, the story “homeobox genes in normal and malignant hematopoiesis” is not at its end but is still growing rapidly since the last review on this topic in this journal nearly 10 years ago.1 In those 10 years dysregulation of homeobox genes has been recognized as one of the molecular hallmarks of human AML. We know today that homeobox genes are key players for determining “stemness” and are essential for initiating and maintaining self-renewal programs on which leukemic stem cell function depends.83 ParaHox genes such as Cdx2 are particularly powerful to transform hematopoietic progenitors into leukemic stem cells, at least partly because it stands upstream of Hox genes and is able to perturb expression of several Hox genes at the same time. It is intriguing to speculate that targeting dysfunctional ParaHox genes such as CDX2 in human leukemia might reverse HOX gene dysfunction on a larger scale, thereby depriving the leukemic stem cell the molecular “environment” it needs to propagate leukemic growth. It is indeed unfortunate that we know so little about the mechanisms that induce aberrant CDX2 expression in patients with AML, because this would open possibilities to develop strategies to re-silence CDX2 expression in leukemic cells. Another hope is to identify signaling cascades that are essential for CDX2-induced transformation. Many questions are still open, and clarification of the role of other members of the ParaHox gene cluster such as GSH2 for leukemogenesis was just initiated. We can be confident that the next years will generate exciting data about the role of ParaHox genes for normal tissue homeostasis and will improve our understanding of how to correct ParaHox dysfunction in human leukemias.
Acknowledgments
The authors apologize to authors whose work could not be cited because of space constraints. They thank all members of their institutes for vivid discussions on this topic. They also thank Silvia Thoene, Shiva Bamezai, and Tamoghna Mandal for assisting in the preparation of figures.
This work is supported by grants from the Else Kröner-Fresenius-Foundation, the Deutsche Forschungsgemeinschaft, and the Bundesministerium für Bildung und Forschung (V.P.S.R. and C.B.).
Authorship
Contribution: V.P.S.R., R.K.H, and C.B. contributed to the writing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christian Buske, Institute of Experimental Cancer Research, University of Ulm, Albert-Einstein-Allee 11, 89081 Ulm, Germany; e-mail: christian.buske@uni-ulm.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal