Abstract
Mutations of genes encoding isocitrate dehydrogenase (IDH1 and IDH2) have been recently described in acute myeloid leukemia (AML). Serum and myeloblast samples from patients with IDH-mutant AML contain high levels of the metabolite 2-hydroxyglutarate (2-HG), a product of the altered IDH protein. In this prospective study, we sought to determine whether 2-HG can potentially serve as a noninvasive biomarker of disease burden through serial measurements in patients receiving conventional therapy for newly diagnosed AML. Our data demonstrate that serum, urine, marrow aspirate, and myeloblast 2-HG levels are significantly higher in IDH-mutant patients, with a correlation between baseline serum and urine 2-HG levels. Serum and urine 2-HG, along with IDH1/2-mutant allele burden in marrow, decreased with response to treatment. 2-HG decrease was more rapid with induction chemotherapy compared with DNA-methyltransferase inhibitor therapy. Our data suggest that serum or urine 2-HG may serve as noninvasive biomarkers of disease activity for IDH-mutant AML.
Introduction
Mutations in genes encoding isocitrate dehydrogenase (IDH1/2) were recently discovered in acute myeloid leukemia (AML).1,2 Their prognostic significance remains under investigation.3-9
IDH proteins catalyze the oxidative decarboxylation of isocitrate to α-ketoglutarate (α-KG). IDH mutations reside in the active site10 and include missense alterations affecting arginine-132 (R132) in IDH1, and the analogous arginine residue (R172), or one at arginine-140 (R140), in IDH2.1-3,5,6,8,11 The altered IDH proteins instead catalyze reduction of α-KG to the metabolite 2-hydroxyglutarate (2-HG).12 2-HG is normally present at low levels in cells,13 readily interconverted by 2-HG dehydrogenase to α-KG,14,15 but IDH mutations promote its accumulation in myeloblasts and sera of affected patients.10
No prior studies have prospectively measured 2-HG in IDH-mutant AML during treatment. We focused on the utility of 2-HG as a potential biomarker of disease burden and sought to assess the effect of treatment on the trajectory and kinetics of 2-HG levels. To accomplish this, we serially measured serum, urine, marrow aspirate, and myeloblast 2-HG during conventional therapy for newly diagnosed AML.
Methods
Adult patients at Massachusetts General Hospital, eligible for treatment of newly diagnosed AML, as defined by World Health Organization criteria, were enrolled. Samples were obtained through a protocol approved by our institution, Dana-Farber Harvard Cancer Center, its institutional review board, and the scientific review committees, with the approved protocol number 11-121. Informed consent was obtained per the Declaration of Helsinki.
Serum, urine, and marrow samples were obtained for 2-HG measurement before therapy. Mononuclear cells from blood and marrow aspirate were isolated using density gradient centrifugation with Ficoll-Hypaque (GE Healthcare). 2-HG measurement was performed by Agios Pharmaceuticals, with methods previously described.13,16 Serum and myeloblast 2-HG levels were considered elevated if > 1000 ng/mL or 1000 ng/2 × 106 cells, respectively, as per previous reports.10 In those with elevated baseline 2-HG, serum and urine were serially obtained for 2-HG measurement, at days 7, 14, 30, 60, and relapse. Bone marrow samples were collected at baseline, days 14, day 30, and relapse.
IDH1/2 mutational assays were performed through single-base extension sequencing using the SNaPshot assay (Applied Biosystems) and through direct sequencing using the Sanger method, as previously described.17 The relative IDH1/2 mutational burden during treatment was determined using next-generation sequencing as number of wild-type to mutant reads. Briefly, IDH1/2 exon 4 PCR amplicons were generated using primers containing Ion Torrent adapters and unique barcodes. Samples were normalized through quantitative PCR quantification and combined. The resulting library was amplified on Ion Sphere particles by emulsion PCR, and sequencing was performed on the Ion PGM Sequencer (Life Technologies).
AML therapies included cytarabine- and idarubicin-based induction, the DNA-methyltransferase inhibitors decitabine or 5-azacitidine, or low-dose cytarabine. Statistical evaluation was performed using a 2-tailed Mann-Whitney test for comparing values between groups and a correlation coefficient to assess relationship between variables.
Results and discussion
Forty-two patients were enrolled, with 10 (24%, 90% exact binomial CI, 14%-37%) displaying baseline elevations in serum or myeloblast 2-HG (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). In 9 of 10 patients, IDH1/2 mutations were discovered: 2 IDH1-R132H, 1 IDH2-R172M, and 6 IDH2-R140Q mutations. In the remaining patient in whom IDH mutations were not found, myeloblast 2-HG was elevated (4040 ng/2 × 106 cells), but serum 2-HG was not (241 ng/mL). No IDH mutations were found in 32 patients without elevated 2-HG. One IDH-mutant patient did not have elevated serum 2-HG but did demonstrate elevated myeloblast levels.
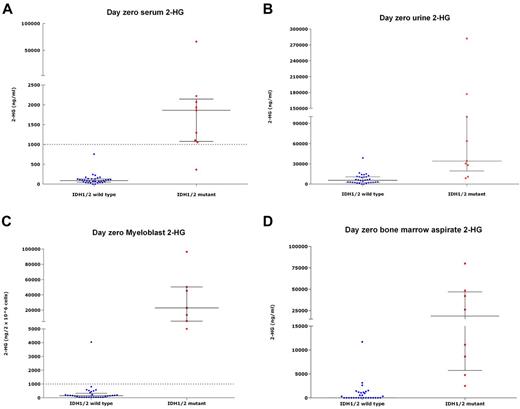
Serum 2-HG for IDH-mutant patients (median, 1863 ng/mL; range, 365.1-66 207 ng/mL) was significantly higher than for IDH-wild-type patients (median, 87 ng/mL; range, below quantitative limit [BQL]-755 ng/mL, P < .001; Figure 1A). Urine 2-HG was higher in IDH-mutant patients (median, 34 100 ng/mL; range, 8620-282 000 ng/mL vs median, 5525 ng/mL; range, BQL-38 800 ng/mL, P < .001), as were marrow aspirate 2-HG (mean, 27 925 ng/mL vs mean, 924 ng/mL, P < .001), and myeloblast 2-HG (median, 22 880 ng/2 × 106 cells; range, 5560-96 400 ng/2 × 106 cells vs median, 148.5 ng/2 × 106 cells; range, BQL-4040 ng/2 × 106 cells, P < .001; Figure 1B-1D). Baseline serum 2-HG was correlated with urine 2-HG levels (r = 0.82, P < .001). We also measured serum 2-HG on 7 samples from healthy persons, all measuring < 100 ng/mL (median, 91 ng/mL; range, 33-93 ng/mL).
2-HG levels in IDH-mutant and IDH-wild-type samples. 2-HG measurements in samples from patients with a wild-type-IDH1/2 (n = 33) versus those harboring IDH1/2-mutations (n = 9), obtained at presentation, showing 2-HG measurement in serum (A), urine (B), myeloblasts (C), and marrow aspirate (D). Each point represents an individual patient sample; blue dots represent IDH-wild-type samples; and red dots, IDH-mutant samples. The left column in each figure represents the wild-type samples; and the right column, the mutant samples. In each column, horizontal bars represent the median; and vertical lines, the lower and upper quartiles. (A,C) Dotted line across graphs indicates 2HG levels of 1000 ng/mL serum or 1000 ng/2 × 106 within cells, respectively, above which 2-HG levels were deemed to be elevated in this study. All figures describe a statistically significant difference in 2-HG levels between IDH1/2-mutant samples relative to wild-type samples (P < .05). One IDH-mutant patient did not have elevated baseline serum levels of 2-HG but did demonstrate elevated myeloblast 2-HG at baseline.
2-HG levels in IDH-mutant and IDH-wild-type samples. 2-HG measurements in samples from patients with a wild-type-IDH1/2 (n = 33) versus those harboring IDH1/2-mutations (n = 9), obtained at presentation, showing 2-HG measurement in serum (A), urine (B), myeloblasts (C), and marrow aspirate (D). Each point represents an individual patient sample; blue dots represent IDH-wild-type samples; and red dots, IDH-mutant samples. The left column in each figure represents the wild-type samples; and the right column, the mutant samples. In each column, horizontal bars represent the median; and vertical lines, the lower and upper quartiles. (A,C) Dotted line across graphs indicates 2HG levels of 1000 ng/mL serum or 1000 ng/2 × 106 within cells, respectively, above which 2-HG levels were deemed to be elevated in this study. All figures describe a statistically significant difference in 2-HG levels between IDH1/2-mutant samples relative to wild-type samples (P < .05). One IDH-mutant patient did not have elevated baseline serum levels of 2-HG but did demonstrate elevated myeloblast 2-HG at baseline.
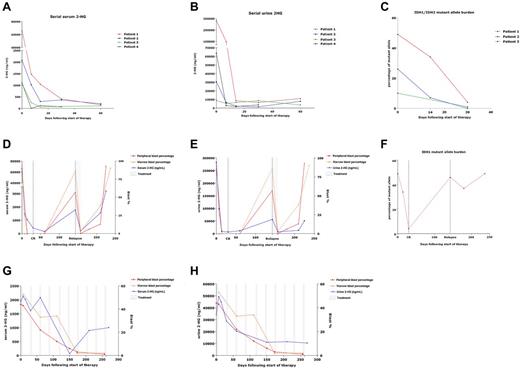
In those with elevated baseline 2-HG, serum and urine 2-HG decreased during therapy, concordantly with decreases in blast counts. Seven patients were treated with induction chemotherapy, 2 with 5-azacitidine, and 1 with decitabine. Serum and urine 2-HG levels during treatment are demonstrated for patients who achieved remission after induction (Figure 2A-B). More rapid and profound decreases were noted with induction chemotherapy versus DNA-methyltransferase inhibitor therapy. For the latter, a slower decrease in 2-HG occurred with response, as demonstrated in a patient with partial response to 5-azacitidine, with a follow-up of ∼ 250 days (Figure 2G-H).
Serial measurement of 2-HG during therapy. Serum (A) and urine (B) 2-HG levels measured serially over the course of the first 60 days of treatment for patients with baseline elevations in 2-HG and who achieved complete remission after induction chemotherapy. (C) IDH1/2-mutant allele burden measured serially in marrow samples over the course of the first 30 days of treatment for 3 of these patients with complete remission after induction chemotherapy. (D-F) Serum (D) and urine (E) 2-HG levels measured serially over time for a patient (patient 1 in panels A-C) who received cytarabine- and idarubicin-based induction, went on to relapse at ∼ day 150, and was then treated with reinduction therapy consisting of mitoxantrone, etoposide, and cytarabine (MEC), to which he responded transiently before displaying refractory disease. (F) IDH1-mutant allele burden for the same patient, measured serially in marrow samples, displaying initial decrease in IDH1-mutant allele burden with remission, with subsequent increase associated with relapse. (G-H) Serum (G) and urine (H) 2-HG levels measured serially over time (∼ 250 days) for a patient who received hypomethylating therapy with 5-azacitidine 75 mg/m2 intravenously on days 1-5 of 28-day cycles.
Serial measurement of 2-HG during therapy. Serum (A) and urine (B) 2-HG levels measured serially over the course of the first 60 days of treatment for patients with baseline elevations in 2-HG and who achieved complete remission after induction chemotherapy. (C) IDH1/2-mutant allele burden measured serially in marrow samples over the course of the first 30 days of treatment for 3 of these patients with complete remission after induction chemotherapy. (D-F) Serum (D) and urine (E) 2-HG levels measured serially over time for a patient (patient 1 in panels A-C) who received cytarabine- and idarubicin-based induction, went on to relapse at ∼ day 150, and was then treated with reinduction therapy consisting of mitoxantrone, etoposide, and cytarabine (MEC), to which he responded transiently before displaying refractory disease. (F) IDH1-mutant allele burden for the same patient, measured serially in marrow samples, displaying initial decrease in IDH1-mutant allele burden with remission, with subsequent increase associated with relapse. (G-H) Serum (G) and urine (H) 2-HG levels measured serially over time (∼ 250 days) for a patient who received hypomethylating therapy with 5-azacitidine 75 mg/m2 intravenously on days 1-5 of 28-day cycles.
For 3 patients, marrow samples were available for IDH mutational analysis at remission, after induction chemotherapy. Using the SNaPshot assay and/or direct sequencing of loci, an IDH1/2 mutation was no longer detectable in 2 samples; but in the remaining sample, an IDH1 R132H mutation was detectable, even with low serum 2HG levels. Interestingly, to date, this is the only patient who has relapsed, at ∼ 150 days after induction. His 2-HG increased precipitously with relapse, and again transiently decreased after reinduction, to which he was ultimately refractory (Figure 2D-E). Intriguingly, this patient's baseline serum and urine 2-HG were the highest measured, 66 207 ng/mL and 282 000 ng/mL, respectively. Through next-generation sequencing of available serial samples from 3 patients who received induction therapy, we also found that IDH1/2 mutational burden (mutant allele as fraction of total IDH1/2) decreased over time concordantly with 2-HG (Figure 2C). The mutational burden reached 0%, 1%, and 4%, at remission in these patients, with the latter patient identified as the sole relapser in whom the mutant burden subsequently rose in concordance with 2-HG (Figure 2F).
Emerging evidence suggests that 2-HG plays a role in promoting malignancy.10,18,19 2-HG, structurally homologous to α-KG, may interfere with α-KG–activated enzymes, like TET, histone demethylases, and prolyl hydroxylases, suggesting roles in epigenetic modulation and HIF-1α down-regulation.10,19-22 Reports of aberrant hypermethylation and inactivation of loci in IDH-mutant AML are intriguing,23 with the underlying mechanism possibly related to α-KG down-regulation or increase in 2-HG.23-25
With this study, we provide the first prospective evidence that elevated serum and myeloblast 2-HG are specific features of IDH1/2-mutant AML. None of the studied patients with non-elevated 2-HG demonstrated any IDH mutations, and 9 of 10 with elevations did have elevated serum or myeloblast 2-HG, suggesting that 2-HG is a discriminatory marker in this setting. The sole sample that demonstrated elevated myeloblast 2-HG, but no IDH mutation, is under investigation to determine the underlying cause. We also report that urine and marrow aspirate 2-HG are similarly elevated in IDH-mutant patients. Most significantly, our data suggest that serum and urine 2-HG may serve as easily measured, noninvasive biomarkers of disease and can be followed during treatment in patients with elevated baseline levels.
Serum and urine 2-HG decreased with response to therapy in a consistent and predictable fashion. Decreases occurred more rapidly with induction chemotherapy than with DNA-methyltransferase inhibitor therapy, as expected given the kinetics of response to these agents. In all evaluated patients with a baseline elevation in serum 2-HG and who subsequently achieved a complete remission, serum 2-HG decreased to < 500 ng/mL by day 30 and < 200 ng/mL by day 60. We also found that the relative burden of IDH1/2-mutant alleles correspondingly decreased with treatment, and this too may serve as a molecular surrogate for disease activity, especially at remission.
Longer follow-up with larger populations will validate the value of 2-HG as a biomarker in monitoring minimal residual disease and predicting clinically meaningful outcomes. In time, 2-HG levels may play an important role in the noninvasive management of patients receiving conventional or IDH-targeted therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Thomas Spitzer and Ronald Takvorian at the Massachusetts General Hospital Cancer Center for their kind support of this study.
This work was supported in part by the Rogers Family Foundation in Boston, MA (philanthropic grant) and the Massachusetts General Hospital Cancer Center (Thematic Priority #3 Grant).
Authorship
Contribution: A.T.F. developed the study design, performed data collection, management, and analysis, and wrote the manuscript; H.S. performed data collection and analysis and helped create figures; K.K.B., P.C.A., and E.C.A. contributed to the study design and patient enrollment and helped edit the manuscript; J.F. enrolled and monitored patients on study and helped with sample and data collection; M.B. and K.M.E. helped with sample and data collection; D.R.B., H.U.L., C.R.M., and A.J.I. performed mutational analysis for patients on study; K.S.S., K.E.Y., S.A., and D.P.S. helped in the performing and reporting of 2-HG assays at Agios Pharmaceuticals, Cambridge, MA, in kind collaboration; C.H. provided sample processing; A.E. provided figures and helped edit the manuscript; D.S.N. helped with study design, provided biostatistical support, and helped edit the manuscript; R.M.S. helped with study design and helped edit the manuscript; and Y.-B.C. helped with study design and development and edited the manuscript.
Conflict-of-interest disclosure: K.S.S., K.E.Y., S.A, and D.P.S. are employees of Agios Pharmaceuticals. R.M.S. is a consultant for Agios Pharmaceuticals. D.R.B. is a paid consultant for Bio-Reference Laboratories Inc (licensee of SNaPshot). The remaining authors declare no competing financial interests.
Correspondence: Amir T. Fathi, Massachusetts General Hospital, Harvard Medical School, Zero Emerson Place, Suite 118, Boston, MA 02114; e-mail: afathi@partners.org.