Abstract
The ability to distinguish clonal B-cell populations based on the sequence of their rearranged immunoglobulin heavy chain (IgH) locus is an important tool for diagnosing B-cell neoplasms and monitoring treatment response. Leukemic precursor B cells may continue to undergo recombination of the IgH gene after malignant transformation; however, the magnitude of evolution at the IgH locus is currently unknown. We used next-generation sequencing to characterize the repertoire of IgH sequences in diagnostic samples of 51 children with B precursor acute lymphoblastic leukemia (B-ALL). We identified clonal IgH rearrangements in 43 of 51 (84%) cases and found that the number of evolved IgH sequences per patient ranged dramatically from 0 to 4024. We demonstrate that the evolved IgH sequences are not the result of amplification artifacts and are unique to leukemic precursor B cells. In addition, the evolution often follows an allelic exclusion pattern, where only 1 of 2 rearranged IgH loci exhibit ongoing recombination. Thus, precursor B-cell leukemias maintain evolution at the IgH locus at levels that were previously underappreciated. This finding sheds light on the mechanisms associated with leukemic clonal evolution and may fundamentally change approaches for monitoring minimal residual disease burden.
Introduction
Pediatric B precursor acute lymphoblastic leukemia (B-ALL) is generally thought to be a clonal disease resulting from malignant transformation and expansion of a single B cell.1-4 Clonality of B-cell populations can be assessed by analysis of gene rearrangements that occur at the immunoglobulin heavy chain (IgH) gene. Early in B-cell development, somatic recombination at the IgH locus gives rise to unique rearrangements of the variable (VH), diversity (D), and joining (JH) gene segments.5,6 In this 2-step process, recombination signal sequences mediate D to JH joining, which is followed by VH to D-JH joining.7-9 During this recombination, nontemplated nucleotides (N-bases) may be added at the junctions between gene segments, and other nucleotides may be deleted from the VH, D, and JH germline sequences.10 The resulting unique VH-D-JH rearrangements are used as clonotypic markers in pediatric B-ALL.
PCR-based methods have shown changes in clonal IgH rearrangements between initial diagnosis and relapse in a significant proportion of pediatric B-ALL cases.11-13 These changes at the IgH locus could represent the persistence of ancestral clones that later expand, or continued evolution of the primary clone in the setting of antineoplastic therapy. Ongoing changes at the IgH locus have been shown to be the result of 2 distinct mechanisms. First, one VH segment can be exchanged for an alternative 5′ VH while retaining the same D-JH, which is known as VH replacement. Alternatively, a partially rearranged IgH gene where only D-JH joining has occurred in the ancestral clone may recombine with multiple VH segments.14-18
Previous studies have investigated the clinical significance of clonal IgH evolution in B-ALL patients.15,19 Clones present at relapse possessing evolved IgH rearrangements can often be traced back to minor clonal populations present at diagnosis that were presumably resistant to chemotherapy.12,20-23 Moreover, these retrospective evaluations have shown that the burden of relapse clone at diagnosis predicts time to relapse in a subset of B-ALL patients.22,24 These studies highlight the need to identify all clonal malignant populations at diagnosis, which will enhance the likelihood of detecting minor, potentially chemoresistant leukemic clones as patients progress through treatment. Despite these significant implications, the extent of clonal evolution at diagnosis and the variation of clonal evolution among B-ALL patients remain largely unstudied.
To further characterize clonal evolution in B-ALL, we performed IgH repertoire sequencing on diagnostic samples from 51 children with B-ALL. Sequencing-based methods have been used previously to characterize B-cell diversity in individual samples with high sensitivity.25-28 In this approach, each rearranged IgH sequence represents a unique tag, or clonotype, enabling identification of cells containing that rearrangement. This method enables quantitative determination of IgH rearrangement frequencies, which is a direct measure of clonotype abundance. This method can also be used to detect evolved IgH rearrangements and distinguish between the 2 mechanisms responsible for generating them. Using this approach, we identified high-frequency leukemic “index” clonotypes and observed distinct but related “evolved” clonotypes in most B-ALL patients.
Methods
Clinical samples
A total of 51 diagnostic bone marrow samples from children with B-ALL diagnosed at Lucile Packard Children's Hospital were collected on a protocol approved by the Stanford University Institutional Review Board as previously described.29 Informed consent was obtained before specimen collection and samples were deidentified before use in studies, in accordance with the Declaration of Helsinki.
Peripheral blood mononuclear cells were obtained from 35 patients with chronic lymphocytic leukemia (CLL) diagnosed at Stanford University Medical Center. All CLL patient samples were obtained with explicit authorization and monitoring by the Stanford University Institutional Review Board.
Flow cytometry and cell sorting
On thawing of cryopreserved bone marrow mononuclear cells, one-third of the vial volume was washed and lysed immediately in RLT plus buffer (QIAGEN) for nucleic acid isolation according to manufacturer guidelines. The remainder of each vial was suspended in PBS containing 2% FBS and washed twice before antibody labeling. See supplemental Methods for a description of these methodologies (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
DNA preparation
DNA was isolated using AllPrep DNA mini and/or micro kits, according to the manufacturer's instructions (QIAGEN).
IgH amplification and sequencing
Genomic DNA was amplified using locus specific primer sets for IgH. The goal of this amplification reaction was to reproducibly amplify all possible rearranged IgH sequences in the sample while appending the necessary sequences for cluster formation and sample indexing. See supplemental Methods for a description of the primer design, and amplification and sequencing reactions.
Analytical methods
Clonotype determination.
A clonotype was defined when at least 2 identical sequence reads were obtained. See supplemental Methods for a description of the clonotype determination analysis methods.
Passed/failed samples.
Flow cytometry: Samples with < 500 starting cells were not analyzed further. One patient had only 1 malignant cell gate (patient 13).
Sequencing.
The number of molecules associated with each clonotype in each sample was estimated based on the frequency of a reference IgH sequence spiked into each sample in known quantities. Clonotypes with less than 1 molecule were discarded.
Disease clone calibration criteria
The frequency of all the clonotypes in each sample was determined by calculating the number of sequencing reads for each clonotype and dividing by the total number of passed sequencing reads in the sample. Individual clonotypes with ≥ 5% frequency were designated index clonotypes.
Evolution criteria
Identification of potentially related clonotypes.
Potentially related clonotypes for further analysis were identified using 3 criteria: (1) identical JH segment sequence to index clonotype, (2) > 1 molecule, and (3) different VH allele from the index clonotype.
Identification of evolved clonotypes.
Potentially related clonotypes were analyzed for the extent of N terminal and D segment (NDN) sharing. The NDN sequence was determined by identifying the location where the consensus sequence does not match the VH and JH segment sequences. In the case of index clones, the JunctionAnalysis tool from IMGT30 was used to increase accuracy and identify which bases belong to the D segment. The location of the VH allele was obtained from the Ensembl database (supplemental Methods).
Results
Clinical characteristics
Bone marrow diagnostic samples from 51 children with B-ALL were evaluated. Patient and sample characteristics are summarized in Table 1 and supplemental Table 1. The patients were a random sampling of children diagnosed with B-ALL at Lucile Packard Children's Hospital over the previous 7 years. Data were collected on prognostic variables, including age, initial white blood cell count, cytogenetics, CNS status (1, 2, and 3), and end of induction minimal residual disease (MRD). Based on the information presented in Table 1, these samples are representative of the general patient population treated at Lucile Packard Children's Hospital and are similar to what is seen in recently published B-ALL studies.31
Summary of patient characteristics
| Characteristic . | No. . | % . |
|---|---|---|
| Age, y | ||
| 0-1 | 1 | 2 |
| 1-10 | 38 | 75 |
| > 10 | 12 | 24 |
| Initial WBC count | ||
| < 50 000 | 38 | 75 |
| > 50 000 | 13 | 25 |
| NCI risk group | ||
| Standard risk | 33 | 65 |
| High risk | 15 | 29 |
| Very high risk | 2 | 4 |
| Infant leukemia | 1 | 2 |
| Cytogenetics | ||
| TEL-AML | 15* | 29 |
| Hyperdiploid without trisomy 4, 10, and 17 | 10 | 20 |
| Hyperdiploid with trisomy 4, 10, and 17 | 7* | 14 |
| MLL-rearranged | 1 | 2 |
| Philadelphia chromosome | 2 | 4 |
| Other | 10 | 20 |
| Normal | 6 | 12 |
| Not done | 1 | 2 |
| CNS stage | ||
| 1 | 38 | 75 |
| 2 | 12 | 24 |
| 3 | 1 | 2 |
| End of induction MRD | ||
| < 0.1% | 36 | 71 |
| > 0.1% < 1% | 4 | 8 |
| > 1% | 4 | 8 |
| Not done | 7 | 14 |
| Current disease status | ||
| Relapse | 5 | 10 |
| Continuous remission | 46 | 90 |
| Characteristic . | No. . | % . |
|---|---|---|
| Age, y | ||
| 0-1 | 1 | 2 |
| 1-10 | 38 | 75 |
| > 10 | 12 | 24 |
| Initial WBC count | ||
| < 50 000 | 38 | 75 |
| > 50 000 | 13 | 25 |
| NCI risk group | ||
| Standard risk | 33 | 65 |
| High risk | 15 | 29 |
| Very high risk | 2 | 4 |
| Infant leukemia | 1 | 2 |
| Cytogenetics | ||
| TEL-AML | 15* | 29 |
| Hyperdiploid without trisomy 4, 10, and 17 | 10 | 20 |
| Hyperdiploid with trisomy 4, 10, and 17 | 7* | 14 |
| MLL-rearranged | 1 | 2 |
| Philadelphia chromosome | 2 | 4 |
| Other | 10 | 20 |
| Normal | 6 | 12 |
| Not done | 1 | 2 |
| CNS stage | ||
| 1 | 38 | 75 |
| 2 | 12 | 24 |
| 3 | 1 | 2 |
| End of induction MRD | ||
| < 0.1% | 36 | 71 |
| > 0.1% < 1% | 4 | 8 |
| > 1% | 4 | 8 |
| Not done | 7 | 14 |
| Current disease status | ||
| Relapse | 5 | 10 |
| Continuous remission | 46 | 90 |
Sample 8 had both TEL-AML and hyperdiploid with trisomy 4, 10, and 17.
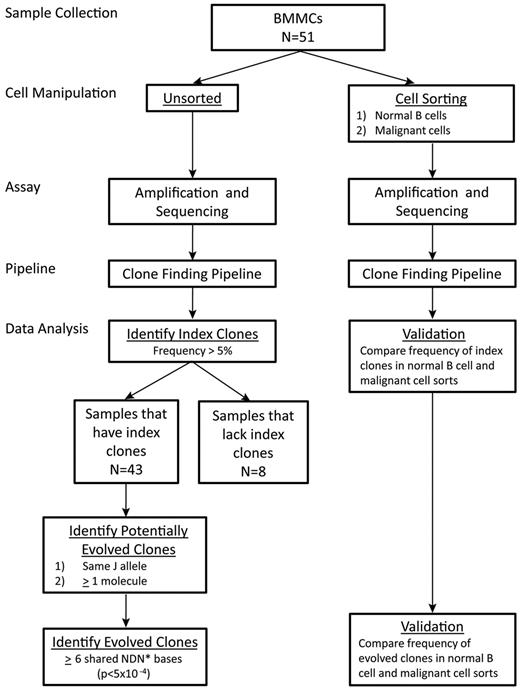
Study schema: sequencing the IgH locus in B-ALL diagnostic bone marrow samples
Clonal evolution in B-ALL bone marrow samples at diagnosis was characterized by amplification and sequencing of the IgH locus. To definitively identify disease (“index”) clonotypes and to determine the frequency of index and related clonotypes in each patient, bone marrow mononuclear cells isolated and cryopreserved from each of the 51 patients at diagnosis were retrieved and split into 2 workflows. In one workflow, DNA from unsorted cells was isolated, and IgH was amplified, sequenced, and analyzed using index clone identification and clonal evolution algorithms. This unsorted sequence data provide the quantitative measure for identification of disease, or index, clonotypes in a sample. The number of input cells and mapped reads are shown in supplemental Table 2.
In a second workflow, flow cytometry was used to sort CD45+CD10−CD19+CD20+ cells consistent with normal B cells (herein referred to as normal B cells), as well as those carrying markers typical of B-ALL cells32 (phenotypes varied among patients; supplemental Table 2). DNA from these sorted cells was isolated, and the IgH gene was amplified and sequenced. Clonotypes identified in the normal B-cell and leukemic sorted populations were used to validate the disease clonotypes identified in the unsorted analysis.
The unsorted and sorted sequencing data provide 2 complementary uses for this study. The unsorted sequencing data, a quantitative measure of the full immune repertoire, enables the identification of index clones in a patient sample. Specifically, we use the unsorted sequence data to identify individual clones that appear at > 5% frequency in the unsorted population (ie, an “index” clone). In contrast, data from the sorted populations are merely used for validation of the index and evolved clones. Using the dataset from the sorted cells, we see enrichment of index and evolved clones in populations sorted for malignant cell markers and de-enrichment of the index and evolved clones in populations sorted for normal B-cell markers. Thus, the unsorted data provide the quantitative measurement that is necessary for the identification of the index clones, whereas the sorted data are merely used for qualitative validation of index and evolved clones. In total, these experiments generated > 145 million mapped IgH sequencing reads, an average of > 750 000 reads per sample. The study schema is summarized in Figure 1.
Schematic of workflow. *See “Methods” for definition of effective NDN bases in which D bases are weighted differently than N bases to account for D segment sharing.
Schematic of workflow. *See “Methods” for definition of effective NDN bases in which D bases are weighted differently than N bases to account for D segment sharing.
Leukemic index clonotypes identified in B-ALL diagnostic and relapse samples
We set a frequency threshold of 5% to designate a clonotype as an index leukemic clone. Using this threshold, we identified 86 leukemic index clonotypes in the diagnostic bone marrow samples (Table 2; Figure 2). Sixteen patients had 1 index clonotype, 16 patients had 2 index clonotypes, and 11 had > 2 index clonotypes (Table 2). For patients with 1 or more index clonotypes, a significant majority of the IgH reads were generated by the index clonotypes (mean 87%; Table 2). For 2 patients, relapse samples were also available and results were very similar to diagnostic samples. In patient 1, neither sample had an index clone. In patient 48, the index clones were 29%, 24%, 24%, and 23% at diagnosis and these same clones were 31%, 24%, 23%, and 22% at relapse.
Per-patient summary of malignant clones in B-ALL samples and sample sorts
| Patient ID . | No. of index clones . | Total frequency of index clones, % . | Individual index clone frequencies, % . | Total no. (%) evolved clones* . | Total frequency of index clones in sort for normal B cells, % . | Total frequency of index clone in gate for malignant cells, % . |
|---|---|---|---|---|---|---|
| 1 | 0 | No index | No index | No index | No index | No index |
| 2 | 0 | No index | No index | No index | No index | No index |
| 3 | 1 | 90.3 | 90† | 14 (0.3) | 0.5 | 98.0 |
| 4 | 0 | No index | No index | No index | No index‡ | No index‡ |
| 5 | 3 | 95.7 | 76†, 10†, 10† | 668 (33.3) | 3.3 | 98.3 |
| 6 | 5 | 80.8 | 33†, 18†, 11†, 9†, 9† | 883 (61.7) | 0.7 | 90.3 |
| 7 | 1 | 88.7 | 89† | 25 (3.4) | Not sorted | Not sorted |
| 8 | 2 | 96 | 79†, 17† | 301 (62.2) | Not sorted | Not sorted |
| 9 | 2 | 98.1 | 63†, 35 | 42 (8.8) | 7.3 | 98.8 |
| 10 | 3 | 99.3 | 47†, 31†, 22† | 18 (6.8) | 0.4 | 99.8 |
| 11 | 0 | No index | No index | No index | No index | No index |
| 12 | 2 | 99.5 | 53†, 46† | 12 (60.0) | NA | 99.5 |
| 13 | 0 | No index | No index | No index | No index | No index |
| 14 | 1 | 92.8 | 93† | 22 (0.3) | 0.2 | 98.0 |
| 15 | 5 | 78.4 | 23†, 21†, 17†, 13†, 5† | 4024 (85.5) | 14 | 79.6 |
| 16 | 2 | 98.4 | 63†, 35† | 4 (0.8) | 6.7 | 99.3 |
| 17 | 3 | 98.8 | 39†, 36, 24† | 13 (2.4) | 0 | 99.0 |
| 18 | 3 | 72 | 59†, 8†, 6† | 368 (43.0) | Not sorted | Not sorted |
| 19 | 1 | 75.2 | 75 | 0 (0.0) | Not sorted | Not sorted |
| 20 | 3 | 89 | 50†, 30†, 9† | 460 (29.9) | Not sorted | Not sorted |
| 21 | 1 | 97.3 | 97 | 0 (0.0) | 0.2 | 96.4 |
| 22 | 1 | 96.4 | 96† | 35 (18.1) | 41.5 | 96.2 |
| 23 | 3 | 64.9 | 30†, 21†, 13† | 2645 (34.6) | 0.03 | 57.2 |
| 24 | 0 | No index | No index | No index | No index | No index |
| 25 | 1 | 99.9 | 100† | 7 (12.7) | 8.4 | 99.9 |
| 26 | 2 | 97.3 | 59†, 38† | 196 (8.1) | 1.5 | 99.6 |
| 27 | 2 | 97.1 | 58, 39† | 22 (1.3) | 0 | 96.6 |
| 28 | 3 | 98.6 | 38†, 36, 25† | 9 (4.9) | NA | 98.5 |
| 29 | 3 | 71.6 | 51, 12†, 8† | 17 (0.2) | 0.5 | 79.7 |
| 30 | 1 | 93.1 | 93† | 18 (2.9) | 5 | 94.5 |
| 31 | 2 | 76.9 | 39, 38† | 4 (0.4) | 0 | NA§ |
| 32 | 2 | 99 | 50†, 49† | 11 (1.4) | 0.3 | 99.9 |
| 33 | 1 | 66.6 | 67† | 3 (0.3) | 0.4 | 99.4 |
| 34 | 2 | 99.5 | 85, 14 | 0 (0.0) | 7.5 | 99.9 |
| 35 | 2 | 63.4 | 49†, 15† | 1149 (27.5) | 1 | 65.8 |
| 36 | 1 | 22.4 | 22 | 0 (0.0) | 0 | 96.2 |
| 37 | 0 | No index | No index | No index | No index | No index |
| 38 | 2 | 97 | 66†, 32 | 154 (13.4) | 4.7 | 97.7 |
| 39 | 1 | 99 | 99 | 0 (0.0) | NA | 99.7 |
| 40 | 1 | 84.9 | 85† | 24 (0.3) | 1.8 | 89.4 |
| 41 | 0 | No index | No index | No index | No index | No index |
| 42 | 2 | 99.1 | 56†, 43† | 5 (4.2) | 2.1 | 99.3 |
| 43 | 2 | 98.9 | 58†, 41 | 2 (8.7) | NA | 98.6 |
| 44 | 2 | 99.1 | 74, 25† | 3 (0.8) | 1.4 | 99.9 |
| 45 | 2 | 79.9 | 72†, 8 | 5 (0.3) | 0.6 | 73.7 |
| 46 | 1 | 94.6 | 95 | 0 (0.0) | 2.1 | 97.8 |
| 47 | 1 | 98.5 | 99† | 60 (5.1) | 1.5 | 99.7 |
| 48 | 4 | 99.8 | 29, 24†, 24†, 23† | 30 (36.1) | 3.2 | 99.9 |
| 49 | 1 | 99 | 99† | 4 (0.6) | 0.7 | 99.5 |
| 50 | 2 | 81.5 | 75†, 7† | 2724 (41.7) | 0.9 | 86.4 |
| 51 | 1 | 5.8 | 6† | 4 (0.5) | 0 | 39.6 |
| Patient ID . | No. of index clones . | Total frequency of index clones, % . | Individual index clone frequencies, % . | Total no. (%) evolved clones* . | Total frequency of index clones in sort for normal B cells, % . | Total frequency of index clone in gate for malignant cells, % . |
|---|---|---|---|---|---|---|
| 1 | 0 | No index | No index | No index | No index | No index |
| 2 | 0 | No index | No index | No index | No index | No index |
| 3 | 1 | 90.3 | 90† | 14 (0.3) | 0.5 | 98.0 |
| 4 | 0 | No index | No index | No index | No index‡ | No index‡ |
| 5 | 3 | 95.7 | 76†, 10†, 10† | 668 (33.3) | 3.3 | 98.3 |
| 6 | 5 | 80.8 | 33†, 18†, 11†, 9†, 9† | 883 (61.7) | 0.7 | 90.3 |
| 7 | 1 | 88.7 | 89† | 25 (3.4) | Not sorted | Not sorted |
| 8 | 2 | 96 | 79†, 17† | 301 (62.2) | Not sorted | Not sorted |
| 9 | 2 | 98.1 | 63†, 35 | 42 (8.8) | 7.3 | 98.8 |
| 10 | 3 | 99.3 | 47†, 31†, 22† | 18 (6.8) | 0.4 | 99.8 |
| 11 | 0 | No index | No index | No index | No index | No index |
| 12 | 2 | 99.5 | 53†, 46† | 12 (60.0) | NA | 99.5 |
| 13 | 0 | No index | No index | No index | No index | No index |
| 14 | 1 | 92.8 | 93† | 22 (0.3) | 0.2 | 98.0 |
| 15 | 5 | 78.4 | 23†, 21†, 17†, 13†, 5† | 4024 (85.5) | 14 | 79.6 |
| 16 | 2 | 98.4 | 63†, 35† | 4 (0.8) | 6.7 | 99.3 |
| 17 | 3 | 98.8 | 39†, 36, 24† | 13 (2.4) | 0 | 99.0 |
| 18 | 3 | 72 | 59†, 8†, 6† | 368 (43.0) | Not sorted | Not sorted |
| 19 | 1 | 75.2 | 75 | 0 (0.0) | Not sorted | Not sorted |
| 20 | 3 | 89 | 50†, 30†, 9† | 460 (29.9) | Not sorted | Not sorted |
| 21 | 1 | 97.3 | 97 | 0 (0.0) | 0.2 | 96.4 |
| 22 | 1 | 96.4 | 96† | 35 (18.1) | 41.5 | 96.2 |
| 23 | 3 | 64.9 | 30†, 21†, 13† | 2645 (34.6) | 0.03 | 57.2 |
| 24 | 0 | No index | No index | No index | No index | No index |
| 25 | 1 | 99.9 | 100† | 7 (12.7) | 8.4 | 99.9 |
| 26 | 2 | 97.3 | 59†, 38† | 196 (8.1) | 1.5 | 99.6 |
| 27 | 2 | 97.1 | 58, 39† | 22 (1.3) | 0 | 96.6 |
| 28 | 3 | 98.6 | 38†, 36, 25† | 9 (4.9) | NA | 98.5 |
| 29 | 3 | 71.6 | 51, 12†, 8† | 17 (0.2) | 0.5 | 79.7 |
| 30 | 1 | 93.1 | 93† | 18 (2.9) | 5 | 94.5 |
| 31 | 2 | 76.9 | 39, 38† | 4 (0.4) | 0 | NA§ |
| 32 | 2 | 99 | 50†, 49† | 11 (1.4) | 0.3 | 99.9 |
| 33 | 1 | 66.6 | 67† | 3 (0.3) | 0.4 | 99.4 |
| 34 | 2 | 99.5 | 85, 14 | 0 (0.0) | 7.5 | 99.9 |
| 35 | 2 | 63.4 | 49†, 15† | 1149 (27.5) | 1 | 65.8 |
| 36 | 1 | 22.4 | 22 | 0 (0.0) | 0 | 96.2 |
| 37 | 0 | No index | No index | No index | No index | No index |
| 38 | 2 | 97 | 66†, 32 | 154 (13.4) | 4.7 | 97.7 |
| 39 | 1 | 99 | 99 | 0 (0.0) | NA | 99.7 |
| 40 | 1 | 84.9 | 85† | 24 (0.3) | 1.8 | 89.4 |
| 41 | 0 | No index | No index | No index | No index | No index |
| 42 | 2 | 99.1 | 56†, 43† | 5 (4.2) | 2.1 | 99.3 |
| 43 | 2 | 98.9 | 58†, 41 | 2 (8.7) | NA | 98.6 |
| 44 | 2 | 99.1 | 74, 25† | 3 (0.8) | 1.4 | 99.9 |
| 45 | 2 | 79.9 | 72†, 8 | 5 (0.3) | 0.6 | 73.7 |
| 46 | 1 | 94.6 | 95 | 0 (0.0) | 2.1 | 97.8 |
| 47 | 1 | 98.5 | 99† | 60 (5.1) | 1.5 | 99.7 |
| 48 | 4 | 99.8 | 29, 24†, 24†, 23† | 30 (36.1) | 3.2 | 99.9 |
| 49 | 1 | 99 | 99† | 4 (0.6) | 0.7 | 99.5 |
| 50 | 2 | 81.5 | 75†, 7† | 2724 (41.7) | 0.9 | 86.4 |
| 51 | 1 | 5.8 | 6† | 4 (0.5) | 0 | 39.6 |
No index indicates that no index clone was identified in the sample; therefore, index clone frequency data were not applicable; Not sorted, only DNA was available for these samples; therefore, no sorting was performed; and NA, no data are included because of the low number of cells in the flow cytometry gate.
Percent evolved clones of total clones in the sample.
Index clone shows evidence of evolution.
No sorting was performed because of the low number of total viable cells in the sample.
Given the flow cytometry marker used (see supplemental Table 2), no apparent malignant population was observed.
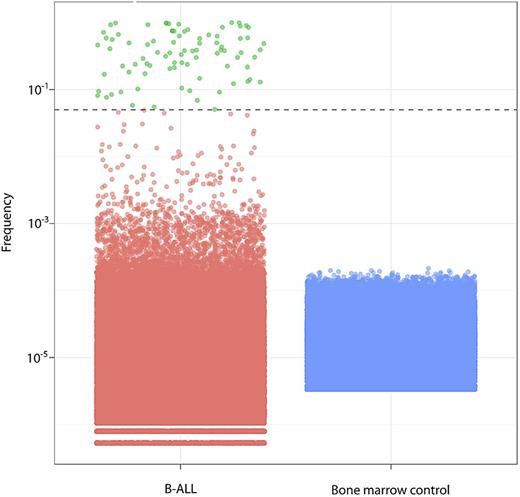
Clone frequencies in B-ALL (green and red) and normal bone marrow controls (blue), with each dot representing a single clone. The dashed line indicates the 5% threshold for index clone selection; green represents B-ALL clones; and red, other clones.
Clone frequencies in B-ALL (green and red) and normal bone marrow controls (blue), with each dot representing a single clone. The dashed line indicates the 5% threshold for index clone selection; green represents B-ALL clones; and red, other clones.
We validated that clonotypes above our threshold are likely to be leukemic by several methods. First, we looked at frequency distributions in healthy bone marrow samples. Individual clonotype frequencies in healthy bone marrow samples from adults were uniformly far below the 5% threshold (Figure 2). Similarly, 99.1% of clonotypes were present at very low frequency (< 0.1%) in the B-ALL diagnostic samples (Figure 2).
We further validated the 5% threshold by confirming that index clonotypes were de-enriched in populations sorted for normal B-cell markers (Table 2). Index clones were found in normal B cells from patients with a mean frequency of 3.4% (median, 1.0%). A low frequency of index clones is expected in the sorted normal B cells because conventional FACS does not permit isolation of pure populations. Moreover, sort purity is lowest when target cells are at very low frequency, as was the case in many of the samples with very low percent of normal B cells. There was at least a 2-fold reduction in index clone frequency in the normal B-cell sort (mean, 27-fold reduction). The reduction of index clone frequency in the normal B-cell sort was consistent throughout the range of index clone frequencies, indicating that even clonotypes just above the threshold (eg, 5%-10% frequency) are leukemic clonotypes. In contrast, within most patients, the sum of the index clone frequencies was enriched or remained similar in cells sorted for leukemic cell markers (CD45−/lowCD10+). The only exception was in patient 31, whose sample had 3% viable cells. These results validated our criteria and threshold for selecting index clonotypes.
Extent of NDN sharing required for identification of evolved clonotypes
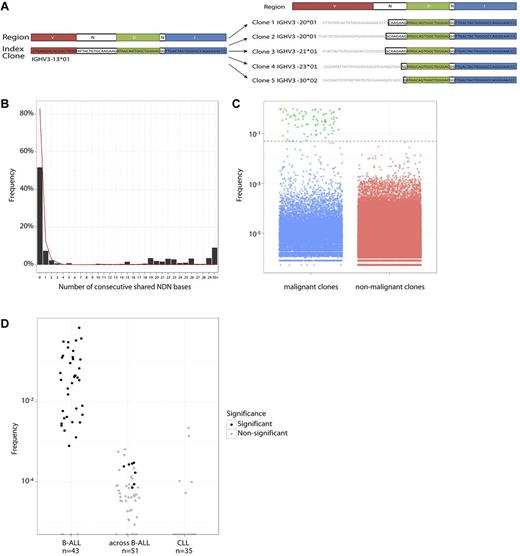
Visual comparison of index and nonindex clonotype sequences within individual patients revealed that many clonotypes shared the same JH segments and a portion of the NDN region adjacent to the JH segment, but differed in their VH segment and the NDN region immediately adjacent to the VH segment. Figure 3A schematically illustrates this in patient 23 where the clonotypes share the JH and a large portion of the NDN regions but differ in their VH segments. This is consistent with a model of VH replacement where VH-VH gene replacement or successive VH to D-JH joinings have occurred.
Evolved clones. (A) Index clone and some high-frequency evolved clones in patient 23. Blue represents JH bases; green, D bases; red, VH bases; and black, shared N bases. These evolved clones share large stretches of bases consistent with common ancestry. (B) Percentage of clones that share JH and NDN bases with index clones. The x-axis represents the number of shared bases. Data from B-ALL samples are shown as the black histogram, indicating a sharp decline that can be explained by random matching, and then an increase in frequency after 10 bases. For comparison, the red line indicates sharing in CLL samples and lacks the increase in sharing after 10 bases. (C) Frequency plots of malignant clones (index, green plus evolved, blue) versus nonmalignant clones (red). Evolved clones are similar in frequency to nonmalignant clones. (D) Per-patient fraction of evolved clones among all clones in B-ALL, CLL, and in patient-permuted B-ALL samples (across B-ALL). Each dot represents a patient, with shading of the dot indicating whether the number of evolved clones in a sample is significant.
Evolved clones. (A) Index clone and some high-frequency evolved clones in patient 23. Blue represents JH bases; green, D bases; red, VH bases; and black, shared N bases. These evolved clones share large stretches of bases consistent with common ancestry. (B) Percentage of clones that share JH and NDN bases with index clones. The x-axis represents the number of shared bases. Data from B-ALL samples are shown as the black histogram, indicating a sharp decline that can be explained by random matching, and then an increase in frequency after 10 bases. For comparison, the red line indicates sharing in CLL samples and lacks the increase in sharing after 10 bases. (C) Frequency plots of malignant clones (index, green plus evolved, blue) versus nonmalignant clones (red). Evolved clones are similar in frequency to nonmalignant clones. (D) Per-patient fraction of evolved clones among all clones in B-ALL, CLL, and in patient-permuted B-ALL samples (across B-ALL). Each dot represents a patient, with shading of the dot indicating whether the number of evolved clones in a sample is significant.
Within each B-ALL sample, we compared other clones to index clones sharing the same JH segment and analyzed the number of consecutive NDN bases shared in the JH-to-VH direction (Figure 3A). In addition to the expected exponential decline in sharing of sequential NDN nucleotides, there were a large number of clones that shared > 15 bases (Figure 3B). In contrast, very few CLL samples exhibited clones sharing more than a few consecutive NDN bases (red line in Figure 3B). Because CLL is a malignancy of B cells at later stages of development, clonal evolution through VH replacement is not anticipated in these samples.33-35 Based on these observations, we developed a bioinformatic algorithm to identify evolved clones based on shared NDN bases. Because D segment lengths vary, yet bases in each D segment are not independent of each other, identical D segments are compressed as 2 effective NDN bases. This modification maintained separation between the level of sharing observed in the B-ALL and CLL samples (supplemental Figure 1). Very few CLL clones (0.03%) with the same JH segment as the index clone shared > 6 effective NDN bases with the index clone, whereas 37% of B-ALL clones with the same JH segment shared 6 or more effective NDN bases with an index clone. This suggests that a threshold of 6 shared effective NDN bases can be used to define evolved clonotypes (equivalent to P < 5 × 10−4 with a geometric distribution). The low detection of evolved clonotypes in CLL samples was not the result of significantly more index clones with short effective NDN length, as the fraction of CLL index clones that have effective NDN length > 6 was larger than that for the B-ALL samples.
Evolved clonotypes identified in B-ALL diagnostic samples
With this algorithm, we identified almost 14 000 evolved clonotypes in the diagnostic B-ALL bone marrow samples (Table 2). The number of evolved clonotypes per B-ALL patient varied widely from 0 to 4024 clonotypes, with 37 of 43 patients having 1 or more evolved clonotypes (Table 2). The number of evolved clonotypes that were associated with each index clonotype is shown in supplemental Table 3. Forty-two percent of clonotypes with a frequency > 0.1% were composed of clonotypes evolved from the ancestral B-ALL clone (Figure 3C).
Validation of evolved clonotype selection criteria
When the clonal evolution selection criteria were applied to the B-ALL patient samples, the majority of samples (37 of 43 patients, 86%) showed the presence of evolved clonotypes (Figure 3D). We validated our algorithm with 3 methods. First, to assess the level of random NDN sharing, we compared the clones from each B-ALL patient with the index clones of all other B-ALL patients (Figure 3D). In this comparison, 0.02% were called evolved clonotypes by our algorithm, similar to the 0.01% frequency expected by chance. The number of called evolved clonotypes in the majority of the across-patient B-ALL comparisons are not significantly higher than zero (P < 10−5, binomial distribution).
To ensure that the observed evolved clonotypes are not generated by amplification artifacts linking the high-frequency index clone sequence with another sequence from a different clone, we assessed 35 CLL samples for the presence of evolved clonotypes using the same definition and thresholds used for B-ALL samples. CLL contains high-frequency disease clones, such as B-ALL, but it is a malignancy of B cells at later stages of development; therefore, no significant evolved clonotypes are expected. A total of 0.01% of clones in CLL samples were called evolved, which is consistent with the number expected by chance with a P value of 10−4 (Figure 3D). In addition, none of the individual samples contained a significant number of evolved clones (P < 10−5, binomial distribution). Along with the across-patient B-ALL comparison, these numbers placed a ceiling on the extent of evolution in B-ALL samples that can be explained by PCR or sequencing artifacts or by random chance sharing.
We further validated the clonal evolution criteria by assessing the presence of the evolved clonotypes in the sorted cell populations. If these clonotypes were indeed leukemic, their frequency in the normal B-cell population was anticipated to be de-enriched compared with the frequency in the initial unsorted sample. Although evolved clones were found in the malignant phenotype sorts, they were virtually absent in the normal B-cell sorts. Evolved clones were found in the normal B-cell sort with a maximum frequency of 0.4% (mean, 0.03%; median, 0%) with an average 58-fold reduction in frequency. These results further validated our selection criteria for identification of evolved clonotypes in the B-ALL diagnostic samples.
Variation in clonal evolution patterns at the patient level
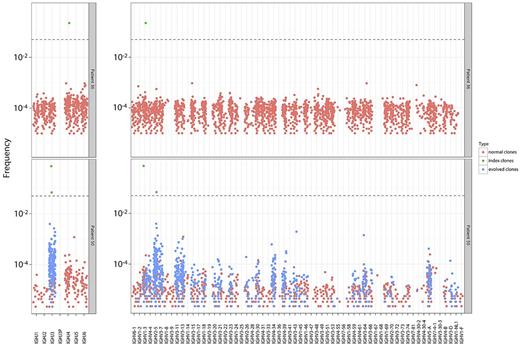
We characterized the clonal evolution pattern in each patient. We found that the percent of evolved clonotypes ranged dramatically from 0% to 86% across the B-ALL patients, with the majority of patients having evolved clonotypes (Table 2). The 43 B-ALL patients who had index clonotypes could be categorized into 3 distinct groups based on the percent of clonal evolution present in their diagnostic sample. Specifically, 6 patients had no evolution (0%), 23 patients had limited evolution (0%-10%), and 14 patients had a high degree of evolution (> 10%; Table 2). Figure 4 shows the contrasting evolution patterns between a patient with no evolution (patient 36) and a patient with a high-degree of evolution (patient 50). In patient 50, 2 higher-frequency clonotypes that share 1 JH segment but have different VH segments are observed. In contrast, a single higher-frequency clonotype is seen in the sample with no evolution (patient 36; Figure 4). The wide variation in clonal evolution patterns between patients may serve as the basis for further exploration of the prognostic significance of such clonal evolution in B-ALL.
Evolution patterns in patients with differing levels of evolution. Samples from 2 representative patients with no evolution (patient 36) and high evolution (patient 50) are shown. JH and VH segments are shown on the x-axis, with clone frequency in logarithmic scale on the y-axis. A single higher-frequency clonotype is seen in the sample with no evolution (patient 36). Two higher-frequency clonotypes that share 1 JH segment but have different VH segments are observed in the patient with a high degree of evolution (patient 50).
Evolution patterns in patients with differing levels of evolution. Samples from 2 representative patients with no evolution (patient 36) and high evolution (patient 50) are shown. JH and VH segments are shown on the x-axis, with clone frequency in logarithmic scale on the y-axis. A single higher-frequency clonotype is seen in the sample with no evolution (patient 36). Two higher-frequency clonotypes that share 1 JH segment but have different VH segments are observed in the patient with a high degree of evolution (patient 50).
We tested for any strong association of evolution with age at diagnosis, initial white blood cell count, CNS involvement, cytogenetics, National Cancer Institute risk group, and day 29 flow MRD. We did not find any significant associations with these clinical variables. However, all 4 evaluable relapse patients showed high levels of evolution at diagnosis, and the association between evolution and relapse was found to be significant (P = .04), suggesting the need for additional validation.
Genesis of evolved clonotypes
The large number of evolved clonotypes observed can be generated by 2 distinct models. The ancestral clonotype may be an incomplete rearrangement (D-JH) of the IgH locus, and its progeny may continuously recombine the VH segment. Alternatively, the original clonotype may, indeed, be a complete VH-D-JH rearrangement subject to continuous VH replacement in progeny cells. The second model would indicate that the VH segments of the evolved clonotypes will reside in a germline location that is 5′ to the ancestral VH clonotype. We therefore assessed whether the VH segment in evolved clonotypes are 5′ or 3′ of the VH segment in index clonotypes, noting that the index clonotypes may not necessarily be the ancestral clonotypes. We observed that, in the majority of samples, there is a strong bias for the VH segment in evolved clonotypes to be 5′ of the VH segment in the index clonotype (supplemental Table 4). In 7 cases, the bias is milder; and in only 2 cases, the bias was for the VH segment in evolved clonotypes to be 3′ of the VH segment in the index clonotype (supplemental Table 4). In patients where there is a strong bias for the VH segment of evolved clonotypes to be 5′of the VH segment of the index clone, the generation of evolved clonotypes is consistent with the second VH replacement model. In these cases, the index clonotype is probably ancestral to most of the evolved clonotypes. In 9 patients, there are substantial numbers of clonotypes whose VH segments are 3′ of the index clonotype, which indicates that these clonotypes were not generated from the index clonotype, but that both were likely generated from a distinct ancestral clonotype. This could be consistent with the first model we outlined, in which the ancestral clone has undergone D-JH rearrangement and its progeny join different VH segments to that D-JH clonotype.
Striking differences in number of evolved clonotypes between independent index clonotypes in the same patients
We also assessed the distribution of evolved clonotypes within samples that contained multiple index clonotypes. Twenty-seven B-ALL patients had multiple index clonotypes in their diagnostic bone marrow samples (Table 2). Our definition of index clones is based on a frequency threshold; therefore, there were several cases where index clones were related to each other by evolution. Similarly, as part of normal B-cell development and in B-ALL, IgH can rearrange in 1 or both chromosomes, potentially leading to more than 1 independent index clonotype (ie, not related to each other by evolution). Altogether, there were 21 samples with more than 1 independent index clone (supplemental Table 3).
In cases where 2 independent index clones were found in a person, we investigated whether evolution occurs equally on both chromosomes or preferentially on 1 chromosome. Altogether, there were 7 samples with 2 independent (ie, not related to each other by evolution) index clonotypes and at least a moderate number (> 20) of evolved clonotypes in the sample. In 3 of 7 cases, all detected evolution occurred for 1 clonotype, whereas no evidence for evolution is seen in the other. In 1 of the 3 patients (patient 38), the effective NDN length of the index clonotype was < 6; therefore, we were not able to detect evolution associated with this clonotype using the algorithm used in this study. In the other 2 patients, evolution could have been detected but was not observed. For example, patient 9 had 2 index clonotypes that were related to 42 (9%) and 0 evolved clonotypes, respectively (supplemental Table 3). In the remaining 4 patients with 2 independent index clonotypes, each of the index clonotypes showed at least some evolved clonotypes. For example, patient 35 had 2 index clonotypes, with 2 different JH alleles, which were related to 1020 (24%) and 129 (3%) evolved clonotypes, respectively (supplemental Table 3).
We evaluated whether there is evidence of an incomplete IgH rearrangement in the 3 cases with exclusive evolution in 1 of the 2 index clonotypes. In all 3 cases, the evolved clonotypes shared several N bases between the VH and D segments with the index clonotype, indicating that they result from a VH-DJH recombination and are not the result of a D-JH recombination.
Different cell surface markers in related clonotypes at the time of diagnosis
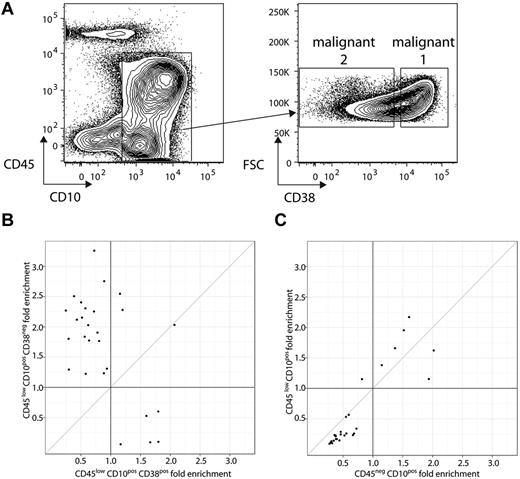
We evaluated whether evolved clonotypes had different cell surface markers than their related index clonotype(s) at the time of diagnosis. This was performed by isolating 2 leukemic cell populations from each sample based on expression of a panel of cell surface markers commonly used for characterizing B-ALL samples, specifically CD10, CD19, CD20, CD34, CD38, CD45, and CD81.32,36,37 Expression of this set of cell surface markers on bone marrow mononuclear cell samples was diverse across patients, and no single set of gating criteria could be applied to all patients. The markers chosen to distinguish the 2 leukemic populations from each patient were determined at the time of sorting on a patient-by-patient basis (see supplemental Table 2 for marker phenotypes of each gated subpopulation from each patient). We compared the frequency of the related clonotypes in the 2 leukemic cell populations and the initial unsorted sample. In some cases, such as patient 23, related clonotypes from the same sample were enriched in 1 leukemic population and de-enriched in the other (Figure 5A-C). For patient 23, we performed flow sorting 2 separate times with 2 different markers (CD38 and CD45) to distinguish each leukemic population. The evolved clonotype frequencies showed no substantial difference in the 2 leukemic populations defined by CD45; however, they showed substantial differences in the leukemic populations defined by CD38 (Figure 5A-C). These results demonstrated that, in some samples, a portion of the evolved clonotypes had different cell surface markers than their related index clonotype(s).
Evolved clone differences between CD45lowCD10+CD38+ and CD45lowCD10+CD38− sorts of patient 23. (A) Flow cytometric plots illustrating CD45 and CD10 (left) and forward scatter (FSC) and CD38 expression (right) on bone marrow mononuclear cells from patient 23. Gate on plot at left indicates CD45−/lowCD10+ “B-ALL” cells. Gates on plot at right indicates malignant 1 and 2 populations (based on the level of CD38 expression) within B-ALL cells identified at left. For this patient, an independent sort was performed to separate another malignant 1 and malignant 2 population based on CD45−CD10+ and CD45lowCD10+. (B) Fold enrichment (compared with unsorted) of the frequency of each evolved clonotype in the 2 malignant populations, with the x-axis showing enrichment (compared with unsorted) in the CD45−/lowCD10+CD38+ population and the y-axis showing enrichment (compared with unsorted) in the CD45−/lowCD10+CD38− population, with each point representing a malignant clone (index/evolved). (C) Fold enrichment (compared with unsorted) of the frequency of each of the evolved clonotypes in each of the 2 malignant populations, with the x-axis showing enrichment (compared with unsorted) in the CD45−CD10+ population and the y-axis showing enrichment (compared with unsorted) in the CD45lowCD10+ population, with each point representing a malignant clone (index/evolved).
Evolved clone differences between CD45lowCD10+CD38+ and CD45lowCD10+CD38− sorts of patient 23. (A) Flow cytometric plots illustrating CD45 and CD10 (left) and forward scatter (FSC) and CD38 expression (right) on bone marrow mononuclear cells from patient 23. Gate on plot at left indicates CD45−/lowCD10+ “B-ALL” cells. Gates on plot at right indicates malignant 1 and 2 populations (based on the level of CD38 expression) within B-ALL cells identified at left. For this patient, an independent sort was performed to separate another malignant 1 and malignant 2 population based on CD45−CD10+ and CD45lowCD10+. (B) Fold enrichment (compared with unsorted) of the frequency of each evolved clonotype in the 2 malignant populations, with the x-axis showing enrichment (compared with unsorted) in the CD45−/lowCD10+CD38+ population and the y-axis showing enrichment (compared with unsorted) in the CD45−/lowCD10+CD38− population, with each point representing a malignant clone (index/evolved). (C) Fold enrichment (compared with unsorted) of the frequency of each of the evolved clonotypes in each of the 2 malignant populations, with the x-axis showing enrichment (compared with unsorted) in the CD45−CD10+ population and the y-axis showing enrichment (compared with unsorted) in the CD45lowCD10+ population, with each point representing a malignant clone (index/evolved).
Discussion
We performed this study to gain deeper insights into clonal evolution in childhood B-ALL. Using a sequencing technology-based platform, we observed ongoing, profound clonal evolution at the IgH locus in diagnostic samples from B-ALL patients. Some diagnostic samples did not contain any high-frequency IgH rearrangements, which is consistent with previous studies revealing a lack of IgH clonality in some B-ALL patients.38 We observed up to 4024 evolved clonotypes per B-ALL patient. This magnitude of clonal evolution has not been described previously, and we anticipate that additional evolved clonotypes may be present that are undetectable using the current algorithmic approach. The majority of evolved clonotypes are present at low frequency at diagnosis, rendering them undetectable by previous investigations. Because we focused our studies on a single locus, we were able to deeply sequence the IgH locus and identify evolved events at extremely low frequency (≤ 10−5), which provides complementary insights to whole genome studies that only reveal high-frequency evolution events. The clinical significance of these low frequency evolved events is unclear and will be the subject of future investigations.
Although this study has very limited power to detect association of evolution with clinical variables (because only 4 patients with index clones and 1 without an index clone relapsed), the association between evolution and relapse was found to be significant. Further studies with sufficient statistical power are needed to determine the clinical significance of the magnitude of evolution in B-ALL diagnostic samples and its correlation with relapse and clinical outcome.
The occurrence of VH replacement in leukemia has previously been investigated, and relapse clones in B-ALL have been traced back to a minor clonal population present at diagnosis that was resistant to remission induction chemotherapy and undetectable by routine monitoring methods.12,20-23 Our data show that, in some diagnostic samples, most of the evolved clonotypes were not derived from the index clone but instead both were generated from some other ancestral clonotype. Presumably, the cells carrying the index clonotype had expanded because of acquisition of an oncogenic mutation, and its biologic behavior and chemoresistance may be significantly different from both the ancestral clone and clonotypes derived from the ancestral clone. We also observed biologic heterogeneity between clones in some samples, where cells bearing some of the evolved clonotypes had different surface markers than their related index clonotypes. The heterogeneity of surface markers within the same sample may have an impact on the detection of MRD using flow cytometric methods. Further studies may ascertain the importance of these distinct clonal populations in leukemic transformation at initial diagnosis as well as at the time of relapse.
Our results also provide insight into the molecular mechanisms responsible for clonal evolution in B-ALL patients. Evolution at the IgH locus has been attributed to multiple mechanisms, including VH replacement or continued recombination of an ancestral incompletely rearranged IgH sequence (ie, VH to D-JH joining).14-18,39,40 Our data suggests that VH replacement is the dominant molecular mechanism responsible for clonal evolution in B-ALL patients, as suggested by predominant usage in evolved clones of VH segments 5′ in the germline configuration to the VH segment used by the index clonotype. In some cases, the extent of clonal evolution was disparate between multiple index clonotypes in the same patient. This can be attributed to allelic exclusion of the evolution process probably resulting from cis-acting factors.41 Our data suggest that IgH locus evolution in leukemic cell clones may be recapitulating some of the mechanisms of allelic exclusion that have been observed in normal lymphocytes, in which the VH to D-JH recombination is activated on one chromosome and inhibited on the other chromosome by mechanisms presumably linked to RAG protein accessibility to the chromatin.7,8
In addition to the biologic significance of this study, our results suggest that the sequencing-based approach holds promise for MRD monitoring in B-ALL. The current standard for childhood B-ALL treatment is risk-modified therapy intensification based largely on the detection of MRD during and after induction therapy. Subsequent MRD monitoring for detection of clinical relapse has shown some clinical utility, such as before bone marrow transplant where MRD positivity has been found to predict increased risk for relapse.42 Despite these encouraging findings, the majority of MRD monitoring studies have shown limited utility after the end of induction. This may be the result of assay sensitivity limitations in the case of flow cytometry or an inability to monitor evolved leukemic clones at relapse that were undetectable at diagnosis in the case of allele-specific oligonucleotide PCR. This study demonstrates that our sequencing-based approach enables quantitative analysis of clonal evolution in diagnostic B-ALL samples, which shows great promise for improved MRD monitoring. However, one limitation of the sequencing approach for MRD evaluation is the requirement of an initial diagnostic sample that contains a high-frequency index clone. Further studies will be necessary to demonstrate the clinical validity of this method for MRD detection and its practical application within current laboratory workflows.
In conclusion, high-resolution IgH repertoire sequencing can be used for the identification and monitoring of the entire population of leukemic clones in diagnostic samples from pediatric B-ALL patients. Our studies also reveal distinct biologic mechanisms that facilitate the ongoing evolution at the IgH locus. Clinical validation is now needed to show that the application of this technology to monitor evolved leukemic clonotypes can fundamentally improve our ability to detect malignant cells before clinical relapse in children with B-ALL, with potential applicability to other lymphoid malignancies.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the Leukemia and Lymphoma Translational Research Award (R618-09), the National Cancer Institute (PO1 CA049605), and the Debra and Andrew Rachleff Endowed Fellowship. F.P., V.E.H.C., M.K., and M.F. were supported in part by Sequenta Inc (research funding).
National Institutes of Health
Authorship
Contribution: C.G., F.P., V.E.H.C., N.L., M.K., and M.F. designed the research; M.K. performed the research; F.P., V.E.H.C., C.G., M.K., and M.F. analyzed the data and wrote the manuscript; N.L., D.B.M., G.D., and A.C.L. provided clinical samples and edited the manuscript; and all authors read and approved the manuscript.
Conflict-of-interest disclosure: F.P., V.E.H.C., M.K., and M.F. are employees of and holders of equity in Sequenta Inc. The remaining authors declare no competing financial interests.
Correspondence: Charles Gawad, Pediatric Hematology/Oncology/Stem Cell Transplantation and Cancer Biology (Mail Code 5798), Stanford University School of Medicine, 1000 Welch Rd, Suite 300, Palo Alto, CA 94304; e-mail: cgawad@stanford.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal