Abstract
Although intrinsic apoptosis defects are causal to the extended survival of chronic lymphocytic leukemia (CLL) B cells, several lines of evidence support a contribution of the peripheral lymphoid organs and BM microenvironment to the extended lifespan of leukemic B cells. Lymphocyte trafficking is controlled by homing signals provided by stromal cell–derived chemokines and egress signals provided by sphingosine-1-phosphate (S1P). In the present study, we show that expression of S1P1, the S1P receptor responsible for lymphocyte egress, is selectively reduced in CLL B cells with unmutated IGHV. Expression of S1P2, which controls B-cell homeostasis, is also impaired in CLL B cells but independently of the IGHV mutational status. We provide evidence herein that p66Shc, a Shc adaptor family member the deficiency of which is implicated in the apoptosis defects of CLL B cells, controls S1P1 expression through its pro-oxidant activity. p66Shc also controls the expression of the homing receptor CCR7, which opposes S1P1 by promoting lymphocyte retention in peripheral lymphoid organs. The results of the present study provide insights into the regulation of S1P1 expression in B cells and suggest that defective egress caused by impaired S1P1 expression contributes to the extended survival of CLL B cells by prolonging their residency in the prosurvival niche of peripheral lymphoid organs.
Introduction
Although progressive accumulation of monoclonal CD5+ B cells in the blood, peripheral lymphoid organs, and BM is the hallmark of chronic lymphocytic leukemia (CLL), the clinical course of this disorder is highly variable, ranging from a stable disease that may only require monitoring over time to a progressive, severe disease.1,2 Several markers have been associated with poor prognosis. Coupled to the cytogenetic abnormalities, the mutational status of immunoglobulin heavy chain variable region (IGHV) genes is the most valuable marker presently available, with unmutated IGHV found in patients who develop aggressive disease.3 Nevertheless, the onset of disease progression and response to treatment are to date largely unpredictable.
At variance with other hematologic malignancies, CLL B cells are usually arrested at G0/G1 and their accumulation is the result of an abnormally prolonged survival rather than uncontrolled proliferation.1,2 Intrinsic defects in the apoptotic machinery underlie the prolonged lifespan of CLL B cells, a major target being the Bcl-2 family, in which overexpression of antiapoptotic members (Bcl-2 and Mcl-1) or impaired expression of proapoptotic members (Bax and Bak) tilts the balance toward cell survival.4 Extrinsic factors consisting mainly of stromal cell–derived chemokines (CXCL12, CXCL13, CCL19, and CCL21) also to contribute to the extended lifespan of CLL B cells by providing survival cues during their transit through peripheral lymphoid tissues and BM.5
Lymphocyte trafficking is tightly controlled by the chemokines present in the lymphoid microenvironment and the chemokine receptors expressed by the lymphocyte itself.6 CLL B cells express increased levels of CXCR4, CCR7, and CXCR5, which has been proposed to enhance their homing to BM and peripheral lymphoid tissues, thereby favoring cognate interactions with resident cells.7 In this context, unique features of CLL B cells with unmutated IGHV are an increased ability to phosphorylate p72Syk in response to sIgM ligation8 and robust signaling by B-cell Ag receptors,9 which have been demonstrated to broadly recognize self-Ags such as modified cytoskeletal proteins and oxidation-specific epitopes.10,11 Therefore, the enhanced homing to the stromal microenvironment contributes to disease progression by favoring not only survival signaling, but also mitogenic signaling by the B-cell Ag receptor in response to self-Ag presented by stromal cells,10 which is likely to account for the small proliferative fraction in CLL clones.3
Sphingosine-1-phosphate (S1P), a sphingolipid metabolite produced by sphingosine kinase-1 and -2 that acts through the activation of the G-protein–coupled receptors S1P1-S1P5, has recently emerged as a major player in lymphocyte trafficking.12 Extracellular S1P, largely derived from erythrocytes and lymphatic endothelial cells, accumulates at high concentrations (high nanomolar to micromolar levels) in circulatory fluids.13 Conversely, the interstitial S1P levels are maintained very low by a battery of S1P-degrading enzymes.14 This results in a gradient that promotes the egress of lymphocytes from their homing sites and their reentry into the bloodstream or lymph, a process mainly controlled by S1P1.15 Therefore, the dwell time of lymphocytes in peripheral lymphoid tissues is tightly controlled by the opposing activities of chemokine and S1P receptors.
We hypothesized that, in addition to their enhanced homing because of increased expression of chemokine receptors,7 a defective response of CLL B cells to S1P would lead to an extended transit time in lymphoid tissues, thereby further favoring their interaction with the prosurvival stromal microenvironment. In the present study, we show that neoplastic B cells from CLL patients with unmutated IGHV, but not from patients with mutated IGHV, have a profound deficiency in S1P1. We also show that the S1P1 defect, which is associated with an up-regulation of CCR7, is selectively caused by impairment in the expression of the pro-oxidant Shc family adaptor p66Shc.
Methods
Patients and healthy donors
Blood samples were collected from 78 patients who satisfied standard morphologic and immunophenotypic criteria for CLL. Normal B cells from 13 buffy coats were used as controls for the healthy adult population. Informed consent was obtained from all patients according to the Declaration of Helsinki. All specimens were collected from patients attending the Hematology and Clinical Immunology Branch, Padua University School of Medicine (Padua, Italy), from 2003-2012. At the time of the collection, patients had never received treatment. The research was approved by the local ethics committee.
B cells (CD19+) were purified by negative selection using the RosetteSep Human B-cell enrichment Cocktail (StemCell Technologies), followed by density gradient centrifugation on Lympholite (Cedarlane Laboratories). The purity of the resulting B-cell population was > 90% as assessed by flow cytometry.
Mice
p66Shc−/− mice in the 129 genetic background were described previously.16 The study was carried out on the p66Shc−/− and 129 mouse colonies bred in the animal facility at the University of Siena (Siena, Italy). Analyses were performed on age- and sex-matched 2- to 9-month-old mice. All animal experiments were carried out in agreement with the Guiding Principles for Research Involving Animals and Human Beings and approved by the local ethics committee.
Mice were killed by cervical dislocation and the spleen, lymph nodes (LNs), blood, and BM were harvested. In some experiments, splenic B or T cells were negatively purified by immunomagnetic sorting using either the Dynabeads Mouse CD43 or the Dynal Mouse T-cell Negative Isolation Kit (Invitrogen), respectively, which resulted in > 85% purity as assessed by flow cytometry.
Cell lines, transfections, and immunoblots
The CLL-derived B-cell line MEC17 was used for the generation of stable transfectants. A mammalian expression vector (pcDNA3; Invitrogen) encoding human full-length p66Shc and the respective mutants p66ShcSA (S→A substitution at position 36)18 and p66ShcQQ (EE→QQ substitutions at positions 132-133)19 were introduced into MEC cells by electroporation. A control line was generated using empty vector. Stably transfected cells were selected in medium containing 1 mg/mL of G418 (Gibco-BRL/Life Technologies). The Jurkat transfectants were described previously.18
Fresh peripheral blood (PB) CLL B cells were transiently cotransfected with 1 μg of green fluorescent protein (GFP) reporter/sample and 5 μg of empty vector (pcDNA3) or the same vector encoding p66Shc (purified using the EndoFree Plasmid Maxi kit from QIAGEN) using the Human B-cell Nucleofector Kit (Amaxa Biosystems). The transfection efficiency was consistently 40% or greater as assessed by flow cytometric analysis of B cells transfected with the GFP reporter, and cell viability at the time of analysis (48 hours posttransfection) was consistently 40%-60%. Surface S1P1 was measured on GFP+ cells, gating on the population with the scatter parameters of live cells. RNA was extracted using the RNA mini kit (QIAGEN) and retrotranscribed using the IScript cDNA synthesis kit (Bio-Rad).
For immunoblot analyses, cells (5 × 106/sample) were lysed in 1% Triton X-100 in 20mM Tris-HCl (pH 8) and 150mM NaCl (in the presence of a protease inhibitor cocktail). Immunoblot analysis of postnuclear supernatants was carried out by chemiluminescence (SuperSignal West Pico Chemiluminescent Substrate kit; Pierce) using as primary Abs anti-ShcA (Upstate Biotechnology), anti-EDG1/S1P1 (Novus Biologicals), or control anti-actin (Millipore) Abs and secondary peroxidase-labeled Abs (Amersham Pharmacia Biotech).
RNA purification and qRT-PCR
Total RNA was extracted from B cells from healthy donors, CLL patients, mice, or MEC cells and retrotranscribed as described previously.20 Two independent reverse transcription reactions were performed on each RNA sample. Real-time quantitative PCR (qRT-PCR) was performed in triplicate on each cDNA on 96-well optical PCR plates (Sarstedt) using SSo Fast EvaGreenR SuperMix (Bio-Rad) according to the manufacturer's instructions and a CFX96 Real-Time system (Bio-Rad). After an initial denaturation for 3 minutes at 95°C, denaturation in the subsequent 42 cycles was performed for 10 seconds at 95°C, followed by 30 seconds of primer annealing at 60°C. Results were processed and analyzed using CFX Manager Version 1.5 software (Bio-Rad). Transcript levels were normalized to GAPDH, which was used as a housekeeping gene. The primers used to amplify the cDNA fragments corresponding to human and mouse transcripts are listed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Flow cytometry
Cells were resuspended at 2 × 106/mL in serum-free medium added with 0.5% fatty acid-free BSA (Sigma-Aldrich) and incubated for 30 minutes at 37°C to allow recycling of intracellular S1P receptors to the cell surface. As a specificity control for S1P1, a sample was incubated with 100nM FTY720 (Santa Cruz Biotechnology) in the same medium for 30 minutes at 37°C to induce receptor down-regulation. Cells were then incubated with anti-human EDG1/S1P1 (Novus Biologicals), anti–human EDG5/S1P2 (R&D Systems), or anti–mouse EDG1/S1P1 (kindly provided by Jason Cyster, Department of Microbiology & Immunology, Howard Hughes Medical Institute, University of California, San Francisco, San Francisco, CA) primary Abs and Alexa Fluor 488 goat anti–mouse IgG or Alexa Fluor 488 goat anti–rat IgG (Invitrogen) secondary Abs. For the mouse experiments, the analysis was carried out on gated CD22+ cells.
For B-cell counts, PB and cell suspensions from LNs, spleen, and BM were stained with fluorochrome-conjugated anti-CD3 and anti-CD22 Abs (BD Biosciences).
Flow cytometry was carried out using a FACScan flow cytometer (BD Biosciences). Data were analyzed and plotted using FlowJo Version 6.1.1 software (TreeStar).
Cell treatments and ROS detection
Cells were incubated with 400μM H2O2 (Sigma-Aldrich) or 0.2mM Trolox (Merck Biosciences), respectively. After 3 hours at 37°C, cells were labeled for 30 minutes at 37°C with 5μM CM-H2DCFDA (Molecular Probes) and an aliquot was used to measure intracellular reactive oxygen species (ROS) by flow cytometry. After 24 hours, RNA was extracted and processed for qRT-PCR. Surface S1P1 was measured by flow cytometry.
Chemotaxis assays
Chemotaxis assays were carried out using 24-well Transwell chambers with 5-μm pore size polycarbonate membranes (Corning Life Sciences) essentially as described previously.21 Filters were soaked overnight in 0.5% fatty acid-free BSA (Sigma-Aldrich) in RPMI 1640 medium. The chemotaxis medium (500 μL of serum-free and 0.5% fatty acid-free BSA medium) with or without 100nM S1P (Sigma-Aldrich) was placed in the lower chamber, and 100 μL of the cell suspension (5 × 105 cells/sample) in chemotaxis medium was placed in the upper chamber. After 3 hours of incubation at 37°C in humidified air with 5% CO2, the upper chamber was emptied, filters were removed, and the cells in the lower chamber were counted by flow cytometry. The migration index was calculated by determining the ratio of migrated cells in treated versus untreated samples.
Statistical analyses
Mean and SD values were calculated using the Student t test (unpaired) and Excel Version 14.0.0 software (Microsoft). The difference between 2 consecutive samples within the same population was determined by the Mann-Whitney rank-sum test if the population was not normally distributed. P < .05 was considered statistically significant.
Results
Defective S1P1 and S1P2 expression in CLL patients with unfavorable prognosis
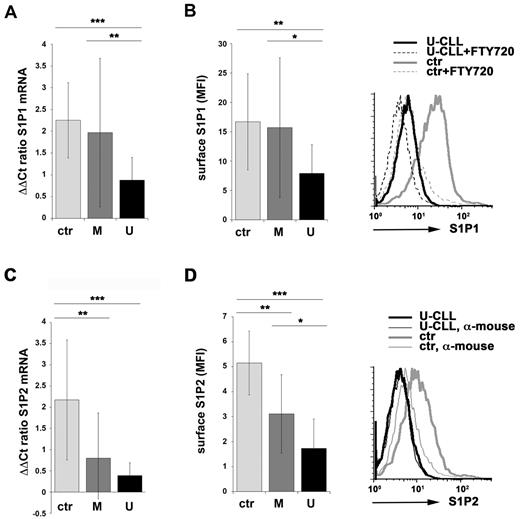
S1P1 expression in PB B cells was measured on a cohort of 78 CLL patients divided according to the IGHV mutational status into 2 subsets of 32 patients with mutated IGHV (M-CLL) and 46 unmutated (U-CLL) patients. Because the levels of surface S1P1 undergo rapid changes depending on the local S1P concentration,12 we first measured the relative abundance of S1P1-specific transcripts in RNA from CLL B cells. B cells purified from PB lymphocytes from 13 healthy donors were used as controls. M-CLL patients showed a large variability in the levels of S1P1 mRNA, as assessed by qRT-PCR, with differences that were not statistically significant compared with healthy controls. Conversely, expression of S1P1 was markedly reduced in B cells from U-CLL patients compared with M-CLL and normal B cells (Figure 1A and supplemental Figure 1).
Impaired S1P1 and S1P2 expression in CLL B cells from patients with unmutated IGHV. (A,C) qRT-PCR analysis of S1P1 (A) or S1P2 (C) mRNA in purified peripheral B cells from either healthy donors (ctr; n = 13) or CLL patients with mutated (M; n = 28 for S1P1; n = 25 for S1P2) or unmutated (U; n = 42 for S1P1; n = 36 for S1P2) IGHV. The relative abundance of gene transcripts was determined on triplicate samples using the ddCt method and is expressed as normalized fold expression (mean ± SD). (B,D) Flow cytometric analysis of surface S1P1 (B) or S1P2 (D) on purified peripheral B cells from either healthy donors (ctr; n = 10 for S1P1; n = 11 for S1P2) or CLL patients with mutated (M; n = 18 for S1P1; n = 16 for S1P2) or unmutated (U; n = 18 for S1P1; n = 16 for S1P2) IGHV. The data in the histograms are expressed as the mean fluorescence intensity (MFI) ± SD. Representative FACS profiles of S1P1 (B) or S1P2 (D) are shown. Specificity controls for each donor/patient included a sample incubated with secondary Ab alone (α-mouse) and, for S1P1 stainings, a sample preincubated with FTY720 to induce receptor down-regulation. ***P < .001; **P < .01; and *P < .05 by Mann-Whitney rank-sum test.
Impaired S1P1 and S1P2 expression in CLL B cells from patients with unmutated IGHV. (A,C) qRT-PCR analysis of S1P1 (A) or S1P2 (C) mRNA in purified peripheral B cells from either healthy donors (ctr; n = 13) or CLL patients with mutated (M; n = 28 for S1P1; n = 25 for S1P2) or unmutated (U; n = 42 for S1P1; n = 36 for S1P2) IGHV. The relative abundance of gene transcripts was determined on triplicate samples using the ddCt method and is expressed as normalized fold expression (mean ± SD). (B,D) Flow cytometric analysis of surface S1P1 (B) or S1P2 (D) on purified peripheral B cells from either healthy donors (ctr; n = 10 for S1P1; n = 11 for S1P2) or CLL patients with mutated (M; n = 18 for S1P1; n = 16 for S1P2) or unmutated (U; n = 18 for S1P1; n = 16 for S1P2) IGHV. The data in the histograms are expressed as the mean fluorescence intensity (MFI) ± SD. Representative FACS profiles of S1P1 (B) or S1P2 (D) are shown. Specificity controls for each donor/patient included a sample incubated with secondary Ab alone (α-mouse) and, for S1P1 stainings, a sample preincubated with FTY720 to induce receptor down-regulation. ***P < .001; **P < .01; and *P < .05 by Mann-Whitney rank-sum test.
To confirm these results, surface S1P1 was quantitated by flow cytometry. Cells were incubated for 30 minutes in serum-free medium in the presence of lipid-free BSA before adding the Ab to allow recycling of receptors that had been internalized in the presence of the high S1P concentrations in the blood.12,22 The pharmacologic agonist FTY720, which when phosphorylated by the endogenous S1P kinases binds to all S1P receptors (with the exception of S1P2), inducing their rapid down-regulation,23 was used as a control. Consistent with the qRT-PCR results, no statistically significant differences were observed between M-CLL and healthy control B cells. Conversely, the levels of surface S1P1 were significantly reduced in U-CLL B cells compared with M-CLL and normal B cells (Figure 1B). Therefore, S1P1 expression was selectively impaired in the U-CLL subset.
Histological analysis of LNs and BM from CLL patients revealed that CLL cells associate with CD40L+ Th cells and follicular dendritic cells in germinal center (GC)–like structures known as pseudo-GCs, where they are believed to receive not only survival, but also proliferation signals.10 Because S1P2 has been recently implicated in the migration and survival of GC B cells,24 we measured the levels of S1P2 mRNA in normal and CLL B cells. Expression of S1P2 was profoundly impaired in CLL B cells compared with normal B cells independently of the IGHV mutational status, with U-CLL cells displaying lower levels than M-CLL cells as assessed by both qRT-PCR (Figure 1C and supplemental Figure 1) and flow cytometry (Figure 1D). This suggests that the behavior of U-CLL B cells during their residency within pseudo-GCs may be selectively affected by their defect in S1P2 expression.
p66Shc controls S1P1 expression in B cells
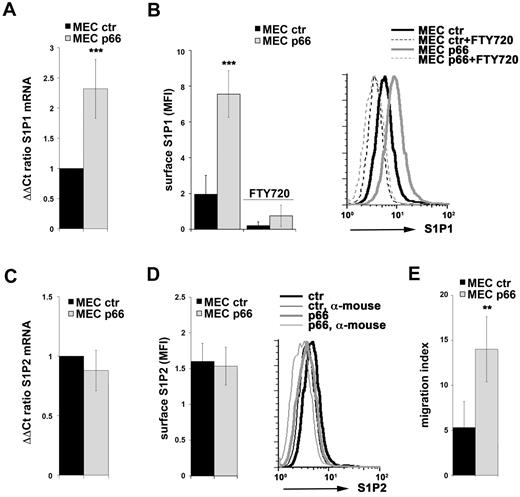
Unlike the situation in T cells,25 the factors that control S1P1 and S1P2 transcription in B cells have as yet not been identified. We have previously reported that the pro-oxidant ShcA isoform p66Shc26 acts as a negative regulator of proliferative and survival signals triggered by the BCR and that p66Shc expression is impaired in CLL B cells, with the lowest levels found in U-CLL patients.20 This impairment in p66Shc expression was found to be causal to the intrinsic apoptosis defects of CLL B cells and resulted from the ability of this adaptor to modulate the Bcl-2 family genes, promoting the expression of proapoptotic members while inhibiting expression of antiapoptotic members.20 Oxidants are known to profoundly affect gene transcription,27 suggesting that p66Shc could control the expression of other genes in addition to Bcl-2 family members through its pro-oxidant activity, including S1P1 and S1P2. To test this hypothesis, we first measured S1P1 and S1P2 expression in MEC cells, a CLL-derived human B-cell line lacking p66Shc, that were stably transfected with either an expression vector encoding p66Shc or empty vector. p66Shc-expressing MEC cells displayed significantly higher levels of both S1P1 mRNA and surface S1P1 compared with the control transfectant (Figure 2A-B). Conversely, p66Shc did not increase S1P2 expression above the very low levels detected in MEC cells (Figure 2C-D). Consistent with the S1P1 defect, the chemotactic response of p66Shc-expressing cells toward S1P was more robust compared with control cells (Figure 2E).
p66Shc controls S1P1 expression in B cells. (A,C) qRT-PCR analysis of S1P1 (A) or S1P2 (C) mRNA in MEC B cells stably transfected with either empty vector (MEC ctr) or an expression construct encoding p66Shc (MEC p66). The relative abundance of gene transcripts was determined on triplicate samples from at least 3 independent experiments using the ddCt method and is expressed as the normalized fold expression (mean ± SD). (B,D) Flow cytometric analysis of surface S1P1 (B) or S1P2 (D) on MEC B cells stably transfected with either empty vector (MEC ctr) or an expression construct encoding p66Shc (MEC p66). The data in the histograms are expressed as the mean fluorescence intensity (MFI) ± SD. Representative FACS profiles of S1P1 (B) or S1P2 (D) are shown. Specificity controls for each experiment included a sample incubated with secondary Ab alone (α-mouse) and, for S1P1 stainings, a sample preincubated with FTY720 to induce receptor down-regulation. (E) Migration of the control and p66Shc-expressing MEC transfectants measured after treatment for 3 hours with S1P (100nM). The data, obtained on duplicate samples from at least 3 independent experiments, are presented as the mean migration index ± SD (ie, the ratio of migrated cells in S1P-treated samples vs untreated samples). ***P < .001; and **P < .01.
p66Shc controls S1P1 expression in B cells. (A,C) qRT-PCR analysis of S1P1 (A) or S1P2 (C) mRNA in MEC B cells stably transfected with either empty vector (MEC ctr) or an expression construct encoding p66Shc (MEC p66). The relative abundance of gene transcripts was determined on triplicate samples from at least 3 independent experiments using the ddCt method and is expressed as the normalized fold expression (mean ± SD). (B,D) Flow cytometric analysis of surface S1P1 (B) or S1P2 (D) on MEC B cells stably transfected with either empty vector (MEC ctr) or an expression construct encoding p66Shc (MEC p66). The data in the histograms are expressed as the mean fluorescence intensity (MFI) ± SD. Representative FACS profiles of S1P1 (B) or S1P2 (D) are shown. Specificity controls for each experiment included a sample incubated with secondary Ab alone (α-mouse) and, for S1P1 stainings, a sample preincubated with FTY720 to induce receptor down-regulation. (E) Migration of the control and p66Shc-expressing MEC transfectants measured after treatment for 3 hours with S1P (100nM). The data, obtained on duplicate samples from at least 3 independent experiments, are presented as the mean migration index ± SD (ie, the ratio of migrated cells in S1P-treated samples vs untreated samples). ***P < .001; and **P < .01.
To further investigate the implication of p66Shc in S1P1 expression, we used splenic B cells from wild-type and p66Shc−/− mice. Cells were purified by immunomagnetic sorting and S1P1and S1P2 expression was measured by qRT-PCR and flow cytometry. In agreement with the results obtained on the MEC transfectants, p66Shc−/− B cells were found to be defective in S1P1 but not S1P2 expression (Figure 3A-B and supplemental Figure 2A). This defect was observed in all peripheral lymphoid tissues analyzed, including spleen, LNs, and PB (Figure 3C and supplemental Figure 2C). The variability in the S1P1 levels in B cells from wild-type mice was correlated with the levels of p66Shc (supplemental Figure 2B), further supporting the implication of p66Shc in the control of S1P1 expression. Interestingly, p66Shc deficiency did not affect S1P1 expression in T cells, as assessed both in Jurkat T cells stably transfected with either empty vector or the p66Shc-encoding construct and in splenic T cells from wild-type and p66Shc−/− mice (supplemental Figure 3A-B). Consistent with the S1P1 defect, migration toward S1P was severely impaired in p66Shc−/− B cells compared with their wild-type counterparts (Figure 3D). Moreover, p66Shc−/− mice showed a reduction in B-cell (but not T-cell) counts in PB, paralleled by an increase in LNs, spleen, and BM (supplemental Figures 3C and 4).
Defective S1P1 expression in B cells from p66Shc−/− mice. (A-B) qRT-PCR analysis of S1P1 (A) or S1P2 (B) mRNA in purified splenic B cells from wild-type (+/+) or p66Shc−/− mice. The relative abundance of gene transcripts was determined on triplicate samples from at least 6 wild-type or p66Shc−/− mice using the ddCt method and is expressed as the normalized fold expression (mean ± SD). (C) Flow cytometric analysis of surface S1P1 on splenic, LN, or PB B cells (analysis on gated CD22+ cells) from wild-type (+/+) or p66Shc−/− mice. The data are expressed as the mean fluorescence intensity (MFI) ± SD (n ≥ 9 mice/group). Specificity controls for each experiment included a sample incubated with secondary Ab alone and a sample preincubated with FTY720 to induce receptor down-regulation. ***P < .001; **P < .01; and *P < .05 by Mann-Whitney rank-sum test. (D) Migration of purified splenic B cells from wild-type (+/+) or p66Shc−/− mice, measured after treatment for 3 hours with S1P (100nM). The data, obtained on duplicate samples from at least 3 independent experiments, are presented as the mean migration index ± SD (ie, the ratio of migrated cells in S1P-treated samples vs untreated samples). ***P < .001.
Defective S1P1 expression in B cells from p66Shc−/− mice. (A-B) qRT-PCR analysis of S1P1 (A) or S1P2 (B) mRNA in purified splenic B cells from wild-type (+/+) or p66Shc−/− mice. The relative abundance of gene transcripts was determined on triplicate samples from at least 6 wild-type or p66Shc−/− mice using the ddCt method and is expressed as the normalized fold expression (mean ± SD). (C) Flow cytometric analysis of surface S1P1 on splenic, LN, or PB B cells (analysis on gated CD22+ cells) from wild-type (+/+) or p66Shc−/− mice. The data are expressed as the mean fluorescence intensity (MFI) ± SD (n ≥ 9 mice/group). Specificity controls for each experiment included a sample incubated with secondary Ab alone and a sample preincubated with FTY720 to induce receptor down-regulation. ***P < .001; **P < .01; and *P < .05 by Mann-Whitney rank-sum test. (D) Migration of purified splenic B cells from wild-type (+/+) or p66Shc−/− mice, measured after treatment for 3 hours with S1P (100nM). The data, obtained on duplicate samples from at least 3 independent experiments, are presented as the mean migration index ± SD (ie, the ratio of migrated cells in S1P-treated samples vs untreated samples). ***P < .001.
The finding that p66Shc controls S1P1 expression suggests that the S1P1 defect observed in U-CLL B cells could be caused by their deficiency in p66Shc. To address this issue, B cells purified from M-CLL and U-CLL patients were nucleofected with the p66Shc-encoding construct and the levels of S1P1 were measured by qRT-PCR. Reconstitution of p66Shc expression in CLL B cells resulted in a significant increase in the levels of S1P1 mRNA, which was confirmed by flow cytometry (Figure 4A-B). Conversely, p66Shc did not affect S1P2 expression (data not shown). Therefore, the defect in S1P1 expression in B cells from U-CLL patients is caused, at least in part, by the impairment in p66Shc expression in these cells.
p66Shc reconstitution in CLL B cells results in enhanced S1P1 expression. qRT-PCR (A) and flow cytometric (B) analysis of S1P1 expression in PB B cells purified from CLL patients with either mutated (M; n = 7) or unmutated (U; n = 9) IGHV nucleofected with either empty vector (vector ctr) or an expression construct encoding p66Shc (p66). The analysis was carried out 48 hours after transfection on GFP+ live cells. All samples were checked for reconstitution of p66Shc expression by qRT-PCR (data not shown). An immunoblot analysis of p66Shc and S1P1 expression on a representative experiment is shown on the right. The relative abundance of gene transcripts was determined on triplicate samples from each patient using the ddCt method and is expressed as the normalized fold expression (mean ± SD; empty vector controls taken as 1 for all CLL samples). The data in the histograms showing the quantification of surface S1P1 are expressed as the mean fluorescence intensity of p66-transfected cells or of vector ctr-transfected cells (normalized to 1 in all empty vector controls) ± SD. ***P < .001; **P < .01; and *P < .05.
p66Shc reconstitution in CLL B cells results in enhanced S1P1 expression. qRT-PCR (A) and flow cytometric (B) analysis of S1P1 expression in PB B cells purified from CLL patients with either mutated (M; n = 7) or unmutated (U; n = 9) IGHV nucleofected with either empty vector (vector ctr) or an expression construct encoding p66Shc (p66). The analysis was carried out 48 hours after transfection on GFP+ live cells. All samples were checked for reconstitution of p66Shc expression by qRT-PCR (data not shown). An immunoblot analysis of p66Shc and S1P1 expression on a representative experiment is shown on the right. The relative abundance of gene transcripts was determined on triplicate samples from each patient using the ddCt method and is expressed as the normalized fold expression (mean ± SD; empty vector controls taken as 1 for all CLL samples). The data in the histograms showing the quantification of surface S1P1 are expressed as the mean fluorescence intensity of p66-transfected cells or of vector ctr-transfected cells (normalized to 1 in all empty vector controls) ± SD. ***P < .001; **P < .01; and *P < .05.
Modulation of S1P1 expression by p66Shc is mediated by its pro-oxidant activity
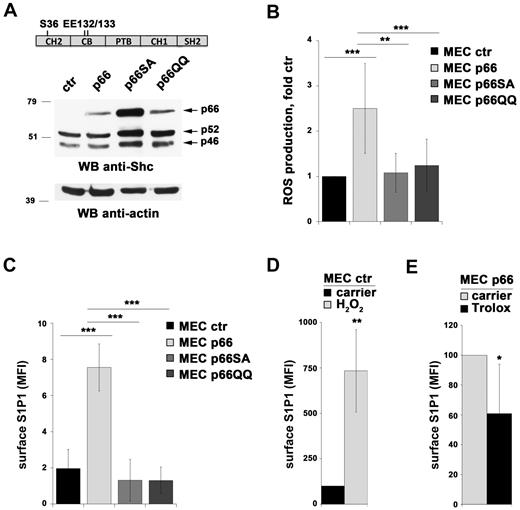
The pro-oxidant activity of p66Shc is mediated by 2 different mechanisms, reflecting its dual intracellular localization in the cytosol and mitochondria. The cytosolic pool enhances both homeostatic and stress-dependent ROS production by inhibiting the expression of ROS-scavenging enzymes through its ability to promote cytosolic translocation of FoxO transcription factors. This activity is dependent on phosphorylation of S36 in the N-terminal collagen homology domain (CH2).28 The mitochondrial pool, which is replenished from the cytosolic pool under conditions of stress,29 increases ROS production by binding cytochrome c, thereby interrupting the respiratory chain.19 This activity requires residues E132Q-E133Q within the cytochrome c–binding domain, which spans a short region adjacent to the CH2 domain (Figure 5A).
p66Shc controls S1P1 expression through its pro-oxidant activity. (A) Immunoblot analysis of Shc expression in MEC B cells stably transfected with empty vector (MEC ctr) or an expression construct encoding either wild-type p66Shc (MEC p66) or the S36A (MEC p66SA) or EE132/133QQ (MEC p66QQ) mutants. A control anti-actin blot of the stripped filter is shown below. The migration of molecular mass markers is indicated. The domain structure of p66Shc highlighting the location of the amino acid residues substituted in the mutants is schematized at the top of the panel. (B) Flow cytometric analysis of ROS production in the MEC B-cell transfectants loaded with the ROS-sensitive probe CM-H2DCFDA. The data are expressed as relative ROS production using as a reference control (empty vector) MEC cells (n = 4). (C) Flow cytometric analysis of surface S1P1 in the MEC B-cell transfectants. The data are expressed as the mean fluorescence intensity (MFI) ± SD (n = 4). Specificity controls for each S1P1 staining experiment included a sample incubated with secondary Ab alone and a sample preincubated with FTY720 to induce receptor down-regulation. (D-E) Flow cytometric analysis of S1P1 mRNA in control MEC cells treated for 24 hours with 400μM H2O2 (D) and in p66Shc-expressing MEC cells treated for 24 hours with 0.2mM Trolox (E). Under these conditions, a 3-hour treatment with H2O2 increased intracellular ROS in control (empty vector) MEC by 2-fold, whereas Trolox reduced intracellular ROS in p66Shc-expressing MEC cells by 65%. ***P < .001; **P < .01; and *P < .05.
p66Shc controls S1P1 expression through its pro-oxidant activity. (A) Immunoblot analysis of Shc expression in MEC B cells stably transfected with empty vector (MEC ctr) or an expression construct encoding either wild-type p66Shc (MEC p66) or the S36A (MEC p66SA) or EE132/133QQ (MEC p66QQ) mutants. A control anti-actin blot of the stripped filter is shown below. The migration of molecular mass markers is indicated. The domain structure of p66Shc highlighting the location of the amino acid residues substituted in the mutants is schematized at the top of the panel. (B) Flow cytometric analysis of ROS production in the MEC B-cell transfectants loaded with the ROS-sensitive probe CM-H2DCFDA. The data are expressed as relative ROS production using as a reference control (empty vector) MEC cells (n = 4). (C) Flow cytometric analysis of surface S1P1 in the MEC B-cell transfectants. The data are expressed as the mean fluorescence intensity (MFI) ± SD (n = 4). Specificity controls for each S1P1 staining experiment included a sample incubated with secondary Ab alone and a sample preincubated with FTY720 to induce receptor down-regulation. (D-E) Flow cytometric analysis of S1P1 mRNA in control MEC cells treated for 24 hours with 400μM H2O2 (D) and in p66Shc-expressing MEC cells treated for 24 hours with 0.2mM Trolox (E). Under these conditions, a 3-hour treatment with H2O2 increased intracellular ROS in control (empty vector) MEC by 2-fold, whereas Trolox reduced intracellular ROS in p66Shc-expressing MEC cells by 65%. ***P < .001; **P < .01; and *P < .05.
To address the role of the pro-oxidant function of p66Shc in the regulation of S1P1 expression, we generated stable MEC transfectants expressing either a mutant carrying a S→A substitution at position 36 (p66ShcSA) or a mutant carrying a double E→Q substitution at positions 132-133 (p66ShcQQ; Figure 5A). The empty vector transfectant lacking p66Shc and the transfectant expressing the wild-type protein were used as controls. We first measured the homeostatic ROS production in all transfectants by flow cytometric analysis of cells loaded with CM-H2DCFDA, a cell-permeant probe that becomes fluorescent after oxidation. The relative levels of ROS were found to be significantly higher in p66Shc-expressing MEC cells compared with controls. Conversely, no difference was observed in the presence of either p66ShcSA or p66ShcQQ (Figure 5B).
Expression of S1P1 was next measured in all transfectants both by qRT-PCR and by flow cytometry. Unlike wild-type p66Shc, the levels of S1P1 mRNA (supplemental Figure 5A) and surface receptor (Figure 5C) were comparable between MEC cells transfected with empty vector and the MEC transfectants expressing either the p66ShcSA or the p66ShcQQ mutant. No effect was observed on S1P2 expression (data not shown). Consistent with these findings, the chemotactic response to S1P of MEC transfectants expressing the p66Shc mutants was comparable to the empty vector transfectant (supplemental Figure 5B). These data suggest that the ability of p66Shc to modulate S1P1 expression is mediated by its ROS-elevating activity. To further assess this possibility, the control MEC transfectant lacking p66Shc was treated for 24 hours with H2O2. The p66Shc-expressing transfectant was treated for the same time with Trolox, a cell-permeable vitamin E analog with strong antioxidant activity. S1P1 expression was found to be correlated with the redox state of the cell, increasing in the presence of high levels of oxidants (Figure 5D and supplemental Figure 5C). Moreover, the increase in S1P1 expression induced by p66Shc was partly abrogated by Trolox (Figure 5E and supplemental Figure 5D), further strengthening the notion that p66Shc controls S1P1 expression by promoting ROS production. Consistent with these findings, homeostatic ROS production in CLL cells was decreased compared with normal B cells, with a statistically significant difference between M-CLL and U-CLL B cells (supplemental Figure 6).
CCR7 expression is regulated by p66Shc
Expression of the homing receptor CCR7, which responds to the chemokine CCL21 that is present at high levels in peripheral lymphoid organs, follows an opposite trend to S1P1, decreasing as the result of ligand-dependent internalization to allow trafficking to proceed in the absence of Ag recognition. In addition, CCR7 expression is shut off at the transcriptional level in activated lymphocytes that have undergone several divisions, with a concomitant increase in S1P1 expression, which allows the egress signal delivered by S1P1 to overcome the CCR7-mediated retention signal.30 CCR7 expression is enhanced in CLL B cells (Figure 6A), with a greater increase in the presence of lymphadenopathy.31
CCR7 expression is negatively regulated by p66Shc through its pro-oxidant activity. (A) qRT-PCR analysis of CCR7 mRNA in purified peripheral B cells from either healthy donors (ctr; n = 8) or CLL patients with mutated (M; n = 29) or unmutated (U; n = 28) IGHV. The relative abundance of gene transcripts was determined on triplicate samples using the ddCt method and is expressed as the normalized fold expression (mean ± SD). ***P < .001 and **P < .01 by Mann-Whitney rank-sum test. (B) qRT-PCR analysis of CCR7 mRNA in MEC B cells stably transfected with empty vector (MEC ctr) or an expression construct encoding either wild-type p66Shc (MEC p66) or the S36A (MEC p66SA) or EE132/133QQ (MEC p66QQ) mutants. **P < .01. (C) qRT-PCR analysis of CCR7 mRNA in purified splenic B cells from wild-type (+/+) or p66Shc−/− mice. The relative abundance of gene transcripts was determined on triplicate samples from at least 5 wild-type or p66Shc−/− mice. **P < .01 by Mann-Whitney rank-sum test. (D) qRT-PCR analysis of CCR7 expression in PB B cells purified from CLL patients with either mutated (M; n = 7) or unmutated (U; n = 6) IGHV nucleofected with either empty vector (vector ctr) or an expression construct encoding p66Shc (p66). The analysis was carried out 48 hours after transfection. All samples were checked for reconstitution of p66Shc expression by qRT-PCR (data not shown). The relative abundance of gene transcripts was determined on triplicate samples from each patient using the ddCt method and is expressed as the normalized fold expression (mean ± SD; empty vector controls taken as 1 for all CLL samples). ***P < .001 and *P < .05.
CCR7 expression is negatively regulated by p66Shc through its pro-oxidant activity. (A) qRT-PCR analysis of CCR7 mRNA in purified peripheral B cells from either healthy donors (ctr; n = 8) or CLL patients with mutated (M; n = 29) or unmutated (U; n = 28) IGHV. The relative abundance of gene transcripts was determined on triplicate samples using the ddCt method and is expressed as the normalized fold expression (mean ± SD). ***P < .001 and **P < .01 by Mann-Whitney rank-sum test. (B) qRT-PCR analysis of CCR7 mRNA in MEC B cells stably transfected with empty vector (MEC ctr) or an expression construct encoding either wild-type p66Shc (MEC p66) or the S36A (MEC p66SA) or EE132/133QQ (MEC p66QQ) mutants. **P < .01. (C) qRT-PCR analysis of CCR7 mRNA in purified splenic B cells from wild-type (+/+) or p66Shc−/− mice. The relative abundance of gene transcripts was determined on triplicate samples from at least 5 wild-type or p66Shc−/− mice. **P < .01 by Mann-Whitney rank-sum test. (D) qRT-PCR analysis of CCR7 expression in PB B cells purified from CLL patients with either mutated (M; n = 7) or unmutated (U; n = 6) IGHV nucleofected with either empty vector (vector ctr) or an expression construct encoding p66Shc (p66). The analysis was carried out 48 hours after transfection. All samples were checked for reconstitution of p66Shc expression by qRT-PCR (data not shown). The relative abundance of gene transcripts was determined on triplicate samples from each patient using the ddCt method and is expressed as the normalized fold expression (mean ± SD; empty vector controls taken as 1 for all CLL samples). ***P < .001 and *P < .05.
We addressed the potential role of p66Shc in the control of CCR7 expression using both the MEC B-cell transfectants and p66Shc−/− B cells. qRT-PCR analysis showed a tight inverse correlation between p66Shc and CCR7 expression (Figure 6B-C and supplemental Figure 2A). Moreover, reconstitution of p66Shc in CLL B cells resulted in a reduction in CCR7 mRNA (Figure 6D), indicating that the p66Shc defect in CLL B cells is responsible at least in part for their up-regulation of CCR7 expression.
To determine whether the impairment in CCR7 expression in the presence of p66Shc is regulated by the same mechanism as S1P1, we measured the levels of CCR7 mRNA in the MEC transfectants expressing the p66Shc mutants devoid of ROS-elevating activity. CCR7 was found to be expressed at levels comparable to control cells in the MEC transfectants expressing either the p66ShcSA or the p66ShcQQ mutant (Figure 6B). Therefore, the genes encoding CCR7 and S1P1 are controlled in opposite directions by the ROS-elevating activity of p66Shc.
Discussion
The levels of S1P receptors are tightly regulated during lymphocyte trafficking, thereby setting their dwell time in peripheral lymphoid organs. S1P receptors are rapidly down-regulated in the S1P-rich circulatory fluids by ligand-dependent internalization and are then effectively recycled to the cell surface in the S1P-poor lymphoid organs, becoming resensitized to promote lymphocyte egress.22 Modulation of the levels of S1P receptors by local S1P pools also controls the location and movements of lymphocytes within specific regions of peripheral lymphoid organs.12,24 In addition, the levels of S1P receptors are regulated by de novo gene transcription, as shown for S1P1 in activated T cells that must leave the lymphoid organs to reach the sites of infection where their effector function is required.15,32
Among the S1P receptors, S1P1 plays a crucial role in regulating the egress of T and B cells from peripheral lymphoid organs toward blood and lymph, overcoming the retention signals provided by CCR7.30 Our data highlight a defect in S1P1 expression in U-CLL B cells and confirm a previous report indicating that CLL B cells overexpress CCR7, which results in enhanced transendothelial migration toward CCL19 and CCL21.31 We hypothesize that the impairment in S1P1 expression in U-CLL B cells results in an abnormal balance between retention (CCR7-mediated) and egress (S1P1-mediated) signals that may increase their dwell time in peripheral lymphoid organs. Indeed, the levels of CCR7 were found to be significantly higher in CLL patients with clinical lymphadenopathy compared with patients lacking this alteration.31 Moreover, although no statistically significant difference in S1P1 expression was observed between M-CLL and control B cells, M-CLL patients with the lowest levels of S1P1 had a higher number of LNs involved compared with high expressers (L.T., unpublished observations, July 2012).
CLL B cells overexpress chemokine receptors such as CXCR4 and CXCR5, the ligands of which are produced at supraphysiological concentrations in the stromal microenvironment in these patients5,33 and provide survival signals that contribute to the prolonged lifespan of CLL B cells.34 An extended dwell time in peripheral lymphoid tissues is expected to favor these interactions. Moreover, a sustained interaction of the signaling-competent BCRs on U-CLL B cells9 with low-affinity self-Ags is expected to result in productive signaling, accounting for the presence of proliferating cells in these tissues.1 This scenario is supported by the results of clinical trials with the Btk inhibitor PCI-32765, which showed a sustained regression of lymphadenopathy accompanied by transient lymphocytosis35 that was also observed in a mouse model of CLL.36 The notion of the potential importance of an extended dwell time in a niche favorable to cell survival and proliferation is consistent with the finding that CCR7 regulates Eμ-Myc lymphoma homing to the LNs and spleen, providing the neoplastic CCR7high cells with a survival advantage.37 Interestingly, p66Shc−/− mice display a moderate B-cell lymphopenia that is correlated with their S1P1 expression defect in B cells. The residual levels of S1P1 in p66Shc−/− cells are likely to be sufficient to delay B-cell egress from the lymphoid organs in these mice. Together with the enhanced BCR signaling in p66Shc−/− B cells,20 an extended dwell time of these cells in the specialized niche of the lymphoid microenvironment may account for the spontaneous GC formation observed in these mice.5 Although p66Shc−/− mice do not develop lymphoid neoplasias, this might provide a selective advantage to B cells that have acquired oncogenic mutations, as was proposed for the priming effect of S1P2 deletion on human diffuse large B-cell lymphoma development.38
The outcome of the defect in S1P2 expression in both M-CLL and U-CLL B cells appears more elusive. S1P2 inhibits the response of GC B cells to follicular chemoattractants in the T-cell zone, thereby favoring their confinement to the S1P-low niche in the follicle center. S1P2 also inhibits Akt activation, thereby restricting GC B-cell survival.24 The B cell–specific S1P2 deletion in the mouse results in the development of a lymphoma with the features typical of human diffuse large B-cell lymphoma, which is preceded by spontaneous GC formation.38 CLL B cells have been shown to form pseudo-GCs both in peripheral lymphoid tissues and BM.10 Although these structures do not have the ordered architecture of true GCs, the defect in S1P2 expression might not only directly contribute to extending their survival in the presence of S1P, but also may increase their opportunities to interact with other cells that have been implicated in delivering prosurvival and/or proliferation cues within pseudo-GC, including T cells (CD40L), FDC (CD44), nurse-like cells (CXCL12, CXCL13, and BAFF), and BM stromal cells (CXCL12).5
The alterations in both S1P1 and CCR7 expression in CLL B cells appear to be causally related to the concomitant impairment in p66Shc expression, because they could be corrected, at least in part, by increasing the levels of p66Shc. This association is further supported by the results obtained on the p66Shc B-cell transfectants and B cells from p66Shc−/− mice. Conversely, p66Shc deficiency does not affect S1P1 expression in T cells. Therefore, p66Shc emerges in B cells as a new player in the coordinate regulation of these 2 key regulators of lymphocyte trafficking. Unlike other immune cell types, the factors controlling expression of S1P1 and CCR7 expression in B cells are as yet largely unknown. The results of the present study identify S1P1 and CCR7 as ROS-responsive genes in B cells and provide evidence that p66Shc participates in the control of their expression through its pro-oxidant activity. This was shown both genetically, using mutants lacking the ability to promote ROS accumulation, and by pharmacologic modulation of intracellular ROS. Quantification of intracellular ROS in B cells from patients who participated in this study showed that CLL B cells had lower levels of homeostatic ROS compared with normal B cells. This finding appears to be at variance with a previous report documenting higher ROS levels in CLL B cells.39 Nevertheless, that study showed that the levels of ROS were correlated with treatment, with levels in untreated patients actually lower than in normal B cells,39 albeit not to the same extent as in our samples. Although our study will need to be extended to a larger number of patients and healthy controls, the fact that we limited our analysis to untreated patients might account for the apparent discrepancy. An intriguing observation is that, in contrast to CCR7, which is overexpressed both in M-CLL and U-CLL B cells (with the highest levels in the latter), the defect in S1P1 can be consistently observed only in U-CLL, notwithstanding the fact that p66Shc expression and ROS production are impaired in M-CLL (albeit to a lesser extent than in U-CLL). These results suggest that CCR7 may be more sensitive to the pro-oxidant activity of p66Shc compared with S1P1.
Despite the recent advances in the characterization of B-CLL, this neoplasia remains incurable. The issue is further confounded by the highly variable clinical course of these patients, with a heterogeneous pattern of clinical features ranging from indolent to aggressive. For this reason, the identification of markers that might predict an unfavorable outcome remains a priority. Several predictive markers have been proposed, some of which are used to characterize discrete CLL subgroups, including cytogenetic abnormalities, IGHV mutational status, ectopic ZAP-70 expression, and surface CD38.3,40 Our data provide evidence of a selective impairment in S1P1 and S1P2 expression in leukemic cells from patients classified as having an unfavorable prognosis based on the lack of IGHV mutations, which suggests the interesting possibility that S1P1 and S1P2 deficiency could be used as new markers for predicting disease behavior. Prospective studies on larger cohorts of patients will be required to assess this possibility.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank John L. Telford and Paolo Bernardi for productive discussions, Jason Cyster for his generous gift of anti–mouse S1P1 and for useful suggestions, and Sonia Grassini for technical assistance.
This work was generously supported by the Italian Association for Cancer Research, Fondo per gli Investimenti della Ricerca di Base and Fondazione CaRiParo and CaRiVerona. N.C. is supported by a Fondazione Italiana per la Ricerca sul Cancro (FIRC) fellowship.
Authorship
Contribution: N.C., L.P., L.T., G.S., and C.T.B. designed the research, analyzed and interpreted the data, and wrote the manuscript; N.C., L.P., O.M.L., and E.C. performed the research; and E.M., F.F., C.G., F.F., and P.G.P. contributed vital reagents.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Corresponding author: Dr Cosima T. Baldari, Department of Evolutionary Biology, Via Aldo Moro 2, 53100 Siena, Italy; e-mail: baldari@unisi.it.
References
Author notes
N.C. and L.P. contributed equally to this work.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal