Abstract
Antiplatelet treatment is of fundamental importance in combatting functions/dysfunction of platelets in the pathogenesis of cardiovascular and inflammatory diseases. Dysfunction of anucleate platelets is likely to be completely attributable to alterations in posttranslational modifications and protein expression. We therefore examined the proteome of platelets highly purified from fresh blood donations, using elaborate protocols to ensure negligible contamination by leukocytes, erythrocytes, and plasma. Using quantitative mass spectrometry, we created the first comprehensive and quantitative human platelet proteome, comprising almost 4000 unique proteins, estimated copy numbers for ∼ 3700 of those, and assessed intersubject (4 donors) as well as intrasubject (3 different blood samples from 1 donor) variations of the proteome. For the first time, our data allow for a systematic and weighted appraisal of protein networks and pathways in human platelets, and indicate the feasibility of differential and comprehensive proteome analyses from small blood donations. Because 85% of the platelet proteome shows no variation between healthy donors, this study represents the starting point for disease-oriented platelet proteomics. In the near future, comprehensive and quantitative comparisons between normal and well-defined dysfunctional platelets, or between platelets obtained from donors at various stages of chronic cardiovascular and inflammatory diseases will be feasible.
Introduction
Platelets play a crucial role in hemostasis and in the pathologic genesis and progression of cardiovascular and inflammatory diseases. As those are the major cause of death in industrialized countries, antiplatelet treatment is recognized as a pharmacologic target of considerable importance. Patients suffering from cardiovascular or cerebrovascular disease benefit from antiplatelet treatment through reduced prevalence of recurrent—mostly fatal—events, and improved safety in case of angiographic intervention.1 In recent years, however, evidence has been accumulating that a significant subset of patients does not respond adequately to certain drugs.2-4 This insufficient responsiveness to antiplatelet treatment imposes an increased risk for severe cardiovascular or other adverse secondary events5 and might be a direct consequence of differences in protein expression patterns and/or posttranslational modification (PTM) states of individual humans. More recently, it has been increasingly recognized that platelets also have major inflammatory functions and affect innate and adaptive immunity and inflammatory diseases.6 The anucleate nature of platelets renders proteomics a valuable tool to help and better understand physiologic as well as pathophysiologic processes related to hemostasis and thrombosis. In various proteomic studies, we and others identified ∼ 2000 proteins,7-13 and more than 1000 PTM in human platelets.12,14,15
However, from qualitative data, conclusions can be inferred only to a limited extent. Deeper understanding of the individual role and contribution of signaling components to the initiation and propagation of platelet activation demands quantitative data for the protein components involved as it allows a weighted appraisal of the relevance of diverse signaling pathways in platelet activation and inhibition, which is mandatory for reconstruction and modeling of pathways. In principle, quantitative global as well as PTM-centric proteomics of reproducibly prepared platelets might allow for the assessment of differences between platelets from healthy individuals or from individual patients or patient cohorts. This appears of interest because platelets are not only a major component of hemostasis but serve as a sensitive monitor of vascular integrity especially in chronic diseases.
To assess the potential of quantitative proteomics to address these issues of utmost relevance, we conducted a systematic study determining the general biologic variance of human platelets, freshly isolated from 4 healthy donors. We identified more than 2500 phosphorylation sites, almost 4000 unique proteins and estimated copy numbers per platelet for ∼ 3700 of those. In addition, we relatively quantified ∼ 1900 proteins between 4 different donors (intersubject variation) and ∼ 1500 proteins between 3 different blood donations of the same volunteer (intrasubject variation), covering a dynamic range of 4 orders of magnitude. Our results clearly demonstrate on a global level that in both cases ∼ 85% of the platelet proteome show no variation.
We calculate a total of ∼ 20 million protein molecules per platelet, corresponding to 1.5 mg of protein/109 platelets which fits well to common laboratory values (1.8 ± 0.2 mg). Based on this and the protein coverage we achieve for human platelet-related pathways taken from the manually curated Reactome website (www.reactome.org), we estimate a coverage of 80%-85% of the entire platelet proteome, concluding that human platelets comprise approximately 5000 rather than the afore-estimated 2000-3000 proteins.16 Taken altogether, this study significantly improves our knowledge of quantitative platelet composition and biologic variance, and demonstrates that quantitative proteomics is a powerful ready-to-use tool for conducting disease-related studies of high clinical relevance,17-19 starting already with small blood donations.
Methods
Platelet isolation and purification
Human platelets were prepared as reported,20 with small modifications. The additional centrifugation step after dilution with buffer was introduced to remove leukocytes and improves sample purity substantially. Blood was obtained from healthy volunteers according to our institutional guidelines and the Declaration of Helsinki. Our studies with human platelets were approved and reconfirmed (September 24, 2008) by the local ethics committee of the University of Würzburg (studies 67/92 and 114/04).
Blood was collected in ACD solution (12mM citric acid, 15mM sodium citrate, 25mM D-glucose), apyrase, and EGTA were added to final concentrations of 0.01 U/mL and 3mM, respectively. Platelet-rich plasma (PRP) was obtained by 5-minute centrifugation at 330g. To reduce leukocyte contamination, PRP was diluted 1:1 with PBS and centrifuged at 240g for 10 minutes. Subsequently, the supernatant was centrifuged for 10 minutes at 430g and pelleted platelets were washed once in CGS buffer (120mM sodium chloride, 12.9mM trisodium citrate, 30mM D-glucose, pH 6.5). A small aliquot was resuspended in HEPES buffer (150mM sodium chloride, 5mM potassium chloride, 1mM magnesium chloride, 10mM D-glucose, 10mM HEPES, pH 7.4) to a final concentration of 3 × 108 platelets/mL for quantifying leukocyte contamination using a leukocount kit (BD Biosciences).
Sample processing for quantitative mass spectrometry
Experimental details for sample preparation/processing are given in supplemental Methods (see the Supplemental Materials link at the top of the article). To assess reproducibility of the entire workflow, proteolytic digests were controlled before LC-MS analysis.21
To assess the intersubject variance of the platelet proteome, samples from 4 donors were independently processed and individually labeled with iTRAQ on the peptide level. Thus, isobaric tags of different stable isotope compositions are attached to all primary amines (free peptide N-termini and Lys residues) which allows pooling of samples and consequently relative quantification based on specific reporter ion signal intensities in corresponding MS/MS spectra.22
Experimental details for iTRAQ labeling, peptide fractionation, TiO2 enrichment, mass spectrometry, spectrum processing, and database searching are given in the supplemental Methods. Search engine results were postprocessed using PeptideShaker software (http://peptide-shaker.googlecode.com) and peptide to spectrum matches (PSM) were exported to the PRIDE repository (http://www.ebi.ac.uk/pride/; accession numbers 22201-22203, 22206).
Protein quantification
Spectrum counting for estimation of protein copy numbers was accomplished using the normalized spectral abundance factor (NSAF)23 chosen for its robustness.24 NSAF relates the number of identified mass spectra to the length of a protein as a measure of its abundance. To estimate copy numbers of the platelet proteome, NSAF were correlated with a set of 24 reference proteins for which copy numbers were found in the literature (Table 1). Relative quantification between donors was conducted using the iTRAQ methodology. Details are given in the supplemental Methods.
Reference set of 24 platelet proteins for which determined copy numbers were found in the literature
| Uniprot accession . | Protein name . | CN/literature . | Estimated CN . | % . | References . |
|---|---|---|---|---|---|
| Actin | ACTB + ACT + ACTA | 2 000 000 | 2 186 000 | 9 | Watson and Harrison25 |
| P02675 | FIBB: fibrinogen beta chain | 100 000 | 88 900 | −11 | Fischer et al26 |
| P50552 | VASP: vasodilator-stimulated phosphoprotein | 78 900 | 44 600 | −43 | Eigenthaler et al27 |
| P08514 | ITA2B: integrin alpha-IIb | 70 000 | 83 300 | 19 | Fischer et al26 |
| P05106 | ITB3: integrin beta-3 | 70 000 | 64 200 | −8 | Fischer et al26 |
| P21926 | CD9: CD9 antigen | 49 000 | 8000 | −84 | Protty et al28 |
| P40197 | GPV: platelet glycoprotein V | 25 000 | 30 200 | 21 | Lopéz and Berndt29 |
| P07359 | GP1BA: platelet glycoprotein Ib alpha chain | 25 000 | 18 900 | −24 | Berndt et al30 |
| P13224 | GP1BB: platelet glycoprotein Ib beta chain | 25 000 | 49 000 | 96 | Berndt et al30 |
| P14770 | GPIX: platelet glycoprotein IX | 25 000 | 32 400 | 30 | Lopéz and Berndt29 |
| P06396 | GELS: gelsolin | 20 000 | 52 900 | 165 | Hartwig31 |
| P16671 | CD36: platelet glycoprotein 4 | 12 000 | 16 700 | 39 | van Schravendijk et al32 |
| P16284 | PECA1: platelet endothelial cell adhesion molecule | 10 000 | 9400 | −6 | Varga-Szabo et al33 |
| P16109 | LYAM3: P-selectin | 10 000 | 8900 | −11 | Shattil et al34 |
| P05771 | KPCB: protein kinase C beta type | 9800 | 9700 | −1 | Eigenthaler et al27 |
| P48509 | CD151: CD151 antigen | 6800 | 3000 | −56 | Protty et al28 |
| Q9HCN6 | GPVI: platelet glycoprotein VI | 3700 | 9600 | 159 | Best et al35 |
| P17301 | ITA2: integrin alpha-2 | 3000 | 4600 | 53 | Cosemans et al36 |
| P13598 | ICAM2: intercellular adhesion molecule 2 | 3000 | 3000 | 0 | Diacovo et al37 |
| P08962 | CD63: CD63 antigen | 2940 | 2200 | −25 | Protty et al28 |
| O75954 | TSN9: tetraspanin-9 | 2800 | 2200 | −21 | Protty et al28 |
| P04156 | PRIO: major prion protein | 2000 | 910 | −55 | Holada et al38 |
| P12318 | FCG2A: low-affinity immunoglobulin gamma Fc region receptor II-a | 1500 | 1000 | −33 | Kelton39 |
| P07942 | LAMB1: laminin subunit beta | 700 | 830 | 19 | Geberhiwot et al40 |
| Uniprot accession . | Protein name . | CN/literature . | Estimated CN . | % . | References . |
|---|---|---|---|---|---|
| Actin | ACTB + ACT + ACTA | 2 000 000 | 2 186 000 | 9 | Watson and Harrison25 |
| P02675 | FIBB: fibrinogen beta chain | 100 000 | 88 900 | −11 | Fischer et al26 |
| P50552 | VASP: vasodilator-stimulated phosphoprotein | 78 900 | 44 600 | −43 | Eigenthaler et al27 |
| P08514 | ITA2B: integrin alpha-IIb | 70 000 | 83 300 | 19 | Fischer et al26 |
| P05106 | ITB3: integrin beta-3 | 70 000 | 64 200 | −8 | Fischer et al26 |
| P21926 | CD9: CD9 antigen | 49 000 | 8000 | −84 | Protty et al28 |
| P40197 | GPV: platelet glycoprotein V | 25 000 | 30 200 | 21 | Lopéz and Berndt29 |
| P07359 | GP1BA: platelet glycoprotein Ib alpha chain | 25 000 | 18 900 | −24 | Berndt et al30 |
| P13224 | GP1BB: platelet glycoprotein Ib beta chain | 25 000 | 49 000 | 96 | Berndt et al30 |
| P14770 | GPIX: platelet glycoprotein IX | 25 000 | 32 400 | 30 | Lopéz and Berndt29 |
| P06396 | GELS: gelsolin | 20 000 | 52 900 | 165 | Hartwig31 |
| P16671 | CD36: platelet glycoprotein 4 | 12 000 | 16 700 | 39 | van Schravendijk et al32 |
| P16284 | PECA1: platelet endothelial cell adhesion molecule | 10 000 | 9400 | −6 | Varga-Szabo et al33 |
| P16109 | LYAM3: P-selectin | 10 000 | 8900 | −11 | Shattil et al34 |
| P05771 | KPCB: protein kinase C beta type | 9800 | 9700 | −1 | Eigenthaler et al27 |
| P48509 | CD151: CD151 antigen | 6800 | 3000 | −56 | Protty et al28 |
| Q9HCN6 | GPVI: platelet glycoprotein VI | 3700 | 9600 | 159 | Best et al35 |
| P17301 | ITA2: integrin alpha-2 | 3000 | 4600 | 53 | Cosemans et al36 |
| P13598 | ICAM2: intercellular adhesion molecule 2 | 3000 | 3000 | 0 | Diacovo et al37 |
| P08962 | CD63: CD63 antigen | 2940 | 2200 | −25 | Protty et al28 |
| O75954 | TSN9: tetraspanin-9 | 2800 | 2200 | −21 | Protty et al28 |
| P04156 | PRIO: major prion protein | 2000 | 910 | −55 | Holada et al38 |
| P12318 | FCG2A: low-affinity immunoglobulin gamma Fc region receptor II-a | 1500 | 1000 | −33 | Kelton39 |
| P07942 | LAMB1: laminin subunit beta | 700 | 830 | 19 | Geberhiwot et al40 |
CN indicates copy number; and %, deviation.
Comparison of quantitative proteomics and transcriptomics data
Results and discussion
Sample purity
In contrast to cell-culture models, for proteome analyses of primary tissues and blood components such as platelets, sample preparation and purity have to be considered thoroughly. In platelet concentrates, changes in activity and degradation can alter proteome patterns, so we decided to analyze platelets directly isolated from fresh blood donations, based on an optimized protocol guaranteeing high purity.20 As determined by the leukocount kit, all platelet samples contained less than 1 leukocyte per 106 platelets and less than 1 erythrocyte per 104 platelets.
The platelet proteome
In this study, we initially identified more than 3700 proteins in human platelets with a false discover rate (FDR) of < 1% using a multipronged approach (Figure 1). We conducted a 2-step enrichment of phosphopeptides with low and high selectivity to identify additional proteins which were not accessible to the global approach—most likely because of low abundance and/or the presence of only few unique peptides within the respective sequences. This led to the identification of 1355 and 1052 phosphorylation sites of high and lower confidence (regarding the unambiguous localization of the phosphoamino acid) from 263 additional proteins at < 1% FDR (supplemental Table 2).
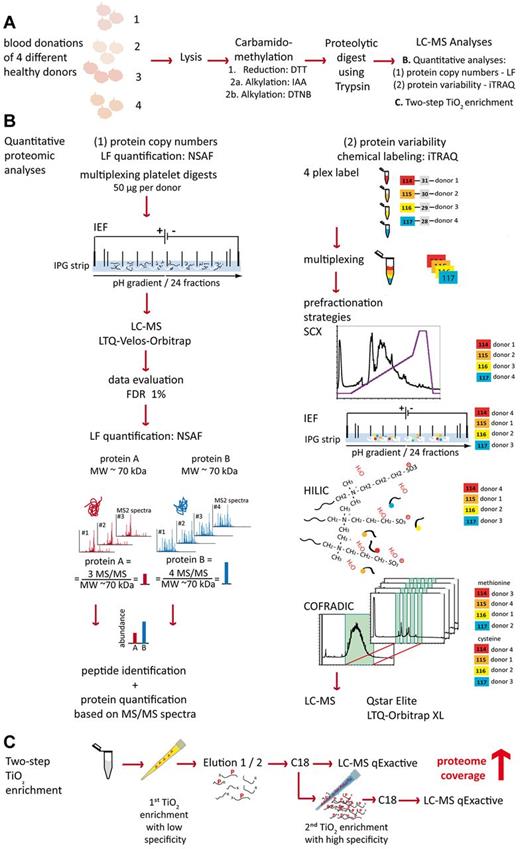
Overview of the analytical strategy. (A) Platelets were isolated from fresh blood donations and lysed, and proteins were carbamidomethylated and digested using trypsin. (B) Quantitative analysis was conducted in a 2-pronged way. (1) For absolute quantification of protein copy numbers, equal amounts of digest from the 4 donors were combined and peptides separated by in-solution isoelectric focusing. Obtained fractions were analyzed by LC-MS/MS on an LTQ Obritrap Velos and copy numbers were calculated based on the NSAF method. (2) For quantifying the biologic variance of the human platelet proteome, 100 μg of each sample were labeled with iTRAQ 114, 115, 116, and 117, respectively. Samples were multiplexed and fractionated using different techiques, namely SCX, IEF, HILIC, and COFRADIC. Obtained fractions were analyzed by LC-MS on Orbitrap XL and Qstar Elite mass spectrometers. (C) To increase the coverage of the human platelet proteome, a 2-step TiO2 enrichment, first with low and then with high specificity for phosphopeptides was conducted and samples analyzed by LC-MS/MS on a q-Exactive mass spectrometer.
Overview of the analytical strategy. (A) Platelets were isolated from fresh blood donations and lysed, and proteins were carbamidomethylated and digested using trypsin. (B) Quantitative analysis was conducted in a 2-pronged way. (1) For absolute quantification of protein copy numbers, equal amounts of digest from the 4 donors were combined and peptides separated by in-solution isoelectric focusing. Obtained fractions were analyzed by LC-MS/MS on an LTQ Obritrap Velos and copy numbers were calculated based on the NSAF method. (2) For quantifying the biologic variance of the human platelet proteome, 100 μg of each sample were labeled with iTRAQ 114, 115, 116, and 117, respectively. Samples were multiplexed and fractionated using different techiques, namely SCX, IEF, HILIC, and COFRADIC. Obtained fractions were analyzed by LC-MS on Orbitrap XL and Qstar Elite mass spectrometers. (C) To increase the coverage of the human platelet proteome, a 2-step TiO2 enrichment, first with low and then with high specificity for phosphopeptides was conducted and samples analyzed by LC-MS/MS on a q-Exactive mass spectrometer.
Estimation of protein copy numbers
Protein abundance estimation was conducted based on NSAF. Since nonunique peptides, shared between different proteins still pose a major challenge to proteomics, these might lead to deviations of estimated copy numbers for isoforms/homologs. Thus, ambiguous peptide sets derived exclusively from different protein isoforms or homologous proteins were considered as protein groups in our final proteome dataset; ambiguous peptide sets derived from nonrelated proteins were omitted. Based on the correlation to 24 reference proteins (Table 1), copy numbers for 3718 identified protein/protein groups were estimated, ranging from 2.2 106 (actin) to less than 500 copies per platelet for low abundant proteins. In general, our data correlate surprisingly well with known literature values yielding a Pearson coefficient of R2 = 0.90. Notably, these copy numbers are estimates which shall provide researchers a general overview of platelet composition—although in good accordance with the current knowledge, in individual cases, copy number estimates can significantly differ from real copy numbers. This can be attributed to general limitations of large-scale spectral counting-based quantification of cells, which comprise sample preparation (eg, tryptic digestion), mass spectrometric detection/identification, and data interpretation. (1) Only unique peptides can be unambiguously assigned to a single protein rendering protein isoforms problematic; (2) hydrophobic proteins can be depleted during sample preparation; (3) proteins with multiple transmembrane domains (TMD) generate comparatively few tryptic peptides accessible for the analysis and may be underestimated; and (4) proteins which are extensively modified (eg, glycosylated) are underrepresented because digestion might, and identification will be, hampered.
In the case of CD9, the reported 49 000 copies in the literature differ extremely from the estimated 8000 copies in our study. This discrepancy might be ascribed to the generation of only 2 fully tryptic peptides < 20 amino acids (and 3 additional ones > 25 amino acids), because of the presence of 4 TMD within only 228 amino acids.
From the obtained copy numbers and the corresponding protein molecular weights, we calculate an approximate protein concentration of 1.5 mg per 109 platelets which is in close agreement with typical laboratory reference values (1.8 ± 0.2 mg). The final list of identified proteins and copy numbers is given in supplemental Table 3.
Comparison to transcriptome data
To compare our proteome to transcriptome and SAGE RNA data, we generated a merged dataset comprising 9505 entries, 8070 of which referring to individual proteins (3598 from proteomics, 6202 from RNAseq, and 2737 from SAGE). The overlap between all 3 datasets is considerably low with 8%, whereas 12%, 38%, and 9% of proteins were exclusively found in proteome, RNAseq, and SAGE data, respectively. From the 20th percentile of the proteins quantified by RNAseq, 71% are shared with proteome data, whereas from the 20th percentile of the MS-derived proteins, 85% are shared with RNAseq data. However, for the most frequent proteins/transcripts, no correlation of RPKM and protein copy number can be observed (R2 < 0.1).
Purity of all datasets was evaluated based on 68 established CD markers (Table 2) selected from UniProt (http://www.uniprot.org/docs/cdlist), HPRD (http://www.hprd.org/), and the BD Biosciences CD marker handbook, of which 28 were shown to be expressed in platelets. Indeed, all of these except for CD23, and also 2 established nonplatelet proteins, CD81 and CD97, were identified in our proteome. CD37, present in ∼ 110 000 copies per leukocyte,46 is also present in platelets (own FACS data). It was also identified by Dowal et al in their recent analysis of platelet palmitoylation,13 and is estimated to ∼ 790 copies per platelet based on our MS data. In comparison, RNAseq data contain 15 nonplatelet CD marker transcripts, whereas in the SAGE data 5 nonplatelet markers were found, yet at significantly higher rates than in the RNAseq data.
Overview of CD expression
| CD . | Cell . | Platelet . | RNA . | SAGE . | MS copy numbers . |
|---|---|---|---|---|---|
| CD19 | B cell | − | + | − | − |
| CD20 | B cell | − | + | + | − |
| CD146 | Endothelial cell | − | − | − | − |
| CD35 | Erythrocyte | − | − | − | − |
| CD44 | Erythrocyte | − | + | − | − |
| CD66b | Granulocyte | − | − | − | − |
| CD49a | Monocyte | − | − | − | − |
| CD117 | Stem cell | − | − | − | − |
| CD243 | Stem cell | − | − | − | − |
| CD33 | Stem cell | − | − | − | − |
| CD34 | Stem cell | − | − | − | − |
| CD90 | Stem cell | − | − | − | − |
| CD133 | Stem cell | − | − | − | − |
| CD135 | Stem cell | − | − | − | − |
| CD4 | T cell | − | − | − | − |
| CD8a | T cell | − | + | − | − |
| CD8b | T cell | − | − | − | − |
| CD81 | T cell | − | + | + | 1000 |
| CD97 | Granulocyte | − | + | + | 580 |
| CD37 | B cell | + | + | − | 790 |
| CD93 | Neutrophils | + | + | − | 750 |
| CD92 | Platelet | + | + | + | 3300 |
| CD41 | Platelet | + | + | + | 83 300 |
| CD61 | Platelet | + | + | + | 64 200 |
| CD62P | Platelet | + | + | − | 8900 |
| CD9 | Platelet | + | + | + | 8000 |
| CD23 | Platelet | + | − | − | − |
| CD31 | Platelet | + | + | − | 9400 |
| CD36 | Platelet | + | + | + | 16 700 |
| CD42a | Platelet | + | + | + | 32 400 |
| CD42b | Platelet | + | − | − | 18 900 |
| CD42c | Platelet | + | + | + | 49 000 |
| CD42d | Platelet | + | + | − | 30 200 |
| CD49b | Platelet | + | + | − | 4600 |
| CD49f | Platelet | + | + | − | 11 500 |
| CD51 | Platelet | + | − | − | 1400 |
| CD84 | Platelet | + | + | − | 2900 |
| CD109 | Platelet | + | − | − | 1700 |
| CD110 | Platelet | + | + | + | 1600 |
| CD147 | Platelet | + | + | − | 2900 |
| CD151 | Platelet | + | + | + | 3000 |
| CD226 | Platelet | + | + | − | 2800 |
| CD107a | Platelet (activated) | + | + | − | 3000 |
| CD107b | Platelet (activated) | + | + | + | 2000 |
| CD63 | Platelet (activated) | + | − | + | 2200 |
| CD111 | Stem cell | + | − | − | 570 |
| CD112 | Stem cell | + | + | − | 750 |
| CD11c | Dendritic cell | o | + | − | − |
| CD123 | Dendritic cell | o | − | − | − |
| CD62E | Endothelial cell | o | − | + | − |
| CD236 | Epithelial cell | o | + | − | − |
| CD235a | Erythrocyte | o | − | − | − |
| CD238 | Erythrocyte | o | − | − | − |
| CD247 | Lymphocyte | o | + | − | − |
| CD62L | Lymphocyte | o | + | − | − |
| CD131 | Megakaryocyte | o | + | − | − |
| CD203c | Megakaryocyte | o | − | − | − |
| CD14 | Monocyte | o | + | − | − |
| CD56 | NK cell | o | − | − | − |
| CD228 | Stem cell | o | − | − | − |
| CD280 | Stem cell | o | − | + | − |
| CD338 | Stem cell | o | − | − | − |
| CD277 | Stem cell | o | + | − | 670 |
| CD318 | Stem cell | o | − | − | − |
| CD227 | Stem cell | o | − | − | − |
| CD3d | T cell | o | + | − | − |
| CD3e | T cell | o | + | − | − |
| CD3g | T cell | o | − | − | − |
| CD . | Cell . | Platelet . | RNA . | SAGE . | MS copy numbers . |
|---|---|---|---|---|---|
| CD19 | B cell | − | + | − | − |
| CD20 | B cell | − | + | + | − |
| CD146 | Endothelial cell | − | − | − | − |
| CD35 | Erythrocyte | − | − | − | − |
| CD44 | Erythrocyte | − | + | − | − |
| CD66b | Granulocyte | − | − | − | − |
| CD49a | Monocyte | − | − | − | − |
| CD117 | Stem cell | − | − | − | − |
| CD243 | Stem cell | − | − | − | − |
| CD33 | Stem cell | − | − | − | − |
| CD34 | Stem cell | − | − | − | − |
| CD90 | Stem cell | − | − | − | − |
| CD133 | Stem cell | − | − | − | − |
| CD135 | Stem cell | − | − | − | − |
| CD4 | T cell | − | − | − | − |
| CD8a | T cell | − | + | − | − |
| CD8b | T cell | − | − | − | − |
| CD81 | T cell | − | + | + | 1000 |
| CD97 | Granulocyte | − | + | + | 580 |
| CD37 | B cell | + | + | − | 790 |
| CD93 | Neutrophils | + | + | − | 750 |
| CD92 | Platelet | + | + | + | 3300 |
| CD41 | Platelet | + | + | + | 83 300 |
| CD61 | Platelet | + | + | + | 64 200 |
| CD62P | Platelet | + | + | − | 8900 |
| CD9 | Platelet | + | + | + | 8000 |
| CD23 | Platelet | + | − | − | − |
| CD31 | Platelet | + | + | − | 9400 |
| CD36 | Platelet | + | + | + | 16 700 |
| CD42a | Platelet | + | + | + | 32 400 |
| CD42b | Platelet | + | − | − | 18 900 |
| CD42c | Platelet | + | + | + | 49 000 |
| CD42d | Platelet | + | + | − | 30 200 |
| CD49b | Platelet | + | + | − | 4600 |
| CD49f | Platelet | + | + | − | 11 500 |
| CD51 | Platelet | + | − | − | 1400 |
| CD84 | Platelet | + | + | − | 2900 |
| CD109 | Platelet | + | − | − | 1700 |
| CD110 | Platelet | + | + | + | 1600 |
| CD147 | Platelet | + | + | − | 2900 |
| CD151 | Platelet | + | + | + | 3000 |
| CD226 | Platelet | + | + | − | 2800 |
| CD107a | Platelet (activated) | + | + | − | 3000 |
| CD107b | Platelet (activated) | + | + | + | 2000 |
| CD63 | Platelet (activated) | + | − | + | 2200 |
| CD111 | Stem cell | + | − | − | 570 |
| CD112 | Stem cell | + | + | − | 750 |
| CD11c | Dendritic cell | o | + | − | − |
| CD123 | Dendritic cell | o | − | − | − |
| CD62E | Endothelial cell | o | − | + | − |
| CD236 | Epithelial cell | o | + | − | − |
| CD235a | Erythrocyte | o | − | − | − |
| CD238 | Erythrocyte | o | − | − | − |
| CD247 | Lymphocyte | o | + | − | − |
| CD62L | Lymphocyte | o | + | − | − |
| CD131 | Megakaryocyte | o | + | − | − |
| CD203c | Megakaryocyte | o | − | − | − |
| CD14 | Monocyte | o | + | − | − |
| CD56 | NK cell | o | − | − | − |
| CD228 | Stem cell | o | − | − | − |
| CD280 | Stem cell | o | − | + | − |
| CD338 | Stem cell | o | − | − | − |
| CD277 | Stem cell | o | + | − | 670 |
| CD318 | Stem cell | o | − | − | − |
| CD227 | Stem cell | o | − | − | − |
| CD3d | T cell | o | + | − | − |
| CD3e | T cell | o | + | − | − |
| CD3g | T cell | o | − | − | − |
Platelet refers to the literature and our own data. Estimated copy numbers per platelet are given.
SAGE indicates presence in SAGE data; MS, presence in MS data; and o, not tested.
Protein stoichiometry
Platelet interaction with subendothelial layer matrix proteins and prothrombotic plasma components is mediated by heterooligomers of glycoproteins and particularly integrins of specific stoichiometry. The integrin heterodimer α2bβ3 is the most relevant integrin for platelet aggregation, acting as fibrinogen receptor. The 2 subunits are found almost equally expressed in the proteome (83 000 and 64 200 copies) and also in the transcriptome (1357 and 1223 RPKM in RNAseq). The subunits of the von Willebrand factor receptor GPIX/Ibα/Ibβ/V are found in stoichiometric ratios of 1.00:0.58:1.51:0.93 in the proteome, while GPIbα is missing in the transcriptome data and GPV is only detected in trace amounts. Collagen interacts with platelets via the integrin α2β1, GPIV, or GPVI/Fc receptor γ complex. Glycoprotein GPIV is the most abundant of these followed by GPVI/FcRg (1.00:0.85 stoichiometry), and α2β1 (2:1 stoichiometry).
In quantitative proteomics, the Iα (P10644), Iβ (P31321), IIα (P13861), and IIβ (P31323) regulatory and α (P17612) and β (P22694) catalytic subunit isoforms of PKA were found. The regulatory to catalytic subunit stoichiometry as derived from proteomics data with 1:0.82 is close to the expected 1:1 ratio and with ∼ 9600 copies of catalytic subunit twice as much as published.27 In contrast, RNAseq data are clearly deviating, not reflecting the expected stoichiometry.
Pathway analysis
We analyzed the coverage of known platelet pathways for G-protein, integrin, calcium, and cyclic nucleotide signaling. Because of the central role of G-protein signaling in platelet regulation, we analyzed proteome, RNAseq, and SAGE data with respect to the presence of G-protein–coupled receptors (GPCR). Whereas the platelet transcriptome contains 20 GPCR (19 in RNAseq and 4 in SAGE), our initial proteome dataset comprised only 4: ADA2A, PAR4, P2Y12, and V1AR, with an additional 7 GPCR present in our previous membrane study (included in the final dataset supplemental Table 3). This discrepancy between the global and the membrane-dedicated study can be mainly attributed to the combination of (1) low abundance, (2) strong hydrophobicity (7 TMD), and (3) sample complexity, impeding generation as well as detection of proteotypic peptides in case of global studies. Amisten et al evaluated the gene expression profile of platelet GPCR47 with PAR1 (292nd highest abundance in our platelet membrane proteome) and P2Y12 (296th) receptors being the prevalent GPCR followed by SUCR1 (not present), P2Y1 (482nd), and LPA (530th) receptors, in agreement with our previous results11 as well as RNAseq data. For G-protein α subunits, the inhibitory Gα subunits (Giα1-3, GαZ) account for 61%, followed by GαQ with 24%, Gα12 with 10%, and GαS with 5% of all subunit copies. The corresponding Gβ and Gγ subunits are almost equal (34 400 vs 42 300 copies) yet less expressed than Gα subunits. In contrast, in RNAseq data, GαS is found almost exclusively (82%) and in SAGE data Gα13 and Gαi2 together account for 72% of all Gα subunit transcripts.
A steep increase of intracellular calcium constitutes the initial and essential signal for platelet activation.48 Two major mechanisms contribute to the fast increase: calcium entry through the nonselective P2X1 ion channel and mobilization of calcium from intracellular stores.49 P2X1 is the only ligand-gated calcium channel identified in the proteome and the transcriptome and found at a moderate copy number (1400). Calcium mobilization occurs through IP3 receptors (inositol 1,4,5-trisphosphate receptor type 1, 2, and 3 with 2400, 1700, 750 copies; IRAG with 3500 copies). IP3 is formed from phosphatidylinositolbisphosphate by phospholipase C which is expressed in several subtypes and isoforms (PLCB2, PLCB3, PLCB4, PLCG2: 2500, 1700, 1000, 2000 copies). The secondary calcium influx can be achieved by 2 major pathways which have been proposed for platelets: the calcium sensor/calcium release activated channel (STIM1/CRAC) and transient receptor potential channels (TRPC).50,51 Apparently, both pathways are present in human platelets (STIM1: 7400, CRACM1: 1700, TRPC6: 1100 copies). However, proposed interaction partners of STIM1 such as TRPC1 and IPLA252 or other TRPC were detected neither in the platelet proteome nor the transcriptome.
For the resting state of platelets, maintaining low intracellular calcium levels is fundamental. While plasma membrane calcium transporters are only present in low copy numbers (CaATPases PMCA4: 640, and ATPase 2C1: 2200, Na+/Ca2+ Exchanger SLC8A3: 580), mitochondrial calcium transporter (MCU, MICU1, 5900, 1400) and particularly ER/SR CaATPases (SERCA2 and 3 with 9000 and 16 300), which also maintain the filling state of Ca stores, are found in exceedingly high quantity. These numbers underline the outstanding importance of intracellular calcium stores for platelet calcium regulation.53
The major endogenous platelet inhibitory pathways are based on cyclic nucleotide regulation. cAMP is formed by adenylyl cyclases which are anchored in the cell membrane by two 6-TMD each. In our proteome data, only adenylyl cyclase 6 (ADCY6) is present, while in our membrane proteome and in the RNAseq data, AC3 and AC5 are present as well. As expected from previous studies from us and others, only soluble guanylyl cyclase could be detected in platelets: in all datasets, solely GC α3 (Q02108) and ββ1 (Q02153) subunits are present, with quantitative proteomics indicating a 1:1 stoichiometry (3500 and 3700 copies). Cyclic nucleotides are degraded by 3′,5′ cyclic phosphodiesterases; in our current understanding, platelets predominantly express 3 phosphodiesterases: PDE2A, PDE3A, and PDE5A, with PDE5A being the most relevant. Deducing PDE stoichiometries from different data sources results in similar ratios: (1) 1.00:0.17:0.04 for PDE5:PDE3:PDE2 from Western blot,54 (2) 1.00:0.07:0.04 from RNAseq, and (3) 1.00:0.12:(n/a) from quantitative proteomics, with PDE2 only detected after phosphopeptide enrichment, most likely because of low abundance. Combining quantitative proteomics and Western blot data < 300 copies per platelet can be estimated. The major effectors of cyclic nucleotides are the cyclic nucleotide-dependent protein kinases PKA and PKG. Of the PKG subtypes, only PKGI is consistently found in all datasets, yet proteome data indicate a 6-fold lower expression (3500 copies) than literature data.27
Generally, the comparison with transcriptome data clearly indicates that platelet transcripts do not reliably reflect protein expression. The lack of correlation with copy numbers, even for highly expressed proteins, and the rather unlikely frequency distribution of transcriptome data suggest that in platelets, the occurrence of proteins is not interrelated to the presence of transcripts.
Global analysis of the platelet proteome and comparison to quantitative proteome data from human cell lines
To further assess the general quality/coverage of the present dataset on a global scale and thus evaluate whether it is in accordance with current knowledge, we analyzed our ∼ 4200 platelet proteins with regard to enriched GO terms and coverage of platelet-related pathways in the Reactome database. Although validity and integrity of the underlying protein-interaction and function databases are a matter of debate, the obtained information can nevertheless help to appraise the quality and reasonableness of proteomic data and furthermore allows for comparing large-scale proteomic data.
Compared with the human proteome (Uniprot human, ∼ 18 000 entries), our platelet proteome is highly enriched in GO terms which can be related to platelet function, such as “endomembrane system,” “actin cytoskeleton,” “platelet α granule,” “platelet dense granule,” “intracellular signal transduction,” or “filopodium.”
Among the 10 most significantly enriched pathways were “platelet activation,” “hemostasis,” “response to elevated platelet cytosolic Ca2+,” “membrane trafficking,” and “platelet degranulation.” Surprisingly, this analysis also suggested an important role for ubiquitination in platelet regulation.
As all the aforementioned data confirm a highly pure and comprehensive analysis of the platelet proteome, we analyzed several platelet-specific human pathways in the Reactome database more in detail, namely “GPVI signaling” (stable identifier React_1695.3), “GP1b-IX-V signaling” (React_23847.2), “platelet homeostasis” (React_23876.2) and “thrombin signaling through PAR” (React_21384.2; all summarized in supplemental Table 4). However, here one of the limitations of global proteome analyses (that, often, isoform resolution and differentiation of highly homolog proteins is not possible because of the absence of sufficient unique peptide sequences) has to be considered. Hence, some small G proteins or PP2A and PDE subunits are missing; we therefore considered these as protein groups as usually several but not all of these members/subunits were unambiguously detected (in analogy to our final protein list). Thus, we achieve 97% (32 of 33 proteins) coverage of the Reactome “GPVI signaling” pathway, 90% (9 of 10) coverage of “GP1b-IX-V signaling,” 90% (18 of 20) coverage of “platelet homeostasis,” and 89% (8 of 9) coverage of “thrombin signaling through PAR.” Concluding, we estimate a coverage of 80%-85% of the complete platelet proteome, even when taking into account missing isoforms.
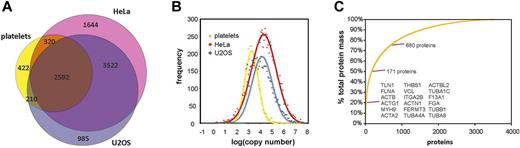
To reveal discrepancy or congruency of protein expression and copy numbers between proteomes of human platelets and human cell lines, we aligned our data to quantitative proteome data from HeLa43 and U2OS44 cells. The combined datasets encompass a total of 9696 individual proteins (disregarding splice variants). The overlap is rather complete with only 4% of the proteins exclusively found in platelets, 69% present at least in 2 sets and 27% in all sets (Figure 2). Thirty-two percent of the platelet proteome overlaps with both sets. The overlap of the highly expressed proteins (95th percentile) of each dataset (platelets, HeLa, U2OS) is clearly higher. Fifty-eight percent of the 671 proteins in this set are present in all 3 cells and 88% in at least 2 cells. A subset of 28 proteins (4%) is exclusively present in platelets and is mainly composed of γ-actin, glycoproteins, and platelet and coagulation factors (supplemental Table 5A).
Comparison of the platelet proteome to proteomes of human HeLa and U2OS cells, encompassing a total of 9696 proteins. (A) All 3 proteomes show a remarkable overlap, which is virtually independent from copy numbers. (B) The distribution of copy numbers is similar for the 3 proteomes. However, because of the smaller cell volume in platelets the mean of the frequency distribution is shifted to lower copy numbers which is in accordance with Lundberg et al who reported a correlation between cell size and protein expression levels.55 (C) Contribution of proteins to the total protein mass in the platelet proteome: only 18 proteins account for 20%, 171 proteins for 50%, and 680 proteins for 75% of the total protein mass in human platelets.
Comparison of the platelet proteome to proteomes of human HeLa and U2OS cells, encompassing a total of 9696 proteins. (A) All 3 proteomes show a remarkable overlap, which is virtually independent from copy numbers. (B) The distribution of copy numbers is similar for the 3 proteomes. However, because of the smaller cell volume in platelets the mean of the frequency distribution is shifted to lower copy numbers which is in accordance with Lundberg et al who reported a correlation between cell size and protein expression levels.55 (C) Contribution of proteins to the total protein mass in the platelet proteome: only 18 proteins account for 20%, 171 proteins for 50%, and 680 proteins for 75% of the total protein mass in human platelets.
According to Engelhard et al,56 the coverage of the total protein mass for the HeLa proteome is ∼ 51%, compared with 80% for the platelet proteome, whereas for U2OS data an appropriate analysis was not possible because of the cutoff thresholds in the data. The frequency distribution of logarithmized copy numbers indicates almost identical maximum and full width at half maximum values for HeLa (μ = 4.4, σ = 1.03) and U2OS (μ = 4.2, σ = 0.85). In contrast, the maximum of the frequency distribution of the platelet proteome is shifted to lower copy numbers and is narrowed (μ = 3.16, σ = 0.55) reflecting the lower protein expression levels and a limited protein inventory (Figure 2). Protein copy numbers of the different cells do neither correlate for the complete proteome nor subsets based on the stratified data. The ratio of copy numbers for the platelet proteome in relation to the HeLa proteome is mainly between 0.02 and 0.1, in relation to the U2OS proteome between 0.05 and 0.5 (supplemental Table 5B). The distribution of protein sizes in the 3 datasets is almost identical to the total human proteome as provided by Swissprot, demonstrating the absence of any bias for protein size in mass spectrometry analysis.
In summary, the qualitative comparison of the human platelet proteome to HeLa and U2OS proteomes indicates considerable similarities which are virtually independent from copy numbers, while the distribution of copy numbers is similar for the 3 cell types. However, for platelets the smaller cell volume shifts the mean of the frequency distribution to lower copy numbers, whereas highly expressed proteins exclusively found in the platelet proteome are typical platelet proteins.
Potential contamination with other blood components
Because estimated copy numbers and stoichiometries in this study substantially reflect and extend present knowledge, we addressed the important issue of contamination with other blood components. In particular, the sponge-like structure of platelets57 leads to an extensive perfusion of the open canalicular system with plasma components. Consequently, a complete removal of plasma components is almost unattainable and it remains questionable to which extent the respective plasma proteins indeed are an actual and vital part of the platelet proteome. We therefore compared our dataset with the so-far most comprehensive plasma proteome study providing concentration estimates for a large set of plasma proteins.58 Indeed, 149 of the 150 most abundant plasma components are also present in our dataset (supplemental Table 3), with albumin estimated to ∼ 53 000 copies per platelet. The established concentration of albumin in plasma (50 mg/mL) corresponds to the occurrence of ∼ 450 000 copies per fL. Considering the mean platelet volume of 9.7 ± 0.5 fL, we conclude that our samples contained approximately 1% by volume (0.1 fL) plasma per platelet. From the occurrence of the second most abundant plasma protein transthyretin (0.77 mg/mL corresponding to 29 000 copies per fL) at 4500 copies per platelet, almost the same share, namely 1.5% by volume (0.15 fL) plasma per platelet can be deduced.
Considering that a significant amount of hemoglobin can be ascribed to marginal hemolysis during platelet preparation a relevant contamination by erythrocytes can almost be ruled out. The absence of one of the most abundant erythrocyte proteins, the band 3 anion transporter (1 000 000 copies per erythrocyte),59 underlines this assumption. However, even if the detected hemoglobin (280 000 000 copies per erythrocyte) resulted from erythrocytes, the estimated 61 000 copies would account for less than 1 erythrocyte per 5000 platelets.
Platelet proteome variation
For the 1900 proteins relatively quantified between 4 different donors and covering a dynamic range of 4 orders of magnitude, we calculated SD over the obtained ratios, with the resulting median SD over all quantified proteins amounting to 0.14. To account for (1) technical variations during sample processing and (2) MS analysis, we classified all proteins with SD 2 times higher than the median SD as potentially differential. Although this is significantly lower than the usually expected biologic variance between humans (≫2×), 85% of the quantified proteins are below this cutoff and therefore show no/almost no variation between the analyzed donors.
Whereas individual deviations increase with lower copy numbers, there is no clear tendency toward variation for low abundant proteins; one-fourth of all proteins and one-fourth of the proteins with SD > 0.28 have less than 1000 copies per platelet.
To assess the impact of a single outlier ratio (ie, 1 donor), for those proteins with SD > 0.28, we determined SD3patients values when omitting the most deviating ratio and set the SD3patients cutoff to 0.14 (1× median SD). Then, still 171 proteins (9%) show potential variation.
The group of differential proteins comprises potential/known contaminants such as hemoglobin, MHC molecules, and apolipoproteins, most likely because of slight differences during platelet isolation. In principle, for individual proteins, a differing degree of PTM as well as single amino acid exchanges might simulate a variation in protein abundance, caused by the reduced recovery of respective peptide sequences.
In contrast, proteins central to platelet function have low SD which predominantly reflect the technical error of iTRAQ-based protein quantification (∼ 20%): GPIbα 0.05; GPIbβ 0.11; GPIV 0.16; GPVI 0.24; GPIX 0.10; PAR4 0.25; platelet basic protein 0.09; platelet factor 4 0.13; STIM1 0.07; P2X1 0.17; VASP 0.11; LASP 0.10; PKA ∼ 0.20; PKG 0.10; actin ∼ 0.10.
In principle, these results agree with Winkler et al who compared platelets from healthy donors using the difference in gel electrophoresis (DIGE) strategy and determined a coefficient of variation (CV) of 18% based on the relative quantification of 500 reproducibly found spots.60 However, 2-dimensional gel spots can easily contain up to 10 or more proteins, rendering accurate and global quantification on the protein level challenging. In contrast, in this study, 1900 proteins were relatively quantified by MS (CV of 13%) yielding a substantial increase of quantitative information.
In addition, we assessed the intradonor variation by quantitatively analyzing platelet samples purified from 3 different blood donations of the same donor. We independently processed 2 technical replicates per sample and analyzed all 6 samples without prefractionation by LC-MS. Since omitting the prefractionation reduced the number of identified spectra per protein, the obtained NSAF values were used only for relative comparison. From the 1505 proteins which were identified in all samples with high confidence (CV 12.2%), ∼ 85% showed no variation (CV > 24.4%). Furthermore, technical variances determined using the 2 replicates per sample were 12.7%, 14.8%, and 13.8%, respectively (supplemental Table 6). Only 7 proteins were considered as regulated in both intrasubject and intersubject comparisons, namely DDX3X, SAMM50, UQCRH, GLTP, EIF5A, BCL2L1, and ACAA1, of which 3 are of mitochondrial origin.
These results confirm that platelet formation as membrane-encapsulated fragments of megakaryocytes is a highly reproducible and regulated process.
Conclusion
We conducted a systematic and thorough proteomic characterization of highly pure human platelets (1) identifying ∼ 4000 platelet proteins, (2) estimating copy numbers per platelet for ∼ 3700 of those, relatively quantifying expression levels of (3) 1900 proteins between 4 healthy donors, and (4) 1500 proteins between 3 different blood samples of the same donor. Based on leukocount and quantitative proteome data, we conclude that the contamination with other blood components is negligible: for example, 1 leukocyte per 106 platelets and 1.0%-1.5% by volume plasma per platelet, which can be mainly attributed to the sponge-like structure of the open canalicular system.
To estimate copy numbers per platelet for the identified proteins, we used the established NSAF approach which we previously demonstrated as a robust method for quantifying proteins in complex mixtures. We related the obtained NSAF values to a set of copy numbers found in the literature and found a surprisingly high degree of correlation (R2 = 0.90). Although absolute quantification based on spectral counting can suffer from errors up to 200% in individual cases (membrane proteins or proteins with an unusually frequent occurrence of PTM), known protein complex stoichiometries can be confirmed and unknown deduced.
To this end, our study provides a first comprehensive insight into the qualitative and quantitative platelet composition, allowing for a weighted appraisal of the physiologic relevance of distinct pathways and thus represents a rich source for the systematic search for novel platelet functions and mechanisms.
In addition, our quantitative proteomic comparisons between 4 healthy donors as well as 3 different blood samples from the same donor demonstrate that, despite the dynamics of the proteome and the high biologic variance between human beings, platelet formation from megakaryocytes is a highly regulated and reproducible process yielding only minor differences in protein expression patterns. As we demonstrate, thorough and monitored isolation of platelets from fresh blood donations, combined with quality controlled sample preparation and proteomic analysis, yields highly reproducible data. Thus, results and analytical strategy of the present study build a foundation for future investigations which can directly address issues of utmost clinical relevance such as differences in the response to antiplatelet treatment or in the pathologic potential for the genesis of cardiovascular diseases. Since between healthy donors > 85% of the proteome show no variation, by combining an elaborate quantitative analytical strategy and the appropriate individual patient and/or patient cohort (and size) with the right scientific questions, novel biomarkers, drug targets or key mediators of platelet function can be identified starting from small blood donations. In principle, already less than 100 μg of protein per donor (∼ 4 × 107 platelets, a few mL of blood) will be sufficient indicating that platelet proteome analyses of individual donors and/or patients is possible.
The quantitative results presented in our manuscript demonstrate the feasibility to assess on a global scale quantitative protein differences between (1) normal and well-defined dysfunctional platelets from individual persons, (2) platelets with normal and pathologic response to antiplatelet drugs, and (3) platelets obtained from patients at various stages of chronic cardiovascular and inflammatory diseases. Some such ambitious studies are currently initiated.
There is an Inside Blood commentary on this article in this issue.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Stefanie Wortelkamp and Claudia Schütz for excellent technical assistance.
This work was supported by the Ministerium für Innovation, Wissenschaft und Forschung des Landes Nordrhein-Westfalen, by grants of the “Bundesministerium für Bildung und Forschung (MedSys Project SARA, 31P5800), and the SFB688/TPA2.
J.M.B. and M.V. are PhD candidates at Universities of Dortmund, Germany and Ghent, Belgium and this work is submitted in partial fulfillment of the requirement of the PhD.
Authorship
Contribution: J.M.B. designed and performed research, analyzed the data, and wrote the manuscript; M.V. developed the data interpretation pipeline, analyzed the data, and wrote the manuscript; S.G. prepared and quality controlled platelet samples, and contributed to study design and the writing of the manuscript; S.R. conducted experiments and contributed to the writing of the manuscript; U.W. contributed to study design and the writing of the manuscript; L.M. contributed to data analysis; J.G. analyzed data, and contributed to study design and the writing of the manuscript; A.S. contributed to study design and the writing of the manuscript; and R.P.Z. conceived of the study, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr René Peiman Zahedi, Leibniz–Institut für Analytische Wissenschaften-ISAS-e.V., Otto-Hahn-Str 6b, D-44227 Dortmund, Germany; e-mail: rene.zahedi@isas.de or Dr Jörg Geiger, Institut für Klinische Biochemie und Pathobiochemie, Universitätsklinikum Würzburg, Grombühlstr 12, D-97080 Würzburg, Germany; e-mail: j.geiger@klin-biochem.uni-wuerzburg.de.
References
Author notes
J.M.B. and M.V. contributed equally.