Abstract
The outcome of older (≥ 60 years) acute myeloid leukemia (AML) patients is poor, and novel treatments are needed. In a phase 2 trial for older AML patients, low-dose (20 mg/m2 per day for 10 days) decitabine, a DNA hypomethylating azanucleoside, produced 47% complete response rate with an excellent toxicity profile. To assess the genome-wide activity of decitabine, we profiled pretreatment and post treatment (day 25/course 1) methylomes of marrow samples from patients (n = 16) participating in the trial using deep-sequencing analysis of methylated DNA captured by methyl-binding protein (MBD2). Decitabine significantly reduced global methylation compared with pretreatment baseline (P = .001). Percent marrow blasts did not correlate with global methylation levels, suggesting that hypomethylation was related to the activity of decitabine rather than to a mere decrease in leukemia burden. Hypomethylation occurred predominantly in CpG islands and CpG island-associated regions (P ranged from .03 to .04) A significant concentration (P < .001) of the hypomehtylated CpG islands was found in chromosome subtelomeric regions, suggesting a differential activity of decitabine in distinct chromosome regions. Hypermethylation occurred much less frequently than hypomethylation and was associated with low CpG content regions. Decitabine-related methylation changes were concordant with those previously reported in distinct genes. In summary, our study supports the feasibility of methylome analyses as a pharmacodynamic endpoint for hypomethylating therapies.
Introduction
Acute myeloid leukemia (AML) is a heterogeneous malignant disease characterized by the accumulation of clonal, undifferentiated hematopoietic cells in BM and blood. Despite progress made in the identification of cytogenetic and molecular genetic aberrations that aid in risk stratification and the understanding of mechanisms of leukemogenesis, the majority of adult patients with AML are not cured when treated with conventional chemotherapy.1,2 Thus, novel therapeutic targets and approaches are needed to improve outcomes for older AML patients.3
Epigenetic silencing of structurally normal genes involved in hematopoiesis has been reported in AML and probably contributes to leukemogenesis.4 The addition of a methyl group to the 5′ carbon position of cytosine bases via DNA methyltransferase (DNMT) activity leads to DNA methylation and silencing of gene expression. In contrast to recurrent structural genomic changes in AML, such as loss-of-function mutations or deletions causing permanent loss of gene activity, gene silencing by DNA hypermethylation can be pharmacologically reversed,5 thereby restoring normal patterns of hematopoietic cell differentiation, proliferation, and survival.
Two azanucleoside DNMT inhibitors, azacitidine (5-azacytidine; Vidaza; Celgene) and decitabine (5-aza-2′-deoxycytidine; Dacogen; Eisai), are now approved in the United States for treatment of patients with myelodysplastic syndromes, a clonal myeloid disorder that may evolve into AML. These agents have also been shown to be effective in AML. In a recent study, we reported the clinical results of a 10-day induction regimen of low-dose decitabine in untreated older (≥ 60 years) AML patients who were not candidates for or refused intensive therapy.6 We showed that decitabine induced a complete remission (CR) rate of 47%, an overall response rate of 64%, and a median overall survival duration of approximately 1 year. This regimen was also associated with an improved toxicity profile compared with that expected in patients treated with more intense chemotherapy induction regimens (ie, cytarabine/anthracyclines). Therefore, this regimen should be considered as a framework on which future trials might build on to improve current treatment outcomes in older AML patients.
To further optimize the therapeutic use of decitabine, however, the pharmacodynamic activity of this agent needs to be fully characterized.4 Thus, to gain insights into the genome-wide localization and extent of methylation changes induced by decitabine, we applied an approach that combined “Methylated DNA Capture” with next-generation sequencing (MethylCap-seq) in pretreatment and posttreatment BM samples from older, AML patients treated with decitabine on our phase 2 clinical trial.6
Methods
Patients and samples
This study includes patients (n = 16) who presented with previously untreated AML, who were diagnosed by World Health Organization criteria, treated with decitabine on a single-center phase 2 protocol, and had both pretreatment (day 0) and posttreatment (day 25 of the first cycle) BM samples available for analysis.6 All 3 of these criteria were required for inclusion in this study. Patients received decitabine at 20 mg/m2 intravenously over 1 hour on days 1 to 10 in each 4-week cycle. CR was defined according to International Working Group published criteria.7 Patients included in this analysis who eventually achieved CR required more than 1 cycle of therapy to achieve disease remission. The study design and the results of the trial for the entire cohort of patients have been previously reported.6 Informed written consent approved by The Ohio State University Human Studies Committee was obtained on all subjects before study entry. All the experiments involving human subjects were conducted according to the principles expressed in the Declaration of Helsinki.
MethylCap-seq assay for measuring DNA methylation
BM mononuclear cells were procured, cryopreserved, and then thawed for analysis as previously reported.6 DNA was extracted as previously reported8 and subjected to fragmentation using a Covaris S2 Adaptive Acoustic instrument. Methylated DNA fragments (150-200 bp) were enriched by MBD2 protein (MethylMiner Methylated DNA Enrichment Kit; Invitrogen) as described by the manufacturer's protocol. Illumina sequencing libraries were generated from the enriched methylated material as previously described.9 Library materials were quantified by fluorometric measurement, and quality of the samples was assessed by Agilent Bioanalyzer High Sensitivity DNA analysis before sequencing on the Illumina GAIIx flow cells. Images were captured from the sequencer and analyzed using the Real Time Analysis Version 1.8 software yielding 36-bp single-end sequenced reads.
Methylome analysis workflow
MethylCap-seq analysis for assessing the methylation status of distinct genome regions started with the alignment of the 36-bp reads that passed the filtering to the reference genome (hg18) using the Bowtie short read aligner.10 The 170-bp (average enriched methylated DNA fragment size) genomic regions corresponding to each unique and nonduplicated 36-bp read aligned to hg18 were identified, and all the sequences parsed into 500-bp bins. As part of the binning step, aligned reads in each bin were normalized by converting them to reads per million of uniquely aligned reads (rpm) to adjust for different total read counts per lane of the Illumina flow cell. It was expected that the higher the rpm, the higher the level of methylation in the region spanned by the bin.
Global DNA methylation comparison
As DNA methylation occurs in CpG dinucleotides and the enrichment-based MethylCap-seq method interrogates the methylome in a CpG density-dependent manner, to assess average changes in global DNA methylation before and after decitabine treatment, we plotted (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article) the CpG density along the entire human genome (x-axis) versus mean rpm (y-axis; the mean of the rpms of all bins in a given CpG density interval) and then calculated the area under the curves for each patient sample. The area under the curve value for each sample then served as the global methylation indicators (GMIs) for that sample. The significance of global methylation differences before and after decitabine treatment was assessed by a paired sample nonparametric test (Wilcoxon signed rank test). Similarly, the significance of global methylation differences between patients who eventually achieved CR and those who never achieved CR was assessed by a Student t test.
Methylation analyses of genomic features
Genomic terms (eg, “genomic features”) used in our data analysis workflow are summarized in supplemental Table 1 as a glossary list. The definition for each genomic feature analyzed in this study is presented in Table 1. Elements within each genomic feature are referred to as “regions.” They share the feature characteristics common to that genomic feature as described in Table 1. The methylation level of a particular genomic feature was calculated in pretreatment and posttreatment samples by summing the rpm values of all of the regions of that particular genomic feature (eg, rpm values for all CpG islands in the genome of each sample were summed to provide the methylation level of the genomic feature called CpG islands). Comparing the mean of the rpm sums obtained from the pretreatment samples with that obtained from the posttreatment samples, we were able to assess DNA methylation changes after decitabine treatment for each of the genomic features (using a paired Wilcoxon test and a statistical significance level of P < .05). The levels of DNA methylation changes for each genomic feature were also compared between patients who eventually achieved CR and those who never achieved CR (using a paired Wilcoxon test and a statistical significance level of P < .05).
Definition of genomic features
| Genomic features . | Definition . |
|---|---|
| CpG islands | Minimum criteria as defined in UCSC Genome Browser: 200 bp; 50% GC content; CpG observed-overexpected ratio of 0.6 |
| CpG island shores | Shores are regions from 200 bp to 2 kb from both ends of CpG island22 |
| CpG inlands | For large CpG islands only: 2 kb inwards from each end of the island23 |
| Promoter-associated CpG islands | CpG islands that fall within 10 kb upstream and 1 kb downstream of a promoter24 |
| Gene promoters | 1 kb upstream and downstream of TSS of RefSeq genes25 |
| Gene deserts | A region of a chromosome that contains few if any genes as defined by UCSC Genome Browser |
| miRNAs | miRNA coding transcripts as defined in the RefSeq gene track on UCSC Genome Browser |
| miRNA-associated CpG islands | CpG island intersecting a miRNA |
| miRNA-associated promoters | Promoter intersecting a miRNA |
| Repeat-masked regions | The UCSC Genome Browser definition of RepeatMasker track was used; repeat-masked regions are elements including DNA sequences found in interspersed repeats and low complexity DNA sequences |
| RefSeq genes | The UCSC Genome Browser definition of RefSeq was used; RefSeq genes were defined as known human protein-coding and non–protein-coding genes collected by NCBI RNA reference sequences |
| RefSeq gene-associated CpG islands | CpG islands intersecting a RefSeq gene |
| Genomic features . | Definition . |
|---|---|
| CpG islands | Minimum criteria as defined in UCSC Genome Browser: 200 bp; 50% GC content; CpG observed-overexpected ratio of 0.6 |
| CpG island shores | Shores are regions from 200 bp to 2 kb from both ends of CpG island22 |
| CpG inlands | For large CpG islands only: 2 kb inwards from each end of the island23 |
| Promoter-associated CpG islands | CpG islands that fall within 10 kb upstream and 1 kb downstream of a promoter24 |
| Gene promoters | 1 kb upstream and downstream of TSS of RefSeq genes25 |
| Gene deserts | A region of a chromosome that contains few if any genes as defined by UCSC Genome Browser |
| miRNAs | miRNA coding transcripts as defined in the RefSeq gene track on UCSC Genome Browser |
| miRNA-associated CpG islands | CpG island intersecting a miRNA |
| miRNA-associated promoters | Promoter intersecting a miRNA |
| Repeat-masked regions | The UCSC Genome Browser definition of RepeatMasker track was used; repeat-masked regions are elements including DNA sequences found in interspersed repeats and low complexity DNA sequences |
| RefSeq genes | The UCSC Genome Browser definition of RefSeq was used; RefSeq genes were defined as known human protein-coding and non–protein-coding genes collected by NCBI RNA reference sequences |
| RefSeq gene-associated CpG islands | CpG islands intersecting a RefSeq gene |
UCSC indicates University of California, Santa Cruz; TSS, transcription start site; and NCBI, National Center for Biotechnology Information.
Identification of methylation status of regions within genomic features
To evaluate the effect of decitabine treatment on the methylation status of regions within genomic features (eg, RefSeq genes, promoters, or CpG islands), we defined and identified differentially methylated regions (DMRs) as those regions that showed a statistically significant difference in methylation levels (expressed as rpm) when pretreatment and posttreatment samples were compared using the paired Wilcoxon test with a significance cut-off at a false discovery rate of .05. A DMR was classified as hypomethylated when the mean of the pretreatment rpm was higher than the mean of the posttreatment rpm, or hypermethylated where the mean of the pretreatment rpm was lower than the mean of the posttreatment rpm in the compared samples.
Chromosomal localization of DMRs
To investigate whether DMRs are distributed randomly or nonrandomly along the chromosomes, we decided to evaluate CpG islands, a genomic feature containing high CpG density, for this localization effect. The proximity of CpG dinucleotides in this feature type enables effective enrichment of methylated DNA fragments by the MBD protein. First, we divided the genome in 400-kb bins. We constructed contingency tables for each 400-kb bin along each of the chromosomes. The tables were populated by classifying individual CpG islands within this genomic feature as either DMRs or unchanged, inside or outside of the selected 400-kb bin. The expected number of DMRs was calculated as the ratio of the number of CpG islands within the selected bin to the number of CpG islands in the chromosome that bin locates, multiplied by the total number of observed DMRs in the chromosome. The expected number of unchanged CpG islands was calculated as the ratio of the number of CpG islands within the bin to the number of CpG islands in the chromosome that bin locates, multiplied by the total number of observed unchanged CpG islands in the chromosome. The P value quantifying the deviation of the observed number of DMRs from the expected number of DMRs was calculated using the Pearson χ2 test applied to these 2 × 2 contingency tables and corrected for multiple testing using the Bonferroni correction with a significance cutoff of .05.
Other statistical analyses
To assess the relationship of blast counts and GMI values, we first evaluated potential patterns of difference using graphical analyses given the limited sample sizes. We evaluated pretreatment and posttreatment sample data across all patients as well as for those who eventually achieved a CR and those who never achieved CR, as well as those classified as “informative” versus “uninformative” (based on percentage blasts and/or cytogenetics). Summary statistics were used to initially assess methylation markers (eg, GMI) and blast counts across all patients as well as by “eventual CR” status. Nonparametric tests were used in an exploratory manner to assess potentially significant relationships and trends worth pursuing in future studies. Wilcoxon signed rank tests were used to assess differences in pretreatment versus posttreatment measures, Wilcoxon rank-sum tests were used to assess differences in markers (eg, baseline GMI levels) between 2 independent groups (CR vs not, informative vs uninformative), and Spearman rank correlation coefficients were used to assess relationships between 2 continuous measures (GMI vs blast counts). Analyses were done across all patients as well as within subgroups (CR vs not, informative vs. uninformative). When looking at correlations between GMI and blast counts, we looked at all measures (pretreatment and posttreatment) as well as within each time point. In this exploratory setting where we were evaluating overall measures (GMI) and clinical measures (blasts) and outcomes (CR vs not), significance was defined as P < .05. More formal modeling was avoided because of the limited numbers of patients available for analysis.
Results
Patients' characteristics
Clinical, cytogenetic, and molecular characteristics of the patients with material available for MethylCap-seq analysis are reported in Table 2. Of the 16 patients with available pretreatment sample and day 25 postdecitabine treatment sample, 9 eventually achieved CR and 7 did not. It should be noted that the patients who eventually achieved CR did so after more than 1 cycle of treatment. Therefore, these patients presented on day 25/course 1 with persistent BM > 5% leukemia blast counts and/or abnormal cytogenetics and/or abnormal blood counts (ie, neutrophils < 1000/μL and platelets < 100 000/μL). In the posttreatment BM samples, the mean percentage of blasts was 20.3% for patients who eventually achieved CR (range, 0-45) and 42.3% for those who never achieved CR (range, 3-90; P = .12). Table 3 summarizes pretreatment (day 0) and posttreatment (day 25/course 1) BM blast counts and cytogenetics.
Patient characteristics and response to decitabine treatment
| Patient no. . | Age, y . | Sex . | Gene mutation . | Eventual clinical outcome . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NPM1 . | FLT3-ITD . | CEBPA . | DNMT3A . | TET2 . | IDH1 . | IDH2 . | ||||
| 1 | 73 | F | MUT | WT | WT | WT | MUT | WT | WT | Non-CR |
| 3 | 69 | F | MUT | WT | WT | MUT | WT | WT | WT | CR |
| 4 | 80 | M | WT | WT | WT | WT | WT | WT | WT | Non-CR |
| 5 | 85 | M | WT | WT | WT | WT | WT | MUT | WT | CR |
| 6 | 71 | M | WT | WT | WT | WT | WT | WT | WT | Non-CR |
| 7 | 84 | F | WT | WT | WT | WT | WT | WT | WT | Non-CR |
| 8 | 66 | F | WT | WT | WT | WT | WT | WT | WT | CR |
| 14 | 80 | F | WT | WT | WT | ND | ND | WT | WT | CR |
| 15 | 75 | F | WT | WT | WT | WT | WT | WT | WT | CR |
| 17 | 74 | F | WT | WT | WT | WT | WT | WT | MUT | CR |
| 18 | 74 | M | MUT | WT | WT | WT | WT | WT | WT | CR |
| 19 | 73 | F | WT | WT | WT | WT | WT | MUT | WT | Non-CR |
| 20 | 81 | M | WT | WT | WT | WT | MUT | WT | WT | CR |
| 21 | 76 | M | WT | WT | WT | WT | WT | WT | WT | Non-CR |
| 23 | 67 | M | WT | WT | WT | WT | WT | WT | WT | Non-CR |
| 24 | 68 | M | MUT | WT | WT | MUT | WT | WT | WT | CR |
| Patient no. . | Age, y . | Sex . | Gene mutation . | Eventual clinical outcome . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NPM1 . | FLT3-ITD . | CEBPA . | DNMT3A . | TET2 . | IDH1 . | IDH2 . | ||||
| 1 | 73 | F | MUT | WT | WT | WT | MUT | WT | WT | Non-CR |
| 3 | 69 | F | MUT | WT | WT | MUT | WT | WT | WT | CR |
| 4 | 80 | M | WT | WT | WT | WT | WT | WT | WT | Non-CR |
| 5 | 85 | M | WT | WT | WT | WT | WT | MUT | WT | CR |
| 6 | 71 | M | WT | WT | WT | WT | WT | WT | WT | Non-CR |
| 7 | 84 | F | WT | WT | WT | WT | WT | WT | WT | Non-CR |
| 8 | 66 | F | WT | WT | WT | WT | WT | WT | WT | CR |
| 14 | 80 | F | WT | WT | WT | ND | ND | WT | WT | CR |
| 15 | 75 | F | WT | WT | WT | WT | WT | WT | WT | CR |
| 17 | 74 | F | WT | WT | WT | WT | WT | WT | MUT | CR |
| 18 | 74 | M | MUT | WT | WT | WT | WT | WT | WT | CR |
| 19 | 73 | F | WT | WT | WT | WT | WT | MUT | WT | Non-CR |
| 20 | 81 | M | WT | WT | WT | WT | MUT | WT | WT | CR |
| 21 | 76 | M | WT | WT | WT | WT | WT | WT | WT | Non-CR |
| 23 | 67 | M | WT | WT | WT | WT | WT | WT | WT | Non-CR |
| 24 | 68 | M | MUT | WT | WT | MUT | WT | WT | WT | CR |
FLT3-ITD indicates internal tandem duplication of the FLT3 gene; CR, complete remission (no disease and full blood count recovery); and Non-CR, noncomplete remission.
Patients' blast counts and cytogenetics on day 0 and day 25
| Patient no. . | Day 0 blast, % . | Day 25 blast, % . | Pretreatment cytogenetics (day 0) . | Cytogenetics after 1 cycle of decitabine (day 25) . |
|---|---|---|---|---|
| AML patients with blast counts > 20% or informative cytogenetics on day 25 | ||||
| 1 | 82 | 71 | 46,XX[20] | 46,XX[20] |
| 4 | 24 | 3 | 45,XY,add(2)(q33),del(5)(q13),−12,der(17)t(11;17)(q13;p12),−18, −20,+r,+mar(4)/46,sl,+21,del(21)(q22)(16)/47,sdl1,+del(5)(q13)[4]/nonclonal w/clonal abnormalities(2).ish del(5)(D5S23+,EGR1−) | 46,XY,add(2)(q33),del(5)(q13),−12,der(17)t(11;17)(q13;p12),−18, −20,+21,del(21)(q22.3),+r,+mar1[cp6]/45,sl,+add(5)(q13), −del(5)(q13),add(15)(p11.2),−16,−del(21)[cp5 one is 4n]/47,sl, +del(5)(q13)[4]/48,sl,+18,+mar2[1]/ 46,XY[4] 46,XY,add(2)(q33),del(5)(q13),−12,der(17)t(11;17)(q13;p12),−18, −20,+21,del(21)(q22.3),+r,+mar1[cp6]/45,sl,+add(5)(q13), −del(5)(q13),add(15)(p11.2),−16,−del(21)[cp5 one is 4n]/47,sl, +del(5)(q13)[4]/ 48,sl,+18,+mar2[1]/ 46,XY[4] |
| 5 | 35 | 31 | 47,XY,+13(2)/46,XY[37] | 46,XY[19]/4n[1] |
| 6 | 24 | 13 | 46,XY,t(4;21)(q33;q22)[13]/46,idem,der(3)t(3;4)(p22;q12), −4,add(8)(p12),+19,add(19)(p13.3)(cp10)/46,XY(3).ish t(4;21)(RUNX1+;RUNX1−) | 46,XY,t(4;21)(q33;q22)[1]/46,idem,der(3)t(3;4)(p22;q12), −4,add(8)(p12),+19,add(19)(p13.3)[14]/48,XY,+21,+21[3]/46,XY[2] |
| 7 | 25 | 54 | 46,XX[20] | 46,XX[28]/nonclonal[2] |
| 8 | 22 | 19 | 43,X,t(X;2)(p11.2;p11.2),del(5)(q11.2q33),−7,−12,add(16)(q11.2), −19(cp9)/43,idem,+12,dic(12;19)(p12;p13.3),−add(16), +19[2]/46,XX[7]/ nonclonal w/clonal abnormalities[2] | 44,X,t(X;2)(p11.2;p11.2),del(5)(q11.2q33),−7,der(12)del(12)(q13.1q13.3) del(12)(q24.1q24.3),dic(12;19)(p12;p13.3),−16[cp5]/44,sl,+mar[3]/46,sl, +11,+12, dic(12;19),+19[5]/46,XX[6]/nonclonal[1] |
| 14 | 39 | 31 | 46,XX[20] | 46,XX[20] |
| 15 | 50 | 44 | 46,X,idic(X)(q13)[11]/46,XX[9] | 46,X,idic(X)(q13)[6]/46,XX[14] |
| 19 | 60 | 90 | 46,XX,del(5)(q22q35)[17]/46,XX[7]/nonclonal[1] | 46,XX,del(5)(q22q35)[18]/46,XX[1]/nonclonal[1] |
| 23 | 74 | 52 | 46,XY,t(11;22)(q23;q11.2)(5)/47,sl,+mar1[5]/47,sl,+mar2[3]/48,sl, +mar3,+mar4[6]/46,XY(1).nuc ish (MLLx2)(5′ MLL sep 3′ MLLx1)(143/221)/(MLLx3)(5′ MLLsep 3′ MLLx1)(29/221) | 46,XY,t(11;22)(q23;q11.2)[1]/47,sl,+mar1[12]/47,sl,+mar2, +mar5[3]/47,sl,+ma r3,+mar4[4] |
| 24 | 82 | 45 | 46,XY[21]/nonclonal[1] | Insufficient metaphases (46,XY[8]) |
| AML patients with blast counts < 20% and uninformative cytogenetics on day 25 | ||||
| 3 | 20 | 9 | 46,XX[20] | 46,XX[20] |
| 17 | 38 | 0 | 46,XX,t(8;21)(q22;q22)[14]/45,idem,−7[6] | 46,XX[20] |
| 18 | 53 | 1 | 46,XY[29]/nonclonal[1] | 46,XY[19]/Nonclonal[1] |
| 20 | 41 | 3 | 46,XY[29]/nonclonal[1] | 46,XY[20] |
| 21 | 35 | 13 | 46,XY[20] | 46,XY[20] |
| Patient no. . | Day 0 blast, % . | Day 25 blast, % . | Pretreatment cytogenetics (day 0) . | Cytogenetics after 1 cycle of decitabine (day 25) . |
|---|---|---|---|---|
| AML patients with blast counts > 20% or informative cytogenetics on day 25 | ||||
| 1 | 82 | 71 | 46,XX[20] | 46,XX[20] |
| 4 | 24 | 3 | 45,XY,add(2)(q33),del(5)(q13),−12,der(17)t(11;17)(q13;p12),−18, −20,+r,+mar(4)/46,sl,+21,del(21)(q22)(16)/47,sdl1,+del(5)(q13)[4]/nonclonal w/clonal abnormalities(2).ish del(5)(D5S23+,EGR1−) | 46,XY,add(2)(q33),del(5)(q13),−12,der(17)t(11;17)(q13;p12),−18, −20,+21,del(21)(q22.3),+r,+mar1[cp6]/45,sl,+add(5)(q13), −del(5)(q13),add(15)(p11.2),−16,−del(21)[cp5 one is 4n]/47,sl, +del(5)(q13)[4]/48,sl,+18,+mar2[1]/ 46,XY[4] 46,XY,add(2)(q33),del(5)(q13),−12,der(17)t(11;17)(q13;p12),−18, −20,+21,del(21)(q22.3),+r,+mar1[cp6]/45,sl,+add(5)(q13), −del(5)(q13),add(15)(p11.2),−16,−del(21)[cp5 one is 4n]/47,sl, +del(5)(q13)[4]/ 48,sl,+18,+mar2[1]/ 46,XY[4] |
| 5 | 35 | 31 | 47,XY,+13(2)/46,XY[37] | 46,XY[19]/4n[1] |
| 6 | 24 | 13 | 46,XY,t(4;21)(q33;q22)[13]/46,idem,der(3)t(3;4)(p22;q12), −4,add(8)(p12),+19,add(19)(p13.3)(cp10)/46,XY(3).ish t(4;21)(RUNX1+;RUNX1−) | 46,XY,t(4;21)(q33;q22)[1]/46,idem,der(3)t(3;4)(p22;q12), −4,add(8)(p12),+19,add(19)(p13.3)[14]/48,XY,+21,+21[3]/46,XY[2] |
| 7 | 25 | 54 | 46,XX[20] | 46,XX[28]/nonclonal[2] |
| 8 | 22 | 19 | 43,X,t(X;2)(p11.2;p11.2),del(5)(q11.2q33),−7,−12,add(16)(q11.2), −19(cp9)/43,idem,+12,dic(12;19)(p12;p13.3),−add(16), +19[2]/46,XX[7]/ nonclonal w/clonal abnormalities[2] | 44,X,t(X;2)(p11.2;p11.2),del(5)(q11.2q33),−7,der(12)del(12)(q13.1q13.3) del(12)(q24.1q24.3),dic(12;19)(p12;p13.3),−16[cp5]/44,sl,+mar[3]/46,sl, +11,+12, dic(12;19),+19[5]/46,XX[6]/nonclonal[1] |
| 14 | 39 | 31 | 46,XX[20] | 46,XX[20] |
| 15 | 50 | 44 | 46,X,idic(X)(q13)[11]/46,XX[9] | 46,X,idic(X)(q13)[6]/46,XX[14] |
| 19 | 60 | 90 | 46,XX,del(5)(q22q35)[17]/46,XX[7]/nonclonal[1] | 46,XX,del(5)(q22q35)[18]/46,XX[1]/nonclonal[1] |
| 23 | 74 | 52 | 46,XY,t(11;22)(q23;q11.2)(5)/47,sl,+mar1[5]/47,sl,+mar2[3]/48,sl, +mar3,+mar4[6]/46,XY(1).nuc ish (MLLx2)(5′ MLL sep 3′ MLLx1)(143/221)/(MLLx3)(5′ MLLsep 3′ MLLx1)(29/221) | 46,XY,t(11;22)(q23;q11.2)[1]/47,sl,+mar1[12]/47,sl,+mar2, +mar5[3]/47,sl,+ma r3,+mar4[4] |
| 24 | 82 | 45 | 46,XY[21]/nonclonal[1] | Insufficient metaphases (46,XY[8]) |
| AML patients with blast counts < 20% and uninformative cytogenetics on day 25 | ||||
| 3 | 20 | 9 | 46,XX[20] | 46,XX[20] |
| 17 | 38 | 0 | 46,XX,t(8;21)(q22;q22)[14]/45,idem,−7[6] | 46,XX[20] |
| 18 | 53 | 1 | 46,XY[29]/nonclonal[1] | 46,XY[19]/Nonclonal[1] |
| 20 | 41 | 3 | 46,XY[29]/nonclonal[1] | 46,XY[20] |
| 21 | 35 | 13 | 46,XY[20] | 46,XY[20] |
Global DNA methylation changes
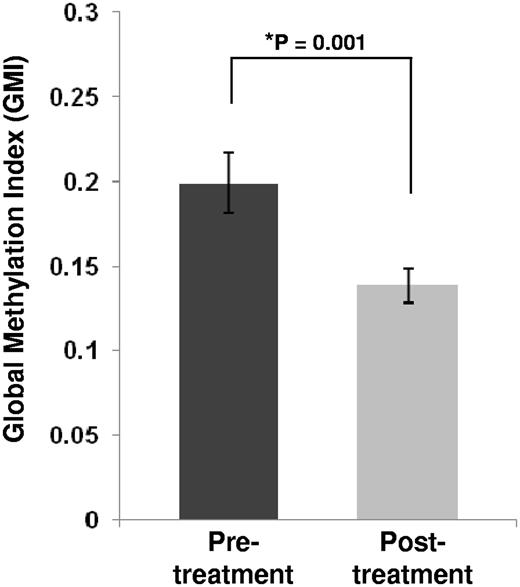
To assess global DNA methylation changes after decitabine treatment, we developed the GMI as an indicator of the level of global methylation in each patient sample (see “Global DNA methylation comparison” for the definition of GMI). comparing pretreatment and posttreatment samples from all patients, we observed a significant decrease in the mean GMI in the posttreatment samples (fold change 1.44; P = .001; Figure 1). Although the number of patients analyzed was limited, we noted that patients who eventually achieved CR had a trend for higher baseline GMI and a more pronounced decrease in posttreatment GMI compared with those who never achieved CR (supplemental Figure 2).
Global methylation levels for all pre- and postdecitabine treatment samples. Global methylation levels were measured for each sample by the global methylation indicator (GMI; see “Global DNA methylation comparison”). The histograms represent the mean GMI values in the sample groups (n = 16). Error bars represent SD within the sample group. The P value for the pretreatment versus posttreatment comparison was calculated with a paired sample nonparametric test (Wilcoxon signed rank test).
Global methylation levels for all pre- and postdecitabine treatment samples. Global methylation levels were measured for each sample by the global methylation indicator (GMI; see “Global DNA methylation comparison”). The histograms represent the mean GMI values in the sample groups (n = 16). Error bars represent SD within the sample group. The P value for the pretreatment versus posttreatment comparison was calculated with a paired sample nonparametric test (Wilcoxon signed rank test).
To address the question of whether GMI values directly correlated with BM blasts and therefore were a mere index of disease burden, we tested for correlation between GMI and percentage blasts (supplemental Figure 3) in all pretreatment and posttreatment samples together (r = 0.14; P = .45); separately for pretreatment samples (r = 0.053; P = .85) and posttreatment samples (r = 0.18; P = .51); in pretreatment (r = 0.067; P = .88) and posttreatment (r = 0.19; P = .62) samples from patients who eventually achieved CR; and in pretreatment (r = 0.25; P = .59) and posttreatment (r = 0.09; P = .85) samples from patients who never achieved CR. We found no significant correlation between blast counts and GMI in all these comparisons.
Furthermore, we identified “informative” patients as those who had persistent abnormal cytogenetics and/or significant disease (ie, BM blast counts > 20%; Table 3) in the posttreatment sample (n = 11). For these patients, we noted that the GMI (P = .024) and not the blast counts (P = .23) decreased significantly at day 25 compared with pretreatment baselines. Taken together, these data indicate that posttreatment changes in BM blasts did not account for all the decrease in DNA methylation observed in posttreatment samples, thereby supporting that the posttreatment decreased GMI was at least partly related to the hypomethylating activity of decitabine on the BM blasts.
DNA methylation changes in distinct genomic features
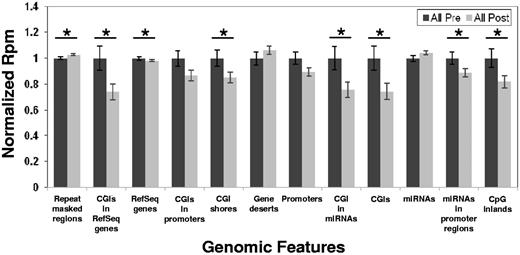
Next, we asked the question of whether the hypomethylating activity of decitabine affected the whole genome or impacted only specific genomic features. To address this question, we examined the methylation status of distinct genomic features as defined in Table 1. We computed the sum of rpm for the regions in each distinct genomic feature and then compared the mean values of the sums (total rpm) for each genomic feature in the pretreatment versus posttreatment samples. When all patients were considered, significant posttreatment hypomethylation was observed only for the following genomic features: CpG islands (CGI, P = .04), CpG island shores (P = .03), CpG inlands (P = .03), miRNA-associated CpG islands (P = .03), miRNA-associated promoters (P = .03), RefSeq genes (P = .03), and RefSeq gene-associated CpG islands (P = .03), whereas significant posttreatment hypermethylation was observed for the repeat-masked regions (P = .04; Figure 2).
Pretreatment and posttreatment methylation changes by genomic features. Error bars represent SEM. *P < .05 using a paired Wilcoxon signed sum.
Pretreatment and posttreatment methylation changes by genomic features. Error bars represent SEM. *P < .05 using a paired Wilcoxon signed sum.
In patients who eventually achieved CR, significant post treatment hypomethylation occurred in the following genomic features (supplemental Figure 4): CpG islands (P = .04), promoter-associated CpG islands (P = .05), promoters (P = .05), and RefSeq gene-associated CpG islands (P = .04), whereas no post treatment hypermethylated genomic features were observed. In contrast, patients who never achieved CR showed no significant methylation changes in any of the genomic features (data not shown).
DMRs
To further characterize how statistically significant methylation changes as a result of decitabine treatment were distributed across the genome, we identified DMRs (see “Identification of methylation status of regions within genomic features” for definition) within each of the genomic features and the direction of their methylation changes (hypomethylated vs hypermethylated). In Figure 2, we presented the overall methylation level in each genomic feature. In Table 4, we presented the percentage of regions identified as DMRs in each genomic feature and whether the DMRs were hypomethylated or hypermethylated after decitabine treatment. When we considered all patients (Table 4), there was a striking difference between CpG island-associated genomic features (CpG islands, shores, inlands, and RefSeq gene-associated islands) and genomic features partially associated with CpG islands (promoters, miRNA-associated promoters, RefSeq genes; following our gene promoter definition listed in Table 1, 43% of promoters overlap with CpG islands) and genomic features not associated with CpG islands (gene deserts and gene deserts). In CpG island-associated genomic features, the majority of the DMRs were hypomethylated after decitabine treatment (range, 94%-99%). For genomic features not fully associated with CpG islands, the percentage of hypomethylated DMRs ranged from 4%-88%; we also observed an increased number of hypermethylated DMRs in these features (Table 4). These results were similar to those obtained from patients who eventually achieved CR (supplemental Table 2). Patients who never achieved CR had no significant changes in global methylation level after decitabine treatment and therefore no DMRs to report (supplemental Figure 2). These results indicate that, although hypomethylated DMRs were the most common type of changes found in patients who were treated with decitabine, these seemed largely limited to CpG island-associated genomic features. The number of observed hypermethylated DMRs, though small in comparison, should be dissected in future studies for their biologic and clinical significance.
Effects of decitabine on 16 AML patient methylomes (CR and non-CR): significant differences in methylation level in various genomic features between day 0 (pretreatment) and day 25 (after first cycle of decitabine treatment)
| Genomic features . | CpG island associated . | Gene associated . | Total regions in genomic features* . | DMRs in genomic features . | ||
|---|---|---|---|---|---|---|
| DMRs, no. (%) total features . | Hypomethylated, no. (%) total DMRs . | Hyperrmethylated, no. (%) total DMRs . | ||||
| CpG islands | Yes | No | 27 639 | 6470 (23) | 6375 (99) | 95 (1) |
| CpG island shores | Yes | No | 55 278 | 8706 (16) | 8033 (92) | 673 (8) |
| CpG inlands | Yes | No | 55 278 | 12 571 (23) | 11 910 (95) | 661 (5) |
| Promoter-associated CpG islands | Yes | Yes | 16 393 | 1457 (11) | 1438 (99) | 19 (1) |
| Gene promoters | Partial† | Yes | 26 374 | 893 (3) | 788 (88) | 105 (12) |
| Gene deserts | No | No | 776 | 519 (67) | 20 (4) | 499 (96) |
| miRNAs | No | No | 925 | 75 (8) | 67 (89) | 8 (11) |
| miRNA-associated CpG islands | Yes | No | 68 | 20 (29) | 20 (100) | 0 |
| miRNA-associated promoters | No | No | 1051 | 86 (8) | 69 (80) | 17 (20) |
| Repeat-masked regions | No | No | 5 016 131 | 0 | 0 | 0 |
| RefSeq genes | Partial‡ | Yes | 35 430 | 10 941 (31) | 6034 (55) | 4907 (45) |
| RefSeq gene-associated CpG islands | Yes | Yes | 20 842 | 5077 (24) | 5005 (99) | 72 (1) |
| Genomic features . | CpG island associated . | Gene associated . | Total regions in genomic features* . | DMRs in genomic features . | ||
|---|---|---|---|---|---|---|
| DMRs, no. (%) total features . | Hypomethylated, no. (%) total DMRs . | Hyperrmethylated, no. (%) total DMRs . | ||||
| CpG islands | Yes | No | 27 639 | 6470 (23) | 6375 (99) | 95 (1) |
| CpG island shores | Yes | No | 55 278 | 8706 (16) | 8033 (92) | 673 (8) |
| CpG inlands | Yes | No | 55 278 | 12 571 (23) | 11 910 (95) | 661 (5) |
| Promoter-associated CpG islands | Yes | Yes | 16 393 | 1457 (11) | 1438 (99) | 19 (1) |
| Gene promoters | Partial† | Yes | 26 374 | 893 (3) | 788 (88) | 105 (12) |
| Gene deserts | No | No | 776 | 519 (67) | 20 (4) | 499 (96) |
| miRNAs | No | No | 925 | 75 (8) | 67 (89) | 8 (11) |
| miRNA-associated CpG islands | Yes | No | 68 | 20 (29) | 20 (100) | 0 |
| miRNA-associated promoters | No | No | 1051 | 86 (8) | 69 (80) | 17 (20) |
| Repeat-masked regions | No | No | 5 016 131 | 0 | 0 | 0 |
| RefSeq genes | Partial‡ | Yes | 35 430 | 10 941 (31) | 6034 (55) | 4907 (45) |
| RefSeq gene-associated CpG islands | Yes | Yes | 20 842 | 5077 (24) | 5005 (99) | 72 (1) |
CR indicates eventual complete remission; and non-CR, eventual noncomplete remission.
Total regions in genomic features with characteristics described in Table 1.
Although not explicitly named as associated with CpG islands, ∼ 43% of gene promoters are overlapped with CpG islands.
Although not explicitly named as associated with CpG islands, gene bodies and 3′UTR regions overlap with CpG islands.
Preferential chromosome location of DMRs
Whether methylation changes associated with the pharmacologic activity of decitabine occur randomly or in specific chromosome regions is unknown. We analyzed this by identifying the localization of CpG island-associated DMRs. In our analysis, we noted that these DMRs appeared to cluster at ends of chromosomes (Figure 3). To assess whether this observation was simply the result of a higher density of CpG islands in chromosomal ends or instead was related to a selective activity of decitabine in these chromosomal regions, we divided each chromosome into 400-kb bins and calculated the expected versus the observed number of CpG island-associated DMRs in each of these bins. We noted that these DMRs significantly clustered (P ≤ .05, Pearson χ2 test) at chromosomal ends except for chromosomes 2, 3, 15, 19, and 21.
Chromosomal locations of DMRs in pre- versus postdecitabine treatment AML samples. The 4 lanes above each chromosome symbol show the number of DMRs in 400-kb bins as a heat map for all genomic features (ALL), CpG islands (CGI), Gene deserts (GD), and RefSeq gene bodies (RG), respectively (intensity in each lane is normalized to the bin with most DMRs in that lane). These 4 features were selected as they depict genomic features with diverse CpG densities (see Table 1 for other features evaluated in this study but not shown here). Comparing the 4 heat map tracks shown here (ALL vs CGI, or GD, or RG), the most significant number of DMRs induced by decitabine treatment is contributed by the CpG islands. This effect of decitabine is most pronounced at the chromosome ends (exception: chromosome 3, chromosome 15, and chromosome Y). The green track below the chromosome symbol shows the distribution of CpG islands (CGIs) across the chromosome (independently of their methylation status). Because CpG islands are also enriched in chromosome ends, we performed Pearson χ2 analysis of observed DMRs versus expected DMRs for every 400-kb bin in the genome (“Chromosomal localization of DMRs”) to test whether enrichment of DMRs within the chromosome ends is related to the higher density of CpG islands in that region or as a nonrandom effect of decitabine treatment. The resulting P values are plotted in the χ2 track in blue. For all chromosomes except 2, 3, 15, 19, and 21, the P values were significant (P ≤ .05) at least at one end of the chromosome, indicating significant clustering of DMRs not accounted for by the dense distribution of CpG islands in that region of the chromosome.
Chromosomal locations of DMRs in pre- versus postdecitabine treatment AML samples. The 4 lanes above each chromosome symbol show the number of DMRs in 400-kb bins as a heat map for all genomic features (ALL), CpG islands (CGI), Gene deserts (GD), and RefSeq gene bodies (RG), respectively (intensity in each lane is normalized to the bin with most DMRs in that lane). These 4 features were selected as they depict genomic features with diverse CpG densities (see Table 1 for other features evaluated in this study but not shown here). Comparing the 4 heat map tracks shown here (ALL vs CGI, or GD, or RG), the most significant number of DMRs induced by decitabine treatment is contributed by the CpG islands. This effect of decitabine is most pronounced at the chromosome ends (exception: chromosome 3, chromosome 15, and chromosome Y). The green track below the chromosome symbol shows the distribution of CpG islands (CGIs) across the chromosome (independently of their methylation status). Because CpG islands are also enriched in chromosome ends, we performed Pearson χ2 analysis of observed DMRs versus expected DMRs for every 400-kb bin in the genome (“Chromosomal localization of DMRs”) to test whether enrichment of DMRs within the chromosome ends is related to the higher density of CpG islands in that region or as a nonrandom effect of decitabine treatment. The resulting P values are plotted in the χ2 track in blue. For all chromosomes except 2, 3, 15, 19, and 21, the P values were significant (P ≤ .05) at least at one end of the chromosome, indicating significant clustering of DMRs not accounted for by the dense distribution of CpG islands in that region of the chromosome.
Among the CpG islands positioned close to chromosome ends, many CpG island-associated DMRs fall into the chromosome subtelomeric regions defined as the terminal 500 kb of each euchromatic chromosome arm.11 More specifically, of the 900 CpG islands in these subtelomeric regions, 323 (36%) were DMRs. The rate of methylation events observed in these regions was significantly higher than the overall genomic rate (P ≤ 10−10, χ2). Table 5 shows the number of CpG island-associated DMRs in the subtelomeric regions of chromosomes.
Number of CpG island-associated DMRs at the subtelomeric regions of chromosomes (∼ 500 kb from end of chromosomes)
| Chromosome . | p arm . | q arm . | P (χ2) . |
|---|---|---|---|
| 1 | 0 | 3 | NS |
| 2 | 5 | 5 | .0001 |
| 3 | 1 | 1 | NS |
| 4 | 1 | 2 | .0017 |
| 5 | 7 | 5 | < .0001 |
| 6 | 1 | 7 | .0009 |
| 7 | 12 | 6 | < .0001 |
| 8 | 3 | 7 | .0022 |
| 9 | 0 | 10 | NS |
| 10 | 6 | 24 | < .0001 |
| 11 | 21 | 2 | < .0001 |
| 12 | 3 | 7 | .0003 |
| 13 | NA | 18 | .0042 |
| 14 | NA | 3 | NS |
| 15 | NA | 1 | NS |
| 16 | 16 | 8 | .0016 |
| 17 | 5 | 28 | < .0001 |
| 18 | 1 | 18 | .0026 |
| 19 | 21 | 15 | .0041 |
| 20 | 2 | 15 | NS |
| 21 | NA | 12 | NS |
| 22 | NA | 21 | < .0001 |
| Chromosome . | p arm . | q arm . | P (χ2) . |
|---|---|---|---|
| 1 | 0 | 3 | NS |
| 2 | 5 | 5 | .0001 |
| 3 | 1 | 1 | NS |
| 4 | 1 | 2 | .0017 |
| 5 | 7 | 5 | < .0001 |
| 6 | 1 | 7 | .0009 |
| 7 | 12 | 6 | < .0001 |
| 8 | 3 | 7 | .0022 |
| 9 | 0 | 10 | NS |
| 10 | 6 | 24 | < .0001 |
| 11 | 21 | 2 | < .0001 |
| 12 | 3 | 7 | .0003 |
| 13 | NA | 18 | .0042 |
| 14 | NA | 3 | NS |
| 15 | NA | 1 | NS |
| 16 | 16 | 8 | .0016 |
| 17 | 5 | 28 | < .0001 |
| 18 | 1 | 18 | .0026 |
| 19 | 21 | 15 | .0041 |
| 20 | 2 | 15 | NS |
| 21 | NA | 12 | NS |
| 22 | NA | 21 | < .0001 |
NS indicates not significant (P > .05); and NA, unannotated heterochromatic arm.
DNA methylation changes in specific genes after decitabine treatment
Lastly, the effect of decitabine treatment was evaluated in genes perturbed by decitabine treatment previously reported by other groups. We extracted from our database methylation data corresponding to 3 loci (CDKN2B, HIST1H2AA, and GAPDH) previously reported by Brenet et al.12 In their study, these authors evaluated the methylation status of each locus by quantitative bisulfite-PCR in AML patients treated with decitabine-priming followed by induction chemotherapy. The authors reported CDKN2B to be methylated in a low percentage of their pretreatment samples. In contrast, HIST1H2AA was hypermethylated in AML patients and significant posttreatment hypomethylation was observed in peripheral blood granulocytes from CR patients but not in their immature BM CD34+ mononuclear cells. GAPDH, a housekeeping gene, showed little to no methylation. In our study, we were able to detect methylation in CDKN2B in all but 1 pretreatment sample. Patients who eventually achieved CR showed higher pretreatment methylation levels (mean methylation level: 1.67 rpm vs 0.63 rpm) and more extensive posttreatment hypomethylation than patients who never achieved CR (62% vs 38%). HIST1H2AA was found to be methylated in all pretreatment samples. Patients who eventually achieved CR had higher pretreatment methylation levels than patients who never achieved CR (mean methylation level: 15.1 rpm vs 10.2 rpm) and more extensive posttreatment hypomethylation (28% vs 3%). GAPDH showed no significant methylation changes in either group of patients. We also examined the methylation changes in 15 genes composed in a methylation classifier predictive of overall survival reported by Figueroa et al.13 As limited information (ie, genomic region and methylation level) was provided for these classifier genes, we were unable to make direct comparisons with the methylation status of the genes in our database. Nevertheless, we noted that the promoter-associated CpG islands for 7 classifier genes showed posttreatment methylation changes (hypomethylation: SMG6, SRR, E2F1, BLR1 [CXCR5], LCK; hypermethylation: BTBD3, NYNRIN) in our patients who eventually achieved CR, whereas the promoter-associated CpG island of only 1 classifier gene, SLC7A6AS, was hypomethylated in our patients who never reached CR after decitabine treatment.
Discussion
Here we report genome-wide methylation profiling in previously untreated older AML patients who received decitabine as a single agent.6 To our knowledge, this is the first report of genome-wide assessment of methylation changes induced by decitabine using a high-throughput sequencing-based approach. Although the MethylCap-seq approach used here does not provide global methylome profiling at single-base resolution, it uncovered differentially methylated features and allowed analyses that were labor-, resource-, and computationally more friendly than approaches, such as whole genome bisulfite sequencing. In contrast to genes and region-specific methods, such as determination of LINE-1 element methylation by pyrosequencing and beads array-based methylation analyses,14 MethylCap-seq provided detailed and unbiased information for all genomic regions, including hard to access regions, such as gene deserts and Repeat-masked regions. Herein, we showed the feasibility of implementing this approach in a clinical setting by a dynamic whole genome analysis of pretreatment and posttreatment BM samples from patients treated with decitabine.
Our methylome analysis reveals several interesting and original observations. First, we demonstrated that decitabine induced significant methylation changes in posttreatment BM. Specifically, our data showed that DNA hypomethylation occurred in distinct genomic features (ie, those associated with CpG islands). The observed methylation changes occurred in many regions within various genomic features, but only a limited number of them achieved posttreatment methylation levels that were significantly different from their pretreatment levels; we designated these regions as DMRs. The types (hypomethylation vs hypermethylation) of DMRs were significantly different across various genomic features. When DMRs occurred in CpG island-associated genomic features, they were mostly hypomethylation in nature. When DMRs occurred in non CpG-island–associated genomic features, they could be either hypermethylated or hypomethylated. Furthermore, we showed that DMRs were densely congregated in the subtelomeric regions of many chromosomes. Our data suggest that DNA hypermethylation in CpG island-associated genomic features is a target for the pharmacologic activity of decitabine and that hypomethylation of these features may be associated with mechanisms of disease remission. However, the biologic implications of high levels of hypomethylation events observed in CpG island-associated genomic features and their localization in the chromosome subtelomeric regions remain to be dissected at the mechanistic level.
Although our study reveals novel and interesting findings, we recognize that these observations require further testing and validation in larger studies before definitive conclusions can be drawn, given the limited number of patients included, the retrospective nature of our analysis, and the absence of methylation analysis from earlier posttreatment time points. Importantly, in this study, CRs were not achieved after the first course of decitabine treatment (day 25 of methylation analysis), suggesting that disease was still present in patients at day 25, even in those who eventually achieved CR. Because we could not directly measure DNA methylation changes in AML blasts, to gain insight into whether the methylation changes merely reflected a decrease in the number of blasts in posttreatment marrow compared with the pretreatment marrow or was instead also related to the pharmacologic activity of decitabine on the leukemia cells, we investigated correlations between percentage blasts and GMI at diagnosis and posttreatment time points and in distinct subsets of patients. We found no significant correlation between GMI and percentage blasts. Indeed, DNA hypomethylation occurred, even in the presence of significant disease and persistent abnormal cytogenetics, thereby suggesting that the global DNA hypomethylation observed after treatment was at least partly the result of the effect of decitabine on the AML blasts. Studies such as the ongoing multi-institutional randomized phase 2 trial of decitabine versus decitabine/bortezomib in older AML patients sponsored by ALLIANCE (formerly known as CALGB) will provide a larger patient population to validate these findings.
With regard to predicting response to decitabine, it would be interesting to assess whether previously identified gene methylation classifiers that have been shown to be predictive of overall survival in AML patients treated with chemotherapy are also predictive of clinical response in patients treated with decitabine.13 In our study, we were unable to compare directly the methylation level of the 15 genes associated with the methylation classifier for AML reported by Figueroa et al with the methylation level of these genes in our pretreatment samples.13 However, we did observe more differentially methylated promoter-associated CpG islands for the 15 classifier genes as a result of decitabine treatment in patients who achieved eventual CR than in those who never achieved CR (7 vs 1, respectively). In future studies, it would be also important to assess how predecitabine and postdecitabine methylation changes may be related to the mutation status of distinct genes involved in epigenetic mechanisms (ie, DNMT3A, TET2, IDH1, IDH2, and ASXL1). These genes have been shown to impact negatively on the clinical outcome of AML patients treated with conventional chemotherapy.8,15-20 However, it is possible that their prognostic significance may vary in patients treated with epigenetic-targeting agents.21 For example, we recently reported preliminary data showing that the presence of DNMT3A mutation21 and the expression of miR-29b,6 which targets and down-regulates DNMT-encoding genes (DNMT1, DNMT3A, and DNMT3B) are associated with better disease response in decitabine-treated AML patients.
In conclusion, we have successfully demonstrated the feasibility of methylome profiling of sequential AML samples, the hypomethylating activity of decitabine, and the potential value of DNA hypomethylation of specific genomic features as a relevant pharmacodynamic endpoint for epigenetic-targeting therapies. Ongoing larger trials of decitabine-based regimens will provide the opportunity for further evaluation and validation of these initial results.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Cancer Institute (CA016058, CA102031, and CA140158), the Coleman Leukemia Research Foundation, and in part by an allocation of computing time from the Ohio Supercomputer Center.
National Institutes of Health
Authorship
Contribution: P.Y., R.B., and G.M. designed the study, monitored study progress, and wrote the manuscript; D.F., M.M., H.-H.T., M.T., S.G., and R.B. analyzed the data; J.C. assisted with figure and table generation; B.R., J.C., and Y.-Z.W. carried out laboratory-based research; P.Y. provided sequencing data; and S.P.W., K.M., A.W., R.K., S.J., M.R.G., J.C.B., C.D.B., M.A.C., W.B., R.G., and G.M. were involved in the care of patients and/or sample procurement.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Guido Marcucci, The Ohio State University, Comprehensive Cancer Center, 898 Biomedical Research Tower, 460 W 12th Ave, Columbus, OH 43210; e-mail: guido.marcucci@osumc.edu; or Ralf Bundschuh, Departments of Physics and Biochemistry, The Ohio State University, 191 W Woodruff Ave, Columbus, OH 43210; e-mail: bundschuh@mps.ohio-state.edu.
References
Author notes
P.Y., D.F., and M.M. contributed equally to this study as first authors.
R.B. and G.M. contributed equally to this study as senior authors.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal