Abstract
The endothelial ETS transcription factor Erg plays an important role in homeostasis and angiogenesis by regulating many endothelial functions including survival and junction stability. Here we show that Erg regulates endothelial cell (EC) migration. Transcriptome profiling of Erg-deficient ECs identified ∼ 80 genes involved in cell migration as candidate Erg targets, including many regulators of Rho- GTPases. Inhibition of Erg expression in HUVECs resulted in decreased migration in vitro, while Erg overexpression using adenovirus caused increased migra-tion. Live-cell imaging of Erg-deficient HUVECs showed a reduction in lamellipodia, in line with decreased motility. Both actin and tubulin cytoskeletons were disrupted in Erg-deficient ECs, with a dramatic increase in tubulin acetylation. Among the most significant microarray hits was the cytosolic histone deacetylase 6 (HDAC6), a regulator of cell migration. Chromatin immunoprecipitation (ChIP) and transactivation studies demonstrated that Erg regulates HDAC6 expression. Rescue experiments confirmed that HDAC6 mediates the Erg-dependent regulation of tubulin acetylation and actin localization. In vivo, inhibition of Erg expression in angiogenic ECs resulted in decreased HDAC6 expression with increased tubulin acetylation. Thus, we have identified a novel function for the transcription factor Erg in regulating HDAC6 and multiple pathways essential for EC migration and angiogenesis.
Introduction
Angiogenesis is a tightly controlled process that involves a cascade of events. A breakthrough in understanding the cellular and molecular mechanisms underlying angiogenesis has been the identification of highly motile, polarized endothelial cells (ECs), named tip cells, which lead the growth of the developing sprout.1 Dynamic reorganization of the cytoskeleton in tip cells supports the projection of lamellipodia and filopodia, which allow the cell to sense the microenvironment for migration cues and guide the sprouting vessel toward an angiogenic stimulus. The functional cross-talk between the actin and tubulin cytoskeletal systems is essential for cell motility.2 Several families of intracellular regulators control these pathways. Among the most studied are the small GTPases of the Rho family, with Rac as the main regulator of lamellipodia and CDC42 as the regulator of filopodia; both of these GTPases have been shown to be involved in angiogenesis.3
The class II histone deacetylase 6 (HDAC6), a known regulator of cell motility,4 has been recently shown to be involved in EC migration and to support angiogenesis.5 HDAC6 is expressed in many cells (including ECs) and localizes predominantly to the cytoplasm, binding to microtubules and deacetylating several cytosolic proteins, including α-tubulin4,6 and cortactin.7 Acetylation of tubulin, characteristic of stabilized microtubules, is thought to contribute to regulating microtubule dynamics,8 while acetylation of cortactin is thought to modulate actin dynamics by regulating the interaction of cortactin with F-actin.7 HDAC6 modulation of both cytoskeletons mediates its role in regulating endothelial migration, cytoskeletal dynamics, and angiogenesis.4,8
We have been interested in the transcriptional regulation of endothelial gene expression and angiogenesis, and have focused on the ETS transcription factor Erg (ETS-related gene). Erg is the most abundantly expressed ETS factor in resting ECs and its expression, although not uniquely endothelial, is restricted mostly to the hematopoietic/endothelial lineage. Erg drives the expression of genes that define the endothelial lineage, such as endoglin, VWF, ICAM-2, and VE-cadherin (reviewed in Randi et al9 ). We and others have shown that Erg is essential for endothelial homeostasis, differentiation, and angiogenesis.10,11 Several Erg target genes have been implicated in angiogenesis (reviewed in Randi et al9 ), and we have shown that Erg-dependent expression of VE-cadherin is essential for endothelial junctional stability and endothelial survival,10 both critical processes in angiogenesis. However, many of the pathways through which Erg exerts its roles are still to be defined.
In this study, we show that Erg regulates endothelial migration and cytoskeletal dynamics. By genome-wide transcriptome profiling of ECs, we identify a network of Erg target genes involved in these pathways. We validate selected hits and focus on HDAC6, one of the most highly regulated genes of this set. We show that Erg drives HDAC6 expression by interacting directly with its regulatory promoter sequences; rescue experiments demonstrate that Erg regulates tubulin acetylation and actin localization via HDAC6. Finally, we confirm these findings in a murine angiogenesis model. Therefore, this study describes a novel pathway in ECs through which Erg regulates angiogenesis and confirms Erg as a master regulator of multiple endothelial functions.
Methods
A supplemental Methods section is available on the Blood Web site (see the Supplemental Materials link at the top of the online article) that includes details of cell culture, transfection conditions, immunofluorescence staining, and video microscopy.
Microarray analysis
Total RNA was prepared from 3 separate HUVEC isolates treated with either control GB or Erg-specific GeneBloc (GB; 100nM) for 24 or 48 hours. Transcript analysis was carried out using an Affymetrix Human Genome U133 Plus 2.0 Array (Almac Diagnostics). The microarray data have been deposited into the NCBI Gene Expression Omnibus portal under accession number GSE32984. Differential gene expression analysis was carried out using the Rosetta Resolver Data Analysis System (Version 7.1; Rosetta Biosoftware). Statistical analysis was performed using false discovery rate (FDR) corrected P values, after Benjamini-Hochberg multiple test correction.
The Database for Annotation, Visualization, and Integrated Discovery (DAVID)12 was used to identify overrepresented gene ontology (GO) categories. The functional clustering tool within DAVID was used to group together GO annotations that have similar gene members and assign an enrichment score (ES). We used an ES > 1.3 (which corresponds to P < .05) to identify genes that may be overrepresented in particular annotation categories.
Gene sets with high enrichment scores were selected from DAVID for further analysis using the ROCK database of genomic and protein interaction datasets.13
Correlation of microarray data from Erg-deficient HUVECs with Erg-overexpressing HEK293 cells14 or with Erg siRNA-treated HUVECs15 were performed using Gene Set Enrichment Analysis software (GSEA, Version 2; http://www.broad.mit.edu/gsea).16
ChIP
ChIP was performed in HUVECs using ChIP-IT (Active Motif) as described.17 Immunoprecipitated DNA was then used as template for real-time PCR, using primers that amplify a region of the HDAC6 promoter18 (GenBank accession NG_012560) containing the putative ETS-binding sites identified using MatInspector software (Genomatrix). See supplemental Table 1 for oligonucleotide sequences and supplemental Figure 6 for the HDAC6 promoter sequence.
Reporter gene assays
An 887-bp region of the HDAC6 promoter proximal to the transcription initiation site, and a smaller nested region of the HDAC6 promoter (465 bp), were PCR amplified from human genomic DNA and cloned into the pGL4 Luciferase Reporter Vector (Promega, and named HDAC6-P1 and P2, respectively; supplemental Figure 6); see supplemental Table 1 for oligonucleotide sequences. Human Erg-2 (p52) cDNA was cloned into pSG5 (Stratagene), as described previously.19 Transactivation assays were performed with the Dual-Luciferase Reporter Assay System (Promega). Cotransfection of HUVECs was performed with GeneJuice transfection reagent (Merck Chemicals), according to the manufacturer's conditions. Briefly, cells were seeded at 5 × 104 per well in a 24-well plate the day before transfection, incubated with 1.5 μL of GeneJuice, 200 ng of reporter DNA, 200 ng of expression plasmid DNA (pSG5Erg or pSG5 empty vector), and 100 ng of pGL4-Renilla Luciferase (Promega) for 48 hours. Luciferase activities were measured with the use of a Synergy HT microplate reader.
siRNA inhibition of Erg in vivo
Inhibition of Erg by incorporating Erg siRNA into in vivo Matrigel plugs was carried out as described previously10 and in more detail in supplemental Methods.
Image acquisition
Images were acquired using a LSM510Meta confocal microscope (Carl Zeiss); for details please see supplemental Methods.
Statistical analyses
Comparisons between groups were made using the Student t test.
Results
Transcriptome analysis of Erg target genes in ECs
Erg is highly expressed in resting, differentiated ECs, and can act both as an activator or a repressor of gene expression.9 Several studies have identified several endothelial genes as transcriptional targets of Erg.10,11,15,17,19 To build a comprehensive picture of how Erg regulates angiogenesis and identify novel pathways regulated by Erg, we carried out a full transcriptome profile by microarray analysis comparing Erg-positive and Erg-deficient ECs. The RNA required for this study was generated from 3 separate single-donor HUVEC cultures, treated with Erg or control antisense (tools we have previously characterized in detail10,11 ) for 24 or 48 hours. Before proceeding to transcriptome analysis, inhibition of Erg expression in these samples was confirmed by real-time PCR (supplemental Figure 1), and the functional efficacy of the inhibition was confirmed by measuring the levels of 2 known Erg target genes (ICAM-2 and VE-cadherin)10,19 (supplemental Figure 1). Gene expression analysis was performed using Affymetrix Human Genome U133 Plus 2.0 Arrays. Data analysis was carried out using the Rosetta Resolver Gene Expression Data Analysis System. After data filtering (1-way ANOVA P < .01, 1.3-fold change cutoff), a total of 1348 genes showed significantly different expression between control GB and Erg GB at 24 hours (930 down-regulated, 418 up-regulated); a total of 2649 genes were differentially expressed by 48 hours (1511 down-regulated, 1138 up-regulated). Figure 1A shows the composite plot of intensity profiles at 24 hours. Known Erg target genes were found in both sets, including ICAM-2 and VE-cadherin in the down-regulated groups and IL-8 and ICAM-1 in the up-regulated groups (data not shown). As an additional validation of our microarray data, we performed Gene Set Enrichment Analysis (GSEA)16 against a microarray dataset published by Yuan et al,15 obtained comparing Erg siRNA-treated to control siRNA-treated HUVECs. As shown in supplemental Figure 2, a positive correlation was found between the down-regulated genes identified in our study and those identified by Yuan et al (normalized enrichment score [NES] 3.64). To identify the major underlying biologic themes within the microarray expression data we used the DAVID bioinformatics resource.12 Changes were identified in the expression of genes associated with angiogenesis, cell-cell adhesion, and apoptosis (not shown), in line with Erg's known roles in ECs. Several potential Erg target genes from the microarray were selected for validation by RT-PCR and some were also validated at the protein level by Western blotting (supplemental Figure 3).
Gene expression profiling after inhibition of Erg in HUVECs. Microarray analysis of differential gene expression in HUVECs was performed at 24 and 48 hours after Erg inhibition by GeneBloc. (A) Composite plot of intensity profiles of the comparison between control and Erg-deficient HUVECs at 24 hours. Genes with > 1.3-fold up-regulation (red) or down-regulation (green) are shown. (B) Analysis of putative gene function for Erg-regulated genes using the DAVID functional annotation and clustering tool reveals enrichment for genes associated with cell migration, organization of the actin and microtubule cytoskeleton, and GTPase regulation. (C) The ROCK software was used to generate a functional interaction network based on the list of Erg-regulated migration hits (see supplemental Table 2). Gene nodes colored in blue are down-regulated, red are up-regulated, and white are nonhit connectors. Black lines represent direct physical interactions; green lines represent complex or indirect interactions. (D) Comparison between microarray datasets was carried out by Gene Set Enrichment Analysis (GSEA).16 The dataset of differentially regulated genes after 24 hours of Erg inhibition in HUVECs was compared with the dataset of genes up-regulated by Erg overexpression in a non-EC line (HEK293).14 GSEA was performed using standard settings with 1000 data permutations. The graphical outputs show enrichment (green curve) of the Erg overexpression gene set along ranked lists of all genes (middle). Below is the full Erg dataset ranked according to intensity. This analysis identified a significant inverse correlation between the genes down-regulated after Erg inhibition in HUVECs and those up-regulated after Erg overexpression in HEK293 cells. Normalized enrichment score (NES) = −1.3540292, P < .001.
Gene expression profiling after inhibition of Erg in HUVECs. Microarray analysis of differential gene expression in HUVECs was performed at 24 and 48 hours after Erg inhibition by GeneBloc. (A) Composite plot of intensity profiles of the comparison between control and Erg-deficient HUVECs at 24 hours. Genes with > 1.3-fold up-regulation (red) or down-regulation (green) are shown. (B) Analysis of putative gene function for Erg-regulated genes using the DAVID functional annotation and clustering tool reveals enrichment for genes associated with cell migration, organization of the actin and microtubule cytoskeleton, and GTPase regulation. (C) The ROCK software was used to generate a functional interaction network based on the list of Erg-regulated migration hits (see supplemental Table 2). Gene nodes colored in blue are down-regulated, red are up-regulated, and white are nonhit connectors. Black lines represent direct physical interactions; green lines represent complex or indirect interactions. (D) Comparison between microarray datasets was carried out by Gene Set Enrichment Analysis (GSEA).16 The dataset of differentially regulated genes after 24 hours of Erg inhibition in HUVECs was compared with the dataset of genes up-regulated by Erg overexpression in a non-EC line (HEK293).14 GSEA was performed using standard settings with 1000 data permutations. The graphical outputs show enrichment (green curve) of the Erg overexpression gene set along ranked lists of all genes (middle). Below is the full Erg dataset ranked according to intensity. This analysis identified a significant inverse correlation between the genes down-regulated after Erg inhibition in HUVECs and those up-regulated after Erg overexpression in HEK293 cells. Normalized enrichment score (NES) = −1.3540292, P < .001.
Erg controls expression of genes involved in actin dynamics and cell migration
Gene ontology analysis of the microarray data identified a significant number of genes belonging to the related categories of cell migration, regulators of the actin and tubulin cytoskeleton and of the small GTPases (Figure 1B). To visualize the relationships between the Erg candidate target genes regulating cell migration, we selected 81 genes (supplemental Table 2) which according to DAVID fell into the categories of cell migration, motility, cytoskeleton, and GTPase regulation, and used the ROCK database of genomic and protein interaction datasets to build molecular interaction networks13 (Figure 1C). The network map generated by ROCK identified several interconnected clusters of candidate Erg target genes (in blue and red, indicating down-regulation or up-regulation after Erg inhibition in HUVECs, respectively); a few genes connected to the Erg hits, but were not themselves candidate Erg targets, are also included (white). Clearly most of the genes identified in this gene subset are positively regulated (directly or indirectly) by Erg, because their expression is decreased after loss of Erg (in blue). Interestingly, most clusters on the network have potential links to angiogenesis. Among the key clusters and Erg candidate targets are regulators of Rho GTPase activity and several tubulin regulators, including the histone deacetylase HDAC6, one of the most highly down-regulated genes (−2.83-fold) to emerge from the analysis. These results clearly show that inhibition of Erg expression in ECs affects a large number of genes involved in the cytoskeleton and in cell motility. We have previously shown that inhibition of Erg expression in HUVECs results in inhibition of angiogenesis in vivo and of endothelial tube formation in vitro,10,11 assays which are strongly dependent on EC migration. Recently, a role for Erg in cell migration was shown in HEK293 cells, a human embryonic kidney cell line, where overexpression of an Erg-tagged protein resulted in increased migration in a transwell assay.14 In this study by Carver et al, a genome-wide analysis of Erg target genes after Erg overexpression in HEK293 cells was also carried out. Because of the significant differences between the 2 approaches (down-regulation of constitutive Erg expression in ECs vs exogenous Erg overexpression in a cell line), we reasoned that any possible similarity between our gene set and that identified in the Carver et al study would considerably support our findings. Correlation of microarray data from Erg-deficient HUVECs with Erg-overexpressing HEK293 cells was performed using GSEA (Figure 1D). Interestingly, a significant inverse correlation (NES −1.35) in the expression profile of Erg-deficient ECs and Erg-overexpressing HEK293 cells was identified, supporting our findings and the hypothesis that many of the pathways identified here are direct targets of Erg. DAVID functional clustering analysis on the common genes identified by GSEA showed that 19% of these were involved in cell migration and motility (Table 1, supplemental Table 3), in line with a wider role for Erg in regulating cell migration.
Putative functional classification of the common genes identified by GSEA
| Gene ontology term . | No. of genes . | Enrichment score . |
|---|---|---|
| Cell migration | 15 | 2.39 |
| Actin cytoskeleton | 10 | 1.57 |
| Microtubule cytoskeleton | 17 | 1.47 |
| Cell adhesion | 11 | 0.95 |
| Small GTPase regulator | 9 | 0.55 |
| Gene ontology term . | No. of genes . | Enrichment score . |
|---|---|---|
| Cell migration | 15 | 2.39 |
| Actin cytoskeleton | 10 | 1.57 |
| Microtubule cytoskeleton | 17 | 1.47 |
| Cell adhesion | 11 | 0.95 |
| Small GTPase regulator | 9 | 0.55 |
DAVID functional clustering analysis was carried out on the common gene dataset obtained from GSEA of the differentially expressed genes at 24 hours after Erg inhibition in endothelial cells and the up-regulated genes identified in HEK293 cells after Erg overexpression.14 The Enrichment Score—the geometric mean (in −log scale) of P values of genes within a corresponding annotation cluster—is used to rank their biological significance.12
GSEA indicates Gene Set Enrichment Analysis Software; and DAVID, Database for Annotation, Visualization & Integrated Discovery.
Erg regulates EC migration
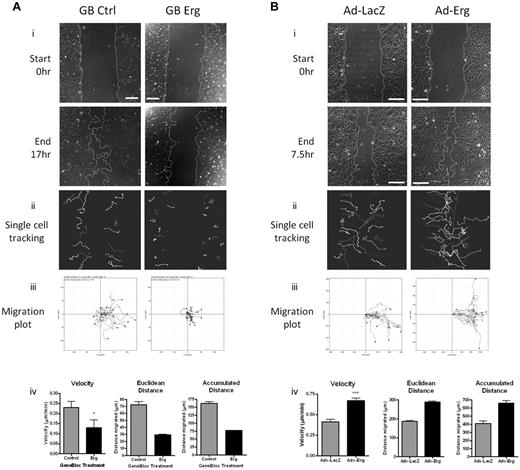
On the strength of these findings, we tested whether Erg regulates EC migration using an in vitro scratch-wound assay. Inhibition of Erg expression in HUVECs resulted in a significant defect in the ability of the cells to migrate into the wounded area compared with controls (Figure 2A, supplemental Videos 1-2). Quantification by single-cell tracking showed that all parameters measured, namely velocity, directionality (Euclidean distance), and accumulated distance, were significantly decreased in Erg-deficient cells (Figure 2Aiv). Erg is a known survival factor for ECs; therefore to rule out the possibility that the defect in cell migration was due to increased cell death, we followed the fate of single cells by time-lapse video microscopy and excluded from quantification those cells which underwent apoptosis within the time of the assay (see supplemental Videos 1-2). To confirm that Erg regulates cell migration, we used the opposite approach and overexpressed human Erg in HUVECs using an adenoviral vector, as previously described.17 As shown in Figure 2B, Erg-overexpressing HUVECs showed increased migration compared with cells infected with control LacZ adenovirus; all the parameters relevant to cell migration were increased in Erg-overexpressing cells compared with control (Figure 2Biv), indicating a dose-dependent effect of Erg on migration. These results indicate that Erg is required for EC migration.
Erg is required for EC migration. (A) HUVECs were transfected with either control-GB FITC or Erg-specific GB FITC for 30 hours. Time-lapse confocal microscopy was used to visualize EC directional migration after a scratch wound. (i) Representative images of scratch wounds at the start and 16 hours after scratching show that compared with control cells (left), directional migration in Erg-deficient cells (right) is reduced. Scale bar, 200 μm. (ii) Single-cell tracking was used to analyze the migratory pathway of individual cells adjacent to the wound. Each track represents the migratory routes of a selected FITC-labeled cell during the course of the 16-hour experiment. (iii) Migration plots showing the migration distance and directionality of single-cell tracks. (iv) Quantification of single-cell velocities, Euclidean distance and accumulated distance were determined from single-cell tracking. Three sets of time-lapse images were captured along each scratch, and 25-40 cells were analyzed per image. *P < .05. Values are mean ± SEM, n = 3 separate experiments. (B) HUVECs were transduced with adenovirus (Ad-LacZ or Ad-Erg) for 48 hours. Endothelial cell directional migration was visualized after a scratch wound by time-lapse confocal microscopy. (i) Representative images showing scratch wounds at the start and at 7.5 hours post-scratch show that compared with Ad-LacZ control cells (left), migration of Erg-overexpressing cells (Ad-Erg, right) is increased. Scale bar, 200 μm. (ii) Single-cell tracking was used to map the migratory pathway of individual cells adjacent to the wound, as above. (iii) Migration plots showing the migration distance and directionality of single-cell tracks. (iv) Quantification of single-cell velocities, Euclidean distance, and accumulated distance were determined from single-cell tracking. Three sets of time-lapse images were captured along each scratch, and 25-40 cells were analyzed per image. ***P < .001. Values are mean ± SEM, n = 3 separate experiments.
Erg is required for EC migration. (A) HUVECs were transfected with either control-GB FITC or Erg-specific GB FITC for 30 hours. Time-lapse confocal microscopy was used to visualize EC directional migration after a scratch wound. (i) Representative images of scratch wounds at the start and 16 hours after scratching show that compared with control cells (left), directional migration in Erg-deficient cells (right) is reduced. Scale bar, 200 μm. (ii) Single-cell tracking was used to analyze the migratory pathway of individual cells adjacent to the wound. Each track represents the migratory routes of a selected FITC-labeled cell during the course of the 16-hour experiment. (iii) Migration plots showing the migration distance and directionality of single-cell tracks. (iv) Quantification of single-cell velocities, Euclidean distance and accumulated distance were determined from single-cell tracking. Three sets of time-lapse images were captured along each scratch, and 25-40 cells were analyzed per image. *P < .05. Values are mean ± SEM, n = 3 separate experiments. (B) HUVECs were transduced with adenovirus (Ad-LacZ or Ad-Erg) for 48 hours. Endothelial cell directional migration was visualized after a scratch wound by time-lapse confocal microscopy. (i) Representative images showing scratch wounds at the start and at 7.5 hours post-scratch show that compared with Ad-LacZ control cells (left), migration of Erg-overexpressing cells (Ad-Erg, right) is increased. Scale bar, 200 μm. (ii) Single-cell tracking was used to map the migratory pathway of individual cells adjacent to the wound, as above. (iii) Migration plots showing the migration distance and directionality of single-cell tracks. (iv) Quantification of single-cell velocities, Euclidean distance, and accumulated distance were determined from single-cell tracking. Three sets of time-lapse images were captured along each scratch, and 25-40 cells were analyzed per image. ***P < .001. Values are mean ± SEM, n = 3 separate experiments.
Erg regulates actin dynamics and lamellipodium formation in ECs
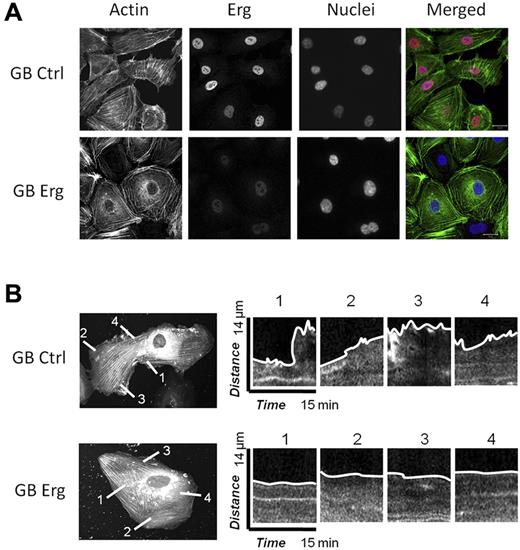
Cell migration and motility rely on the dynamic regulation of the actin cytoskeleton,20 and several of the hits from the Erg microarray study pointed to regulators of actin dynamics. In the migration assay, we observed that Erg-deficient cells appeared rounded and with limited cytoplasmic movement. Staining of control or Erg-deficient HUVECs for actin confirmed that Erg affects actin distribution (Figure 3A): in control, subconfluent HUVECs, actin appeared in stress fibers stretched across the cell body, and also concentrated in the lamellipodia at the edge of the cells. In Erg-deficient cells, however, actin fibers appeared concentrated in bundles at the periphery of the cell, with less intense staining in cytoskeletal protrusions, suggesting dysregulation of the actin cytoskeleton. The effect on actin distribution was quantified by counting the numbers of cells that showed a predominance of either peripheral actin bundles or stress fibers after GB treatment. In control-GB cells, 8% showed actin bundles mostly at the periphery, compared with the majority of cells where actin stress fibers were mostly distributed across the entire cell body. In Erg-GB cells, 78% showed peripheral actin bundles. We then focused on the lamellipodia, which appear as flat, flap-like structures that move around the cell body and at the leading edge of a migrating cell. To visualize single-cell movements, we carried out interference reflection microscopy (IRM) which focuses on the contact surface between the cell and the extracellular matrix. Time-lapse IRM on control or Erg-deficient HUVECs (treated with GB as above for 48 hours) confirmed that the dynamic protrusions of lamellipodia and membrane ruffling, clearly visible in control cells, were significantly reduced in the absence of Erg (supplemental Videos 3-4). To further investigate this effect, we carried out live-cell imaging of individual HUVECs transfected with actin–red fluorescent protein (RFP) and either control or Erg GB. Time-lapse images were collected every 15 seconds for 15 minutes. As shown in Figure 3B, lamellipodia protruded from the control cells. In contrast, in Erg-deficient cells very few lamellipodia were observed; the cells looked less dynamic and less mobile. Kymograph analysis of the plasma membrane showed a significant decrease in membrane movement in Erg-deficient cells compared with control cells (Figure 3B). These results demonstrate that Erg is required for actin dynamics and lamellipodium formation.
Erg regulates the actin cytoskeleton and lamellipodia dynamics. (A) HUVECs were transfected with control-GB or Erg-specific GB for 40 hours. Cells were trypsinised and replated onto fibronectin-coated glass coverslips for 6 hours before being fixed, labeled and visualized for actin (green), Erg (red), and nuclear marker DAPI (blue). Scale bar, 20 μm. After Erg inhibition, actin stress fibers are redistributed into cortical rings around the cell periphery with loss of stress fibers across the cell body. In addition, Erg-deficient cells exhibit a pronounced angular shape. (B) HUVECs were transfected with actin-RFP and allowed to adhere to fibronectin-coated glass chamber slides for 24 hours before transfection with FITC-conjugated control or Erg-specific GB for an additional 24 hours. GeneBloc-transfected cells were identified by the presence of FITC fluorescence in the nucleus (not shown). In these cells, some actin stress fibers are present across the cell body, probably because of actin overexpression. Actin-RFP dynamics and lamellipodia formation were visualized using time-lapse confocal microscopy with images captured every 15 seconds over a period of 15 minutes. Representative images show dynamics of lamellipodia formation in control (top) and Erg cells (bottom). Kymographs show changes in actin-RFP fluorescence intensity at selected points across the cell membrane. In control cells, dynamic actin-rich lamellipodia and membrane protrusions are observed at multiple points around the cell membrane, while inhibition of Erg results in a dramatic reduction in the formation and collapse of actin-containing lamellipodia and the membrane appears static.
Erg regulates the actin cytoskeleton and lamellipodia dynamics. (A) HUVECs were transfected with control-GB or Erg-specific GB for 40 hours. Cells were trypsinised and replated onto fibronectin-coated glass coverslips for 6 hours before being fixed, labeled and visualized for actin (green), Erg (red), and nuclear marker DAPI (blue). Scale bar, 20 μm. After Erg inhibition, actin stress fibers are redistributed into cortical rings around the cell periphery with loss of stress fibers across the cell body. In addition, Erg-deficient cells exhibit a pronounced angular shape. (B) HUVECs were transfected with actin-RFP and allowed to adhere to fibronectin-coated glass chamber slides for 24 hours before transfection with FITC-conjugated control or Erg-specific GB for an additional 24 hours. GeneBloc-transfected cells were identified by the presence of FITC fluorescence in the nucleus (not shown). In these cells, some actin stress fibers are present across the cell body, probably because of actin overexpression. Actin-RFP dynamics and lamellipodia formation were visualized using time-lapse confocal microscopy with images captured every 15 seconds over a period of 15 minutes. Representative images show dynamics of lamellipodia formation in control (top) and Erg cells (bottom). Kymographs show changes in actin-RFP fluorescence intensity at selected points across the cell membrane. In control cells, dynamic actin-rich lamellipodia and membrane protrusions are observed at multiple points around the cell membrane, while inhibition of Erg results in a dramatic reduction in the formation and collapse of actin-containing lamellipodia and the membrane appears static.
Erg regulates tubulin acetylation in ECs
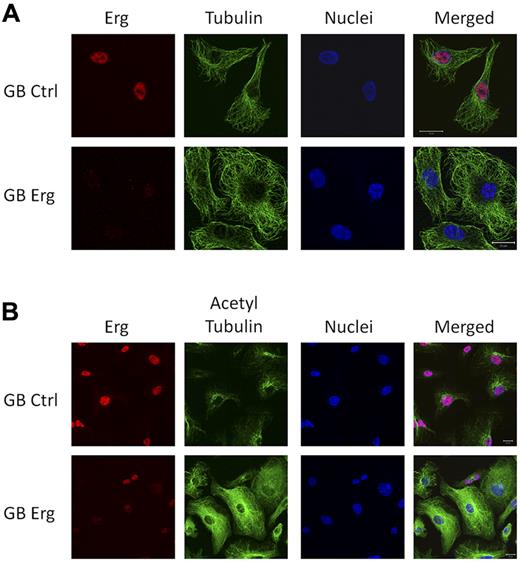
The tubulin cytoskeleton is also crucially involved in cell migration.2 Microtubules are filamentous structures, composed of α- and β-tubulin heterodimers which undergo rapid cycles of assembly and disassembly. Tubulin acetylation has been linked with stabilization of the microtubules (reviewed in Hammond et al21 ). Several of the genes identified in the Erg microarray are known regulators of microtubules (Figure 1B). We therefore investigated the expression, distribution, and acetylation state of tubulin after Erg inhibition in HUVECs. The distribution of tubulin showed no significant difference between control and Erg-deficient HUVECs (Figure 4A). However, tubulin acetylation was dramatically increased in Erg-deficient cells compared with control (Figures 4B, 5B), and was present throughout the cytoplasm, in striking contrast with the distribution in control HUVECs, where acetylated tubulin concentrates in the perinuclear region (Figure 4B, supplemental Figure 4). Tubulin levels were not affected by loss of Erg, as shown by Western blotting (Figure 5B). Thus, Erg regulates tubulin acetylation in ECs.
Erg regulates tubulin acetylation. HUVECs grown on gelatin-coated coverslips were transfected with either control-GB or Erg-specific GB for 48 hours. (A) Cells were labeled and visualized for Erg (red), tubulin (green), and nuclear marker TOPRO3 (blue). The microtubule network appears similar in control and Erg-deficient cells. Scale bar, 20 μm. (B) Cells were labeled and visualized for Erg (red), acetylated tubulin (green), and nuclear marker TOPRO3 (blue). Scale bar, 20 μm. In control cells, acetylated tubulin concentrates at the perinuclear region, as expected, whereas in Erg-deficient cells, acetylated tubulin is found throughout the cytoplasm.
Erg regulates tubulin acetylation. HUVECs grown on gelatin-coated coverslips were transfected with either control-GB or Erg-specific GB for 48 hours. (A) Cells were labeled and visualized for Erg (red), tubulin (green), and nuclear marker TOPRO3 (blue). The microtubule network appears similar in control and Erg-deficient cells. Scale bar, 20 μm. (B) Cells were labeled and visualized for Erg (red), acetylated tubulin (green), and nuclear marker TOPRO3 (blue). Scale bar, 20 μm. In control cells, acetylated tubulin concentrates at the perinuclear region, as expected, whereas in Erg-deficient cells, acetylated tubulin is found throughout the cytoplasm.
Erg regulates HDAC6 expression. (A) HUVECs were transfected with either control-GB or Erg-specific GB for 48 hours. Message RNA levels for HDAC6, determined using real-time PCR, are normalized to GAPDH and expressed relative to control-GB. ***P < .001. Values are mean ± SEM, n = 3. (B) HUVECs were treated with control or Erg-specific GB, as above. Whole-cell protein extracts were analyzed by SDS-PAGE immunoblotting with Abs to Erg, HDAC6, and acetylated tubulin. In Erg-deficient cells, HDAC6 protein expression was reduced while acetylated tubulin was increased compared with controls. Total tubulin levels appear unchanged between treatments. *P < .05, ***P < .001. Values are mean ± SEM, n = 3. (C) ChIP from a confluent HUVEC monolayer was performed as described in “Methods.” Genomic DNA obtained after immunoprecipitation, using a rabbit anti-Erg polyclonal Ab or a negative control rabbit IgG, was used as template in real-time PCR with primers spanning a region of the HDAC6 promoter containing 6 ETS-binding sites (see supplemental Figure). Enhanced enrichment of Erg-immunoprecipitated DNA compared with IgG-immunoprecipitated DNA, relative to total chromatin, indicates that Erg binds to the HDAC6 promoter. Primers annealing in the HDAC6 locus, outside the promoter region, were used as negative control. *P < .05. Values are mean ± SEM, n = 6. (D) An Erg cDNA expression plasmid (pSG5Erg) or an empty expression plasmid (pSG5) were cotransfected with HDAC6 promoter-luciferase constructs (HDAC6-P1 or HDAC6-P2) in HUVECs, and luciferase activity was measured. Values are represented as the fold change in relative luciferase activity over the empty pGL4 vector alone. Erg transactivates both HDAC6 promoter constructs, although to a greater extent using the longer sequence (HDAC6-P1), *P < .05. Values are mean ± SEM, n = 3. (E) HUVECs were transfected with either wtHDAC6-EGFP or an enzymatically dead HDAC6 mutant (mutHDAC6-EGFP) and allowed to adhere to gelatin-coated glass chamber slides for 24 hours before transfection with control GB or Erg-specific GB for an additional 24 hours. Immunofluorescence confocal microscopy was used to visualize cells expressing GFP (green); cells were labeled for acetylated tubulin (red), Erg (purple), and nuclear marker DAPI (blue). Scale bar, 20 μm. For all images, the same intensity settings were used for acetylated tubulin detection which results in some saturation of the signal in the 2 bottom rows. (F) HUVECs were transfected as above. Immunofluorescence confocal microscopy was used to visualize cells expressing GFP (green); cells were labeled for actin (red), Erg (purple), and nuclear marker DAPI (blue). Scale bar, 20 μm. In the Erg-deficient cells, note the cell adjacent to the wtHDAC6-EGFP–expressing one; this untransfected cell still exhibits the peripheral actin redistribution after Erg inhibition as observed in the mutHDAC6-expressing cells and in the Erg GB-treated cells in Figure 3A.
Erg regulates HDAC6 expression. (A) HUVECs were transfected with either control-GB or Erg-specific GB for 48 hours. Message RNA levels for HDAC6, determined using real-time PCR, are normalized to GAPDH and expressed relative to control-GB. ***P < .001. Values are mean ± SEM, n = 3. (B) HUVECs were treated with control or Erg-specific GB, as above. Whole-cell protein extracts were analyzed by SDS-PAGE immunoblotting with Abs to Erg, HDAC6, and acetylated tubulin. In Erg-deficient cells, HDAC6 protein expression was reduced while acetylated tubulin was increased compared with controls. Total tubulin levels appear unchanged between treatments. *P < .05, ***P < .001. Values are mean ± SEM, n = 3. (C) ChIP from a confluent HUVEC monolayer was performed as described in “Methods.” Genomic DNA obtained after immunoprecipitation, using a rabbit anti-Erg polyclonal Ab or a negative control rabbit IgG, was used as template in real-time PCR with primers spanning a region of the HDAC6 promoter containing 6 ETS-binding sites (see supplemental Figure). Enhanced enrichment of Erg-immunoprecipitated DNA compared with IgG-immunoprecipitated DNA, relative to total chromatin, indicates that Erg binds to the HDAC6 promoter. Primers annealing in the HDAC6 locus, outside the promoter region, were used as negative control. *P < .05. Values are mean ± SEM, n = 6. (D) An Erg cDNA expression plasmid (pSG5Erg) or an empty expression plasmid (pSG5) were cotransfected with HDAC6 promoter-luciferase constructs (HDAC6-P1 or HDAC6-P2) in HUVECs, and luciferase activity was measured. Values are represented as the fold change in relative luciferase activity over the empty pGL4 vector alone. Erg transactivates both HDAC6 promoter constructs, although to a greater extent using the longer sequence (HDAC6-P1), *P < .05. Values are mean ± SEM, n = 3. (E) HUVECs were transfected with either wtHDAC6-EGFP or an enzymatically dead HDAC6 mutant (mutHDAC6-EGFP) and allowed to adhere to gelatin-coated glass chamber slides for 24 hours before transfection with control GB or Erg-specific GB for an additional 24 hours. Immunofluorescence confocal microscopy was used to visualize cells expressing GFP (green); cells were labeled for acetylated tubulin (red), Erg (purple), and nuclear marker DAPI (blue). Scale bar, 20 μm. For all images, the same intensity settings were used for acetylated tubulin detection which results in some saturation of the signal in the 2 bottom rows. (F) HUVECs were transfected as above. Immunofluorescence confocal microscopy was used to visualize cells expressing GFP (green); cells were labeled for actin (red), Erg (purple), and nuclear marker DAPI (blue). Scale bar, 20 μm. In the Erg-deficient cells, note the cell adjacent to the wtHDAC6-EGFP–expressing one; this untransfected cell still exhibits the peripheral actin redistribution after Erg inhibition as observed in the mutHDAC6-expressing cells and in the Erg GB-treated cells in Figure 3A.
Erg drives expression of histone deacetylase HDAC6
The remarkable phenotypes observed in Erg-deficient cells combined with the large number of microarray hits suggest that multiple pathways involved in cell migration and cytoskeletal dynamics are regulated by Erg. To begin to determine the molecular basis of this process, we focused on one of the most highly down-regulated genes identified in the microarray study, namely HDAC6, a cytoplasmic enzyme constitutively expressed in ECs which acetylates α-tubulin and cortactin, regulating cell motility and angiogenesis.5-7 HDAC6 mRNA and protein levels were significantly decreased in Erg-deficient HUVECs, and this correlated with a significant increase in tubulin acetylation compared with control cells (Figure 5A-B). Similar regulation of HDAC6 levels and tubulin acetylation was observed after Erg inhibition in primary human dermal microvascular ECs (HMVECs; supplemental Figure 5). To test whether Erg drives HDAC6 transcription directly, we examined the HDAC6 promoter18 for ETS-binding sites, using MatInspector software. ETS transcription factors bind specifically to the core DNA consensus motif GGA-A/T22 ; oligonucleotide screens and ChIPSeq analysis identified a longer consensus motif, [A/C]GGAA[G/A] or AGGA[A/T][G/A], which has been proposed as a specific Erg-binding site.23,24 Nine putative ETS-binding sites (EBS) are located in a 1.2-kb region upstream from the HDAC6 transcription start site; 6 EBS are clustered in an ∼ 200-bp region and 2 of these EBS match the Erg consensus binding motif (supplemental Figure 6). ChIP assay showed that Erg binds to this ∼ 200-bp region of the HDAC6 promoter (Figure 5C), suggesting that Erg regulates HDAC6 expression. To test directly the ability of Erg to drive HDAC6 expression, we used a luciferase reporter system. An 887-bp region of the HDAC6 promoter proximal to the transcription initiation site, which contained the region covered by the ChIP analysis, was cloned into the pGL4-luciferase vector (HDAC6-P1; supplemental Figure 6). In addition, a smaller nested fragment of the HDAC6 promoter (465 bp), containing the ∼ 200-bp region and the cluster of EBS, was also cloned into pGL4-luciferase (HDAC6-P2; supplemental Figure 6). HDAC6 promoter luciferase constructs were coexpressed in HUVECs along with a plasmid encoding Erg-2, or control plasmid. As shown in Figure 5D, both HDAC6 luciferase promoter constructs were transactivated by Erg-2 overexpression in HUVECs, with HDAC6-P1 showing a 4-fold and HDAC6-P2 a 3-fold increase. These results demonstrate that Erg drives transcription of HDAC6 in ECs.
Erg regulates cytoskeletal dynamics and tubulin acetylation via HDAC6
As discussed, HDAC6 has been shown to regulate cell migration and cytoskeletal dynamics. We therefore proceeded to test whether HDAC6 mediates some of the Erg-dependent phenotypes described here. As shown above (Figure 4B), loss of Erg caused increased tubulin acetylation in HUVECs. To test whether this was due to the loss of HDAC6 deacetylating activity, we overexpressed wild-type (WT) HDAC6 or an enzymatically dead HDAC6 mutant in these cells.25,26 As shown in Figure 5E, tubulin acetylation in Erg-deficient cells was normalized by overexpression of WT HDAC6 but not mutant HDAC6. This confirms that Erg-dependent regulation of tubulin acetylation is mediated by HDAC6.
HDAC6 has also been shown to regulate the acetylation of the F-actin–binding protein cortactin, which regulates actin-dependent cell motility.7 To investigate whether the defect in actin polymerization after Erg inhibition in HUVECs (Figure 3A) was due, at least in part, to loss of HDAC6 activity, we overexpressed WT or mutant HDAC6 in these cells. As shown in Figure 5F, overexpression of WT but not mutant HDAC6 in Erg-deficient cells was able to partly normalize the distribution of actin stress fibers throughout the cell body.
Together, these data identify HDAC6 as a novel endothelial target of the transcription factor Erg and a mediator of Erg's role in regulating the microtubule and actin cytoskeleton and therefore cell migration.
Erg regulates HDAC6 expression and tubulin acetylation during angiogenesis in vivo
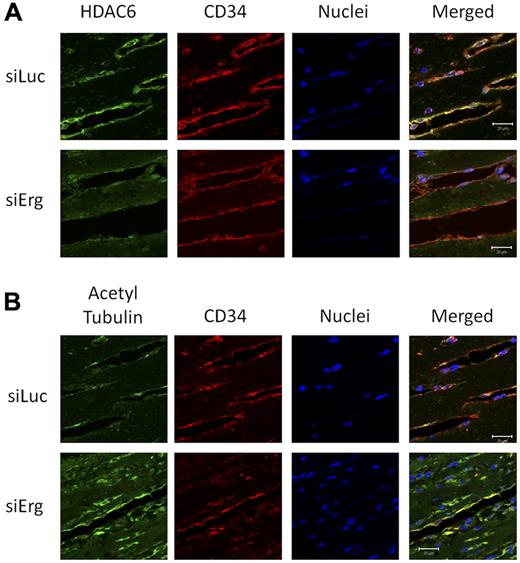
The results described so far identify a novel target for Erg and a novel pathway in the regulation of angiogenesis. To validate our results in angiogenic vessels in vivo, we studied the expression of HDAC6 and its target, acetylated tubulin, in a murine angiogenesis model. Using Erg siRNA in vivo in a Matrigel plug model of angiogenesis, we have previously shown that Erg inhibition results in a dramatic decrease in neovascularization of the plugs (supplemental Figure 7 and Birdsey et al10 ). To test whether Erg regulates HDAC6 expression and tubulin acetylation in angiogenic ECs in vivo, plug sections were immunostained with Abs to HDAC6 and acetylated tubulin. Blood vessels were visualized by immunostaining for CD34. In control siRNA (Luc-siRNA)–treated mice, strong HDAC6 immunostaining was found localized to ECs lining blood vessels (Figure 6A). However, in the Erg-siRNA–treated mice, HDAC6 staining in the plug sections was reduced. In parallel, a marked increase in acetylated tubulin in blood vessels was observed in the Erg-siRNA–treated mice compared with the Luc-siRNA controls (Figure 6B). These data demonstrate that the regulation by Erg of HDAC6 expression and tubulin acetylation occurs in vivo.
Inhibition of HDAC6 and hyperacetylation of tubulin in vivo by Erg siRNA. Matrigel mixture containing either luciferase control siRNA or Erg-specific siRNA (2μM) with bFGF (80 ng/mL) was injected subcutaneously into C57BL/6 mice. Matrigel plugs were harvested 7 days after implantation, fixed, and sectioned. (A) Sections from Matrigel plugs treated with siRNA for luciferase (top) or Erg (bottom) were stained with Abs to HDAC6 (green), CD34 (red), and with the nuclear marker TOPRO3 (blue) and visualized by immunofluorescence confocal microscopy. Scale bar, 20 μm. HDAC6 staining appears less intense in ECs lining the neovessels in the Erg-siRNA–treated samples compared with the control siRNA sample. (B) Sections from Matrigel plugs containing siRNA for luciferase (top) or Erg (bottom) were stained with Abs to acetylated tubulin (green), CD34 (red), and with the nuclear marker TOPRO3 (blue) and visualized by immunofluorescence confocal microscopy. Scale bar, 20 μm. In Matrigel sections containing Erg siRNA, ECs show an increased acetylated tubulin staining compared with the cells from control siRNA samples.
Inhibition of HDAC6 and hyperacetylation of tubulin in vivo by Erg siRNA. Matrigel mixture containing either luciferase control siRNA or Erg-specific siRNA (2μM) with bFGF (80 ng/mL) was injected subcutaneously into C57BL/6 mice. Matrigel plugs were harvested 7 days after implantation, fixed, and sectioned. (A) Sections from Matrigel plugs treated with siRNA for luciferase (top) or Erg (bottom) were stained with Abs to HDAC6 (green), CD34 (red), and with the nuclear marker TOPRO3 (blue) and visualized by immunofluorescence confocal microscopy. Scale bar, 20 μm. HDAC6 staining appears less intense in ECs lining the neovessels in the Erg-siRNA–treated samples compared with the control siRNA sample. (B) Sections from Matrigel plugs containing siRNA for luciferase (top) or Erg (bottom) were stained with Abs to acetylated tubulin (green), CD34 (red), and with the nuclear marker TOPRO3 (blue) and visualized by immunofluorescence confocal microscopy. Scale bar, 20 μm. In Matrigel sections containing Erg siRNA, ECs show an increased acetylated tubulin staining compared with the cells from control siRNA samples.
Discussion
The ETS transcription factor Erg is increasingly recognized as a master regulator of endothelial biology. Erg drives expression of genes that define the endothelial lineage, such as VWF, VE-cadherin, and ICAM-2 and is a key regulator of vascular development and angiogenesis (reviewed in Randi et al9 ). Erg also acts as a repressor of gene expression and controls endothelial homeostasis (Yuan et al15 and Sperone et al17 ), thus possibly representing an exciting novel target for the prevention of vascular diseases. Understanding its full role in ECs is essential to be able to consider strategies to modulate its function in the endothelium. Here we describe a novel function for Erg as regulator of EC motility and migration, processes which are critical to angiogenesis and wound healing, and we identify a new molecular pathway linking Erg to the histone deacetylase HDAC6.
Cell migration is a complex process involving dynamic rearrangement of the cytoskeletons.27 The multiple phenotypes observed in Erg-deficient cells indicate a widespread defect in both actin and tubulin cytoskeletons. The results of the transcriptome analysis of Erg-deficient cells show that multiple pathways are likely to be involved in these defects. The challenge in such studies is, as ever, to identify direct-versus-indirect molecular effects of the intervention, in our case loss of Erg expression. We chose to focus on HDAC6, one of the most highly down-regulated hits in the study, because of the multiple roles this protein plays in regulating cell motility and the cytoskeletons, its high expression in ECs, and its recently identified role in angiogenesis. In this study, we show that HDAC6 mediates the Erg-dependent regulation of actin and tubulin cytoskeletons. HDAC6 was originally cloned as a member of the histone deacetylase family,28,29 but was later shown to be a predominantly cytoplasmic deacetylase of nonhistone proteins, including α-tubulin and cortactin.6,7 Tubulin acetylation affects its polymerization, which is involved in directional cell migration and lamellipodia formation. Whether microtubule deacetylation alone is sufficient to promote cell migration is still controversial; however, microtubule stability has been shown to affect actin-dependent cell motility through the small GTPases Rac-1 and Rho.30 Possibly through multiple mechanisms, HDAC6 regulates cell migration: overexpression of HDAC6 in NIH-3T3 cells leads to increased motility,6 while inhibition of HDAC6 activity using tubacin, a small molecule HDAC6 inhibitor that binds to its tubulin deacetylation catalytic domain, decreases migration and tumor cell invasiveness.31
Based on the results of the microarray study and our own previous data, we propose that HDAC6 is one of several pathways and target genes involved in Erg's regulation of angiogenesis. Clearly HDAC6 does not recapitulate all of Erg's function in ECs. The HDAC6-deficient mouse is viable with no overt defect in vascular development,32 while an Erg transgenic mouse (ErgMld2) where mutation in the DNA-binding domain abolishes its activity as a transactivator (likely to cause the loss of some but not all of Erg's functions) has been reported to die in utero and to display severe defects in hematopoiesis, as well as partial disruption of the vascular network.33 In support of these observations, unpublished data from our laboratory show that complete deletion of Erg in ECs in mice results in embryonic lethality (G.M.B. and A.M.R., manuscript in preparation).
Other candidate Erg target genes identified in this study, some of which were independently validated, provide a clear picture of the fundamental role that Erg plays in regulating the ability of the cells to maintain their structure and move in response to stimuli. The connections between clusters in the network analysis highlight the widespread effect that Erg inhibition has on the cell's machinery regulating movement, shape and adhesion. Although the classic GTPases known to be involved in angiogenesis (Rac, Rho, and CDC42) were not in the list of hits, the presence of crucial regulators of their activity suggests that Erg may regulate, possibly indirectly, small GTPases activation and downstream function. The complex network of genes that are regulated by Erg suggests that some but possibly not all will be direct transcriptional targets; further studies will address this question.
The main limitation of this study is the focus on one single Erg microarray hit, HDAC6, when other hits are possibly also involved in the phenotypes under investigation. This is a common issue with genomic studies, because of the complexity and time requirements of multiple target analysis. Extended studies and crucially advanced bioinformatic analysis will be required to validate multiple pathways and therefore determine the contribution of the Erg-HDAC6 axis within the wider context.
The findings reported in this study are clearly relevant to angiogenesis and wound healing. The molecular mechanisms that regulate the ability of endothelial tip cells to move toward the proangiogenic gradient and thus drive the growth of new sprouting vessels are the focus of intense research. Interestingly, a recent report on tip cell–enriched genes in DLL4+/− mouse mutants identified Erg as up-regulated in tip cells.34 The role of Erg in the migration of tip cells is currently under investigation.
These findings are of interest not only in the context of endothelial homeostasis and angiogenesis, but also for some malignancies. In recent years, the importance of Erg in cancers has become clear. Erg overexpression or increased activity has been identified in various cancers, including sarcoma, acute myeloid leukemia, and prostate cancer—indeed Erg gene rearrangements are found in ∼ 80% of primary prostate cancer (reviewed in Clark and Cooper35 ). Erg acts as an oncogene, promoting tumor cell growth and cell migration14,36 ; however, the targets mediating these effects in cancer are still unknown. Comparison of the gene expression profile of Erg-overexpressing cells (a nonendothelial transformed line) with Erg-deficient ECs using the GSEA software showed a remarkable negative correlation, supporting the model that the pathways identified here may have important implications not only for endothelial migration, and therefore angiogenesis and wound healing, but also for the invasiveness of cancer cells. Further studies will determine the overlap between the candidate targets identified here and in prostate cancer transcriptome studies.
As well as regulating angiogenesis, it is increasingly clear that a crucial function of Erg in ECs is to maintain homeostasis. Recently, we and others have shown that Erg can act to maintain the anti-inflammatory phenotype of resting ECs by repressing inflammatory gene expression, and can actively inhibit TNF-induced inflammation in vivo.15,17 Regulation of cytoskeletal dynamics is crucial for the ability of a cell to move, but is also essential to maintain cell shape, respond to stimuli and divide, and in general for homeostasis. Therefore, these results identify another mechanism though which Erg controls endothelial function, and stress the central role that Erg plays in endothelial biology in health and disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Note added in proof.
While this manuscript was under review, 2 articles were published that support our findings. Yuan et al reported that Erg regulates endothelial expression of RhoJ, one of the genes also identified in our study, and that this pathway was involved in lumen formation.37 Kaluza et al described in detail the role of HDAC6 in regulating angiogenesis and cell migration, and its link to regulation of actin dynamics.38
Acknowledgments
The authors are grateful to Prof Francisco Sanchez-Madrid (Universidad Autónoma de Madrid, Madrid, Spain) for the gift of the HDAC6 plasmids; Dr Richard Starke (Imperial College London, London, United Kingdom) and Dr Vicky Heath (University of Birmingham, Birmingham, United Kingdom) for helpful comments and discussion; Dr Laurence Game (Medical Research Council, London, United Kingdom) for advice on the gene expression profiling study; and Ms Lorraine Lawrence (Imperial College London, London, United Kingdom) for technical support.
This study was supported by grants from the British Heart Foundation.
Authorship
Contribution: G.M.B. developed tools and assays, performed research, data analysis, and interpretation, generated reagents, expanded the original research design, and contributed to manuscript writing; N.H.D. developed assays to perform specific aspects of the research, and contributed to data analysis and interpretation; A.V.S., R.H., and M.D.H. performed specific aspects of research; M.P. contributed reagents and techniques, and contributed to data analysis and interpretation and to manuscript writing; D.O.H. and J.C.M. contributed to data interpretation and manuscript writing; M.Z. and B.G. contributed bioinformatic expertise and tools to carry out data analysis and interpretation; A.J.R. provided reagents and expertise, and contributed to the experimental plan, data analysis, and manuscript writing; and A.M.R. conceived the idea, designed the experimental plan, supervised research, carried out data analysis and interpretation, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anna M. Randi, MD, PhD, NHLI Vascular Sciences, Hammersmith Hospital, Imperial College London, Du Cane Rd, London W12 0NN, United Kingdom; e-mail: a.randi@imperial.ac.uk.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal