A polymorphic variant of the phosphatase PTPN22 has been associated with increased risk for multiple autoimmune diseases. The risk allele is thought to function by diminishing antigen-receptor signals responsible for negative selection of autoreactive lymphocytes. We now show that PTPN22 is markedly overexpressed in chronic lymphocytic leukemia (CLL), a common malignancy of autoreactive B lymphocytes. We also show that overexpression of PTPN22 significantly inhibits antigen-induced apoptosis of primary CLL cells by blocking B-cell receptor (BCR) signaling pathways that negatively regulate lymphocyte survival. More importantly, we show that PTPN22 positively regulates the antiapoptotic AKT kinase, which provides a powerful survival signal to antigen-stimulated CLL cells. This selective uncoupling of AKT from other downstream BCR signaling pathways is a result of inhibition of a negative regulatory circuit involving LYN, CD22, and SHIP. Finally, we show that PTPN22 can be effectively down-regulated by the PKC inhibitors ruboxistaurin and sotrastaurin, resulting in enhanced killing of CLL cells exposed to proapoptotic BCR stimuli. Collectively, these data suggest that PTPN22 overexpression represents a protective mechanism that allows autoantigen-activated CLL cells to escape from negative selection and indicate that this mechanism could be exploited for therapeutic purposes by targeting PTPN22 with PKC inhibitors.
Introduction
Chronic lymphocytic leukemia (CLL) is a common lymphoid malignancy characterized by the expansion and progressive accumulation of mature B lymphocytes that coexpress the T-cell antigen CD5 and B cell surface antigens CD19, CD20, and CD23. The disease has a highly variable clinical course, ranging from rapid progression with fatal outcome to a relatively indolent behavior with normal life expectancy.1
Several lines of evidence suggest that chronic antigen drive plays an important role in the pathogenesis of CLL.1,2 First, the malignant B cells from different patients frequently express similar or identical B-cell receptors (BCRs), suggesting that they recognize the same antigens and that these antigens drive the initial expansions of the malignant clones.3 Second, freshly isolated CLL cells show increased expression of BCR target genes and reduced expression of surface IgM, indicating that they are continuously triggered by antigen in vivo.4,–6 Third, there is a strong correlation between clinical course and certain BCR-related features, such as the mutational status of the immunoglobulin heavy-chain variable (IGHV) genes and ZAP-70 expression, suggesting that BCR signals also play a role during disease progression.7,–9 Lastly, early clinical trials with agents that target the BCR signaling pathway, such as inhibitors of SYK, BTK, and PI3Kδ, are showing considerable activity in patients with CLL, further suggesting that the leukemic cells rely on BCR signals for growth and survival.10,–12
Despite all this evidence, the malignant B cells also display certain features that appear contradictory to the concept that the disease is antigen-driven. These include the frequent autoreactivity of the leukemic cell BCRs,13,,,–17 which in principle would be expected to lead to negative rather than positive selection, and the reduced capacity of the leukemic cells to transduce BCR signals, as evidenced by the less efficient activation of various downstream signaling molecules, including SYK, PLCγ2, NF-κB, JNK, and p38MAPK.6,18,,–21
BCR engagement by antigen in normal and CLL cells triggers a signaling cascade, which, depending on signal intensity, signal duration, and availability of costimulatory signals, can induce a wide range of responses, including proliferation, differentiation, survival, anergy, and apoptosis.21,22 The BCR signal is initially propagated by SRC-family kinases, such as LYN, FYN, and BLK, which phosphorylate the immunoreceptor tyrosine-based activation motifs in the Ig-α and Ig-β chains of the BCR. The kinase SYK is subsequently recruited to the phosphorylated immunoreceptor tyrosine-based activation motifs and becomes activated through SRC-family kinase-dependent phosphorylation and autophosphorylation. SYK further propagates the signal by activating or interacting with various signaling intermediates, including BLNK, BTK, PI3K, PLCγ2, VAV, and RAS. These intermediates then activate downstream signaling molecules, such as the kinases AKT, PKC, ERK, JNK, and p38MAPK, and the transcription factors NF-κB and NFAT.
The intensity and duration of the BCR signal are controlled by various negative regulators, including inhibitory receptors, phosphatases, and ubiquitin ligases. Importantly, some of these negative regulators are also activated by LYN, which functions as both a positive and negative regulator of BCR signaling. This dual role of LYN stems from its unique ability to phosphorylate the immunoreceptor tyrosine-based inhibitory motifs in the inhibitory receptors CD22, FcγRIIb, CD5, and CD72.23 Phosphorylation of these receptors brings the phosphatases SHP-1 and SHIP in the vicinity of the antigen-stimulated BCR, where they terminate the signal by dephosphorylating various activated components of the BCR signaling pathway.
In this study, we investigated whether the reduced capacity of the leukemic cells to transduce BCR signals and undergo negative selection are possibly related to inappropriate expression or function of a negative regulator of antigen-receptor signaling. We focused primarily on the phosphatases SHP-1 and PTPN22, as they are considered principal negative regulators of antigen-receptor signaling in normal B and T lymphocytes, and both have been implicated in the pathogenesis of various lymphocyte disorders. In particular, SHP-1 has been shown to directly dephosphorylate several BCR proximal signaling molecules24,25 and is often down-regulated in lymphoid malignancies.26 PTPN22, on the other hand, has been less extensively studied in B cells but has been shown to terminate T-cell receptor signals by dephosphorylating and inactivating LCK, which is the principal SRC-family kinase in T cells.27 In addition, a polymorphic PTPN22 variant (PTPN22 R620W) has recently emerged as a major risk factor for the development of multiple autoimmune diseases, including insulin-dependent diabetes mellitus, rheumatoid arthritis, and systemic lupus erythematosus,28,–30 suggesting that aberrant activity of this phosphatase can perturb normal lymphocyte functions and homeostasis.
We now report that PTPN22 is markedly overexpressed in CLL and show that overexpression of this phosphatase is at least in part responsible for the inefficient activation of selected BCR signaling pathways, particularly those pathways that negatively regulate the survival of the malignant cells. More importantly, we demonstrate that PTPN22 has a positive effect on the activity of the AKT kinase, which provides critical survival signals to antigen-stimulated CLL cells. This dual activity of PTPN22 could provide a protective mechanism that allows autoreactive B cells to escape negative selection in CLL and possibly other lymphocyte disorders. Importantly, we show that this dual activity of PTPN22 can be exploited for therapeutic purposes to allow selective targeting of the malignant clone in CLL.
Methods
CLL and normal B-cell samples
Blood samples were collected from patients who satisfied standard morphologic and immunophenotypic criteria for B-cell CLL. Informed consent was obtained from all patients according to the Declaration of Helsinki, and approval for the study was obtained from the institutional human research committee at the Catholic University Hospital A. Gemelli. Clinical and laboratory features of the studied patients are provided in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Normal B cells were obtained from tonsils (n = 7) or from peripheral blood (PB) of healthy donors (n = 7).
Mononuclear cells were isolated by Ficoll gradient centrifugation. CLL and normal B cells from 7 tonsillar and 2 PB samples were purified by negative selection with anti-CD3, anti-CD14, and anti-CD16 mouse monoclonal antibodies (kindly provided by Prof Fabio Malavasi, University of Turin, Turin, Italy) and Dynabeads coated with pan anti–mouse IgG antibody (Invitrogen Dynal). Normal B cells from the 5 remaining PB samples were purified by positive selection using CD19 MicroBeads (Miltenyi Biotec). The purity of the selected B-cell populations was evaluated by staining with anti-CD5 R-PE–conjugated and anti-CD19 FITC-conjugated antibodies (BD Biosciences), followed by flow cytometric analysis on a FACSCalibur flow cytometer using CellQuest Version 3.3 software (BD Biosciences). The purity of the negatively selected CLL B cells was more than 97%, negatively selected tonsillar B cells 75% to 85%, negatively selected PB B cells 65% to 75%, and positively selected PB B cells more than 98%.
Cell lines and culture conditions
Freshly isolated CLL B cells, normal B cells, normal T cells, and the lymphoma B-cell lines B104 and BJAB were cultured in RPMI 1640 supplemented with 10% heat-inactivated FBS, 100 U/mL penicillin, 0.1 mg/mL streptomycin, 2mM l-glutamine, and 1mM sodium pyruvate (Invitrogen), at 37°C in the presence of 5% CO2. Stimulations were performed at a cell density of 1 × 107/mL with the following reagents: 10 μg/mL goat F(ab′)2 anti–human IgM, 2 × 107/mL Dynabeads M-450 Epoxy (Invitrogen Dynal) coated with 20 μg goat anti–human IgM (Southern Biotechnology Associates), 100 ng/mL soluble recombinant human CD40L plus 1 μg/mL of enhancer (Alexis Biochemicals), 7.5 μg/mL complete phosphorothioate CpG oligonucleotide 2006 (Microsynth), 1 × 105/mL M2-10B4 bone marrow stromal cells (ATCC-LGC Standards), 250 ng/mL recombinant human B-cell activating factor (R&D Systems), 100 ng/mL recombinant human vascular endothelial growth factor (R&D Systems), 5 μg/mL anti-CD3 mouse monoclonal antibody, and 10 ng/mL 12-O-tetradecanoylphorbol-13-acetate (Sigma-Aldrich) with or without 1μM ionomycin (Invitrogen). The calpain inhibitor calpeptin (Calbiochem-Merck), the proteasome inhibitor MG-132 (Calbiochem-Merck), the pancaspase inhibitor Z-VAD-fmk (BD Biosciences), the p38MAPK inhibitor SB 203580 (Sigma-Aldrich), the PTPN22 inhibitor I-C11, and the protein kinase C inhibitors ruboxistaurin and sotrastaurin (both from Axon Medchem) were used as indicated in the figures or figure legends.
Sequencing analysis
Total cellular RNA was isolated from PBMCs using the Trizol reagent (Invitrogen) according to the manufacturer's instructions. RNA was reverse-transcribed using random hexamers and then PCR amplified with primers reported in supplemental Figure 1. PCR products were purified with the QIAquick PCR purification kit (QIAGEN) and sequenced directly. Sequencing was done with the BigDye Terminator Version 3.1 Cycle Sequencing kit and ABI 3100 genetic analyzer (Applied Biosystems).
Immunoblotting analysis
Cell pellets were lysed in RIPA lysis buffer (10mM Tris-HCl, pH 7.4, 5mM EDTA, 150mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate) containing protease and phosphatase inhibitors (Sigma-Aldrich). The protein concentration of each cell lysate was determined with the RC DC Protein Assay (Bio-Rad). The protein samples were separated by SDS-PAGE and transferred on Immobilon-P polyvinylidene difluoride membranes (Millipore). Membranes were blotted at 4°C with the following antibodies: PTPN22 (R&D Systems), phospho-SHIPY1020, PARP, phospho-SykY352, phospho-SrcY416, phospho-p38MAPKT180/Y182, phospho-SAPK/JNKT183/Y185, phospho-PKCδY505, phospho-AktS473, Akt, phospho-ERKT202/Y204, ERK, phospho-GSK3α/βS21/9, phospho-FoxO1T24/FoxO3aT32, rabbit IgG-HRP, and mouse IgG HRP-linked (Cell Signaling Technology), phospho-SHP-1Y536 (ECM Bioscience), phospho-BLNKY84 (BD Biosciences), SHP-1 (Santa Cruz Biotechnology), phospho-C22Y822 and CD22 (Millipore), or β-actin (Sigma-Aldrich). Immunodetection and quantification were done on a Gel Logic 2200 Imaging System (Eastman Kodak), using ECL Plus enhanced-chemiluminescence detection reagents (GE Healthcare).
PTPN22 knockdown and overexpression experiments
The PTPN22 knockdown experiments were performed using PTPN22 Stealth Select RNAi and Stealth RNAi Negative Control (Invitrogen). The pEF-PTPN22-Myc-His vector (kindly provided by Dr Arthur Weiss) and the control pCDNA3 plasmid (Invitrogen) were purified with the Plasmid Maxi Kit (QIAGEN). Capped and poly(A) tailed mRNAs were prepared using the mMESSAGE mMACHINE T7 Ultra Kit and purified with the MEGAclear kit (both from Ambion). Nucleofection of siRNAs or mRNAs in primary CLL B cells and in the B104 cell line was performed with the Nucleofector system (Amaxa Biosystems) using Nucleofector Solution V or L and the U-013 and C-005 programs, respectively.
Apoptosis assay
The percentages of viable and apoptotic cells were determined by flow cytometric analysis of cells stained with propidium iodide (PI) and annexin-A5–FITC conjugate (Nexins Research). Viable cells are annexin V/PI double negative.
Statistical analysis
Differences in PTPN22 expression between CLL and normal B cells or between the various CLL subsets were evaluated with the Mann-Whitney rank-sum test. The paired t test and Wilcoxon signed-rank test were used to evaluate the significance of differences in leukemic cell survival after PTPN22 down-regulation, overexpression, or inhibition. The relationship between PTPN22 expression and the rate of apoptosis induced by anti-IgM in combination with ruboxistaurin or sotrastaurin was evaluated through the Pearson rank correlation. All statistical analyses were performed using the SigmaStat Version 3.1 program (Systat Software). P values are indicated in the figures.
Results
PTPN22 is significantly overexpressed in CLL B cells
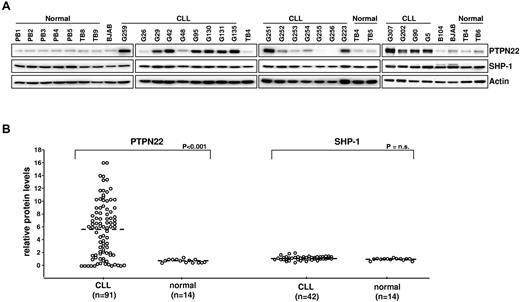
We initially evaluated the levels of PTPN22 and SHP-1 protein in a series of 42 purified PB CLL B cell samples (Figure 1). An additional series of 49 CLL B samples was subsequently analyzed for PTPN22 only. Purified normal B cells from 7 tonsillar and 7 PB samples were analyzed as controls. As shown in Figure 1, tonsilar and PB B cells uniformly expressed small amounts of PTPN22. In contrast, most CLL B-cell samples (> 70%) expressed considerably greater amounts of PTPN22 protein, which ranged from 2-fold to more than 10-fold higher levels than normal B cells (median PTPN22 levels: CLL, 6.1, normal B cells, 0.9, P < .001). Expression of SHP-1 was similar in CLL and normal B cells.
Expression of PTPN22 and SHP-1 in CLL and normal B cells. (A) Cellular extracts from normal PB B cells (PB1, PB2, PB3, PB4, and PB5), normal tonsillar B cells (TB4, TB5, TB6, TB8, and TB9), lymphoma B-cell lines (B104 and BJAB), and CLL B cells were analyzed by immunoblotting with the indicated antibodies. Actin was used as a loading control. (B) Comparison of relative PTPN22 and SHP-1 protein levels in CLL (n = 91 and n = 42, respectively) and normal B cells (n = 14). The BJAB cell line was used as a calibrator. The amount of PTPN22 and SHP-1 in BJAB was arbitrarily set to 1. Mann-Whitney rank-sum test was used to evaluate the significance of differences in PTPN22 and SHP-1 expression between CLL and normal B cells.
Expression of PTPN22 and SHP-1 in CLL and normal B cells. (A) Cellular extracts from normal PB B cells (PB1, PB2, PB3, PB4, and PB5), normal tonsillar B cells (TB4, TB5, TB6, TB8, and TB9), lymphoma B-cell lines (B104 and BJAB), and CLL B cells were analyzed by immunoblotting with the indicated antibodies. Actin was used as a loading control. (B) Comparison of relative PTPN22 and SHP-1 protein levels in CLL (n = 91 and n = 42, respectively) and normal B cells (n = 14). The BJAB cell line was used as a calibrator. The amount of PTPN22 and SHP-1 in BJAB was arbitrarily set to 1. Mann-Whitney rank-sum test was used to evaluate the significance of differences in PTPN22 and SHP-1 expression between CLL and normal B cells.
We next correlated PTPN22 expression with various prognostic markers, including IGHV mutation status, ZAP-70 expression, CD38 expression, cytogenetic abnormalities, and TCL1 expression (supplemental Figure 2A). In addition, we correlated in a smaller series of cases (n = 22) PTPN22 expression with BCR signal competence, which was defined as the capacity to induce ERK phosphorylation on stimulation with anti-IgM (supplemental Figure 2A-B). Median PTPN22 levels were somewhat higher in IGHV-unmutated, ZAP-70–positive, BCR signal competent and CD38-positive cases, but the difference was statistically significant only for the latter subset (P = .031; supplemental Figure 2A). No differences were observed in time to treatment between cases with high and low PTPN22 levels (supplemental Figure 2B).
We also performed nucleotide sequence analysis of the complete PTPN22 coding region in 29 CLL cases and the SHP-1 coding region in 15 cases. Only one mutation was identified, a C1366G substitution in PTPN22 leading to a Q456E amino acid replacement (supplemental Figure 3). The PTPN22 R620W autoimmune-disease risk allele was also detected in only 1 case (3.4%), consistent with the expected frequency of this allele for the same healthy population (3.7%).31 Another less frequent PTPN22 variant (R263Q), which has been associated with reduced risk for autoimmune disease, was observed in another patient.32 Overall, the sequencing analysis revealed that PTPN22 and SHP-1 are not frequently mutated in CLL and that the PTPN22 autoimmune-disease risk variant is not more prevalent in CLL patients than in the general population.
Overexpression of PTPN22 blocks anti-IgM–induced apoptosis
To determine the functional consequences of PTPN22 overexpression, we investigated survival of CLL cells after down-regulation of PTPN22 by RNA interference. The effects of PTPN22 down-regulation were evaluated in unstimulated CLL cells as well as CLL cells stimulated through the BCR. Both soluble and immobilized anti-IgM antibodies were used for BCR stimulation, as these 2 BCR crosslinking agents have been shown to have opposing effects on CLL cell survival. Whereas soluble anti-IgM antibodies usually induce apoptosis, immobilized anti-IgM antibodies typically increase the viability of the leukemic cells.20
As shown in Figure 2, down-regulation of PTPN22 had no effect on the viability of unstimulated CLL cells, as no differences were observed in the percentage of annexin V/PI-negative cells after transfection of 11 different samples with PTPN22-specific or control siRNA (% viable cells: siControl 49 ± 14, siPTPN22 48 ± 14, P = not significant). However, a greater reduction in the percentage of viable leukemic cells was observed after stimulation with soluble anti-IgM in siPTPN22-transfected samples compared with samples transfected with siControl (% viable cells: siControl 44 ± 13, siPTPN22 31 ± 20, P < .001). In addition, no change in leukemic cell viability was observed after stimulation with immobilized anti-IgM in siPTPN22-transfected samples, whereas increased viability was observed in samples transfected with siControl (% viable cells: siControl 55 ± 14, siPTPN22 48 ± 14, P = .001). Altogether, these experiments demonstrated that PTPN22 protects CLL cells from the proapoptotic effect of soluble anti-IgM and enhances the antiapoptotic effect of immobilized anti-IgM.
Silencing of PTPN22 in primary CLL cells enhances proapoptotic and inhibits antiapoptotic BCR signals. (A) CLL cells were transfected with control (siCTRL) or PTPN22 (siPTP) siRNA and placed in culture with bone marrow stromal cells to inhibit spontaneous and nucleofection-induced apoptosis. After 48 hours, CLL cells were removed from the layer of stromal cells and were cultured for additional 48 hours without (unstim.) or with soluble or immobilized anti-IgM (sol-aIgM and imm-aIgM, respectively). The percentage of viable cells was determined by annexin V/PI staining and flow cytometry. One representative experiment of 11 is shown. Viable cells bound with beads coated with imm-aIgM are seen as a separate population. PTPN22 protein levels in the same sample are shown in the bottom panel. Normal B cells (TB9) were used as a standard to evaluate the efficiency of PTPN22 down-regulation. (B) Summary of the data obtained with 11 different CLL samples. Significance of differences in leukemic cell viability was evaluated with the paired t test.
Silencing of PTPN22 in primary CLL cells enhances proapoptotic and inhibits antiapoptotic BCR signals. (A) CLL cells were transfected with control (siCTRL) or PTPN22 (siPTP) siRNA and placed in culture with bone marrow stromal cells to inhibit spontaneous and nucleofection-induced apoptosis. After 48 hours, CLL cells were removed from the layer of stromal cells and were cultured for additional 48 hours without (unstim.) or with soluble or immobilized anti-IgM (sol-aIgM and imm-aIgM, respectively). The percentage of viable cells was determined by annexin V/PI staining and flow cytometry. One representative experiment of 11 is shown. Viable cells bound with beads coated with imm-aIgM are seen as a separate population. PTPN22 protein levels in the same sample are shown in the bottom panel. Normal B cells (TB9) were used as a standard to evaluate the efficiency of PTPN22 down-regulation. (B) Summary of the data obtained with 11 different CLL samples. Significance of differences in leukemic cell viability was evaluated with the paired t test.
To further validate the observation that PTPN22 overexpression protects from BCR-induced apoptosis, we transfected in vitro transcribed PTPN22 mRNA in the lymphoma B-cell line B104. This cell line was chosen because it does not express PTPN22 (Figure 1A) and undergoes massive apoptosis after stimulation with soluble anti-IgM. As shown in Figure 3, overexpression of PTPN22 at levels similar to those in CLL cells resulted in a significantly lesser reduction in the viability of anti-IgM–stimulated B104 cells compared with cells transfected with control mRNA (% viable cells: control 22 ± 10, PTPN22 35 ± 8, P = .017). Consistent with the results from the RNA interference experiments, PTPN22 overexpression had no effect in the absence of BCR engagement, as the viability of unstimulated B104 cells remained unchanged (% viable cells: control 52 ± 9, PTPN22 51 ± 8, P = not significant).
Enforced expression of PTPN22 in B104 cells blocks anti-IgM–induced apoptosis. (A) B104 cells were transfected with control (CTRL) or PTPN22 (PTP) mRNA, left in culture for 3 hours to achieve maximal PTPN22 protein expression, and then cultured for an additional 22 hours with or without soluble anti-IgM. The percentage of viable cells was determined by annexin V/PI staining and flow cytometry. One representative experiment of 6 is shown. Rectangle represents viable cells. Levels of PTPN22 protein 3 hours after transfection of PTPN22 mRNA are shown in the bottom panel. A CLL sample with high PTPN22 expression was loaded on the same gel for comparison. (B) Graph represents summary of data from the 6 independent experiments. Significance of difference in leukemic cell viability was evaluated with the paired t test.
Enforced expression of PTPN22 in B104 cells blocks anti-IgM–induced apoptosis. (A) B104 cells were transfected with control (CTRL) or PTPN22 (PTP) mRNA, left in culture for 3 hours to achieve maximal PTPN22 protein expression, and then cultured for an additional 22 hours with or without soluble anti-IgM. The percentage of viable cells was determined by annexin V/PI staining and flow cytometry. One representative experiment of 6 is shown. Rectangle represents viable cells. Levels of PTPN22 protein 3 hours after transfection of PTPN22 mRNA are shown in the bottom panel. A CLL sample with high PTPN22 expression was loaded on the same gel for comparison. (B) Graph represents summary of data from the 6 independent experiments. Significance of difference in leukemic cell viability was evaluated with the paired t test.
PTPN22 enhances activation of antiapoptotic and inhibits activation of proapoptotic BCR signaling pathways
The previous experiments suggested that PTPN22 functions as a molecular switch that enhances antiapoptotic and inhibits proapoptotic BCR signals. Earlier work by our group had shown that the antiapoptotic BCR signal in CLL cells is transduced primarily by the PI3K/AKT pathway, but the identity of the downstream signaling molecules that transduce the propapoptotic BCR signal was not established.33 Because p38MAPK had been shown to mediate BCR-induced apoptosis in murine splenic B cells and various lymphoma cell lines, including B104,34,35 we investigated whether the selective p38MAPK inhibitor SB203580 will prevent apoptosis induced by soluble anti-IgM in CLL cells. Although p38 MAPK is rather inefficiently activated in CLL cells,20 pretreatment with SB203580 significantly inhibited the cytotoxic effect of soluble anti-IgM, suggesting that this MAPK is at least in part responsible for transducing the proapoptotic BCR signal in CLL cells (% viable anti-IgM–treated cells: control 51.2 ± 15.5, SB203580 62.3 ± 12.9, P = .012; % viable untreated cells: control 72 ± 9.5, SB203580 71.8 ± 9.3, P = not significant; supplemental Figure 4).
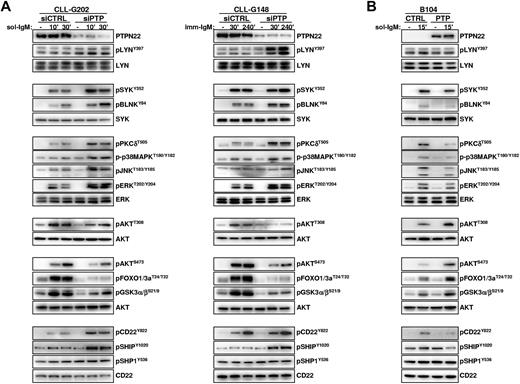
To determine how PTPN22 overexpression affects the activity of AKT, p38MAPK, and other downstream BCR signaling molecules, we analyzed BCR signal transduction in primary CLL cells transfected with control or PTPN22-specific siRNAs. Down-regulation of PTPN22 had no obvious effect on the basal activity of any of the investigated molecules but resulted in a marked increase in the levels of phosphorylated LYN, SYK, BLNK, PKCδ, ERK, JNK, and p38MAPK in CLL cells stimulated with soluble or immobilized anti-IgM (Figure 4A left and right panel, respectively). Paradoxically, down-regulation of PTPN22 had an opposite effect on the activity of AKT, as evidenced by substantially reduced phosphorylation of the activating Thr308 and Ser473 residues and reduced phosphorylation of GSK3 and FOXO, which are both direct targets of AKT.
PTPN22 inhibits activation of proapoptotic and enhances activation of antiapoptotic pathways in anti-IgM–stimulated CLL and B104 cells. (A) CLL cells were transfected with control (siCTRL) or PTPN22 (siPTP) siRNA as described in the legend to Figure 2. After 48 hours, cells were stimulated with soluble (left panel) or immobilized (right panel) anti-IgM for the indicated times. Cellular extracts were prepared and analyzed by immunoblotting with the indicated antibodies. Three independent experiments with similar results were performed. (B) B104 cells were transfected with control (CTRL) or PTPN22 (PTP) mRNA and stimulated with soluble anti-IgM as indicated. Cellular extracts were prepared and analyzed by immunoblotting with the indicated antibodies. One representative experiment of 3 is shown.
PTPN22 inhibits activation of proapoptotic and enhances activation of antiapoptotic pathways in anti-IgM–stimulated CLL and B104 cells. (A) CLL cells were transfected with control (siCTRL) or PTPN22 (siPTP) siRNA as described in the legend to Figure 2. After 48 hours, cells were stimulated with soluble (left panel) or immobilized (right panel) anti-IgM for the indicated times. Cellular extracts were prepared and analyzed by immunoblotting with the indicated antibodies. Three independent experiments with similar results were performed. (B) B104 cells were transfected with control (CTRL) or PTPN22 (PTP) mRNA and stimulated with soluble anti-IgM as indicated. Cellular extracts were prepared and analyzed by immunoblotting with the indicated antibodies. One representative experiment of 3 is shown.
To explore the mechanism responsible for this dual activity of PTPN22, we investigated the consequences of PTPN22 down-regulation on the activity of molecules that negatively regulate BCR signal transduction. As shown in Figure 4, PTPN22 down-regulation resulted in increased anti-IgM–induced phosphorylation of the inhibitory receptor CD22 on Y822, which serves as a docking site for the phosphatases SHIP and SHP-1. In addition, phosphorylation of SHIP on the activating Y1020 residue was increased, suggesting that the recruitment and activation of this principle negative regulator of the PI3K/AKT pathway are impaired when PTPN22 is overexpressed.36 In contrast, phosphorylation of SHP-1 on the activating Y536 residue was only marginally affected. Collectively, these data suggest that the mechanism through which PTPN22 increases signaling through AKT is by selectively preventing the activation of a negative regulatory circuit involving CD22 and SHIP.
To further validate the data obtained from the PTPN22 knockdown experiments, we investigated the effects of PTPN22 overexpression in B104 cells (Figure 4B). Consistent with the knockdown experiments, PTPN22 overexpression resulted in a substantial increase in the amount of phosphorylated AKT, GSK3 and FOXO in anti-IgM stimulated cells, which was associated with a concomitant decrease in the levels of phosphorylated LYN, SYK, BLNK, PKCδ, ERK, JNK, p38MAPK, CD22 and SHIP. These results further establish that PTPN22 is a positive regulator of the AKT pathway and a negative regulator of most other pathways downstream of the BCR.
PKC regulates PTPN22 expression in CLL cells
PTPN22 has a relatively short half-life in T cells and is subject to calpain and proteasome degradation.37 In resting normal T cells, the levels of this protein are low but can be induced by antigen-receptor engagement or treatment with the phorbol ester 12-0-tetradecanoylphorbol-β-acetate (TPA).38,39 To understand the mechanisms responsible for PTPN22 overexpression in CLL, we investigated whether this phosphatase is regulated in a similar manner in CLL cells as in T cells. As shown in Figure 5, the levels of PTPN22 in cultured CLL cells declined by approximately 60% to 80% over 24 hours, consistent with the rapid turnover of this protein in T cells.37 This decline was partially reversed by the calpain inhibitor calpeptin and the proteosomal inhibitor MG132, suggesting that PTPN22 degradation is influenced by both calpain and proteosomal activity (Figure 5A). PTPN22 expression could be further rescued by coculture with M2-10B4 bone marrow stromal cells or stimulation with immobilized anti-IgM, CD40L, or CpG-ODN, although not to the level seen in freshly isolated CLL cells (Figure 5B). A greater effect was seen with TPA, which not only maintained PTPN22 expression to the same extent as in freshly isolated CLL cells but was also capable of inducing PTPN22 in CLL cells that did not express this protein (Figure 5B bottom panel). These effects were specific for PTPN22 because no such changes were observed in the expression of SHP-1. Interestingly, none of the investigated stimuli, including TPA, could induce PTPN22 in normal PB or tonsillar B cells (Figure 5C; and data not shown). In normal B cells, only the combination of TPA with ionomycin induced PTPN22, although the levels were considerably lower than in TPA-activated CLL or normal T cells.
Regulation of PTPN22 expression in normal and CLL B cells. (A) Western blot analysis of cellular extracts obtained from CLL cells immediately on isolation or after 24 hours in culture without or with the caspase inhibitor Z-VAD, the calpain inhibitor calpeptin (Calp.), or the proteasome inhibitor MG132. Because MG132 induces apoptosis in CLL cells, this inhibitor was used both alone and in combination with Z-VAD. Normalized PTPN22 levels are expressed as fold change relative to the level in freshly isolated CLL cells. (B) Expression of PTPN22 and SHP-1 was evaluated in freshly isolated CLL cells, CLL cells cultured for 24 hours in the absence of any stimuli (unst.) or CLL cells cultured for 24 hours with sol-aIgM, imm-aIgM, CD40L + enhancer, CpG, M2-10B4 bone marrow stromal cells, B-cell activating factor, vascular endothelial growth factor, or TPA. Actin was used as a loading control. Shown are 2 representative experiments of 8 performed with sol-IgM, imm-IgM, CpG, stromal cells, and TPA, and 4 performed with CD40L, vascular endothelial growth factor, and TPA. Changes in PTPN22 expression during culture with sol-aIgM, imm-aIgM, CpG stromal cells, and TPA are shown separately for the 5 samples with high PTPN22 levels in freshly isolated CLL cells (top panel) and the 3 samples with low/absent PTPN22 expression (bottom panel). Values in graphs represent mean ± SD. The BJAB cell line was used as an internal standard. (C) PTPN22 expression in normal T and B cells after 24 hours in culture with the indicated stimuli. Ion. indicates ionomycin. (D) Effects of ruboxistaurin and sotrastaurin on PTPN22 and SHP-1 expression in CLL cells cultured for 24 hours with or without stromal cells.
Regulation of PTPN22 expression in normal and CLL B cells. (A) Western blot analysis of cellular extracts obtained from CLL cells immediately on isolation or after 24 hours in culture without or with the caspase inhibitor Z-VAD, the calpain inhibitor calpeptin (Calp.), or the proteasome inhibitor MG132. Because MG132 induces apoptosis in CLL cells, this inhibitor was used both alone and in combination with Z-VAD. Normalized PTPN22 levels are expressed as fold change relative to the level in freshly isolated CLL cells. (B) Expression of PTPN22 and SHP-1 was evaluated in freshly isolated CLL cells, CLL cells cultured for 24 hours in the absence of any stimuli (unst.) or CLL cells cultured for 24 hours with sol-aIgM, imm-aIgM, CD40L + enhancer, CpG, M2-10B4 bone marrow stromal cells, B-cell activating factor, vascular endothelial growth factor, or TPA. Actin was used as a loading control. Shown are 2 representative experiments of 8 performed with sol-IgM, imm-IgM, CpG, stromal cells, and TPA, and 4 performed with CD40L, vascular endothelial growth factor, and TPA. Changes in PTPN22 expression during culture with sol-aIgM, imm-aIgM, CpG stromal cells, and TPA are shown separately for the 5 samples with high PTPN22 levels in freshly isolated CLL cells (top panel) and the 3 samples with low/absent PTPN22 expression (bottom panel). Values in graphs represent mean ± SD. The BJAB cell line was used as an internal standard. (C) PTPN22 expression in normal T and B cells after 24 hours in culture with the indicated stimuli. Ion. indicates ionomycin. (D) Effects of ruboxistaurin and sotrastaurin on PTPN22 and SHP-1 expression in CLL cells cultured for 24 hours with or without stromal cells.
Because TPA is a potent PKC activator, we next investigated whether PKC inhibitors will down-regulate PTPN22 in CLL cells. These experiments were performed with unstimulated CLL cells as well as CLL cells cocultured with M2-10B4 bone marrow stromal cells, which after TPA were the stimulus that induced greatest PTPN22 expression in vitro. As shown in Figure 5D, the PKC inhibitors ruboxistaurin and sotrastaurin induced a dose-dependent reduction in PTPN22 levels, which was especially evident in CLL cells cocultured with M2-10B4 cells. These data further suggest that PKC regulates PTPN22 expression in CLL.
Targeting PTPN22 with PKC inhibitors enhances anti-IgM–induced CLL cell apoptosis
The capacity of PTPN22 to positively regulate antiapoptotic and negatively regulate proapoptotic BCR signals suggested that this phosphatase could represent a potential therapeutic target in CLL. To evaluate this possibility, we first tested the activity of the PTPN22 inhibitor I-C1140 against unstimulated and anti-IgM–stimulated CLL cells. The expectations from this experiment were that I-C11 will display similar effects as the PTPN22 siRNAs (ie, it will potentiate leukemic cell killing by soluble anti-IgM without affecting the viability of unstimulated CLL cells). However, only marginal changes in leukemic cell viability were observed in the presence of I-C11, with no significant increase in anti-IgM–induced apoptosis (data not shown). We therefore investigated the effects of ruboxistaurin and sotrastaurin, as these PKC inhibitors were shown in the previous experiments to effectively down-regulate PTPN22 expression. As shown in Figure 6A and B, both ruboxistaurin and sotrastaurin further reduced the viability of CLL cells stimulated with soluble anti-IgM, despite having no effect on the viability of unstimulated CLL cells. The increase in anti-IgM–induced apoptosis was directly correlated to basal PTPN22 levels and was not seen in the 3 samples with low PTPN22 expression (Figure 6C; supplemental Table 2). These data indicate that PKC inhibitors modulate the proapoptotic BCR signal by targeting PTPN22 expression and suggest that these compounds could represent novel agents for targeted therapy of CLL.
PKC inhibitors enhance anti-IgM induced apoptosis in CLL cells. (A-B) CLL cells were cocultured with bone marrow stromal cells in the presence and absence of 4μM ruboxistaurin (Rub.) or 4μM sotrastaurin (Sotr.). After 24 hours, an aliquot was taken for Western blot analysis of PTPN22 expression, and the remaining cells were split and cultured for an additional 48 hours with or without 10 μg/mL goat F(ab′)2 anti–human IgM (sol-aIgM). Percentage of viable and dead cells was determined by annexin V/PI staining. (A) One representative experiment. (B) A summary of the results from 18 different experiments. Wilcoxon signed-rank test was used to evaluate significance of differences in leukemic cell viability. (C) The relationship between PTPN22 expression and the capacity of PKC inhibitors (PKCi) to enhance the cytotoxic effect of sol-aIgM was evaluated through Pearson rank correlation. The capacity of PKCi to enhance the cytotoxic effect of sol-aIgM (% PKCi + sol-aIgM cytotoxicity) was defined as the increase in the percentage of apoptosis induced by sol-aIgM + PKCi relative to sol-aIgM alone. The values were normalized against the percentage of viable cells at 48 hours to account for spontaneous apoptosis. The following formula was used: % PKCi + sol-aIgM cytotoxicity = [(%ViablePKCi − %Viablesol-aIgM+PKCi) / %ViablePKCi] × 100 − [(%Viableunst − %Viablesol-aIgM) / %Viableunst] × 100. The exact values are provided in supplemental Table 2. The percentage PKCi + sol-aIgM cytotoxicity was plotted against the amount of PTPN22 at 24 hours in cells without PKCi.
PKC inhibitors enhance anti-IgM induced apoptosis in CLL cells. (A-B) CLL cells were cocultured with bone marrow stromal cells in the presence and absence of 4μM ruboxistaurin (Rub.) or 4μM sotrastaurin (Sotr.). After 24 hours, an aliquot was taken for Western blot analysis of PTPN22 expression, and the remaining cells were split and cultured for an additional 48 hours with or without 10 μg/mL goat F(ab′)2 anti–human IgM (sol-aIgM). Percentage of viable and dead cells was determined by annexin V/PI staining. (A) One representative experiment. (B) A summary of the results from 18 different experiments. Wilcoxon signed-rank test was used to evaluate significance of differences in leukemic cell viability. (C) The relationship between PTPN22 expression and the capacity of PKC inhibitors (PKCi) to enhance the cytotoxic effect of sol-aIgM was evaluated through Pearson rank correlation. The capacity of PKCi to enhance the cytotoxic effect of sol-aIgM (% PKCi + sol-aIgM cytotoxicity) was defined as the increase in the percentage of apoptosis induced by sol-aIgM + PKCi relative to sol-aIgM alone. The values were normalized against the percentage of viable cells at 48 hours to account for spontaneous apoptosis. The following formula was used: % PKCi + sol-aIgM cytotoxicity = [(%ViablePKCi − %Viablesol-aIgM+PKCi) / %ViablePKCi] × 100 − [(%Viableunst − %Viablesol-aIgM) / %Viableunst] × 100. The exact values are provided in supplemental Table 2. The percentage PKCi + sol-aIgM cytotoxicity was plotted against the amount of PTPN22 at 24 hours in cells without PKCi.
Discussion
A genetic variant of the phosphatase PTPN22 has been implicated in the pathogenesis of multiple autoimmune diseases. We now show that this phosphatase is likely to play an important role in the pathogenesis of CLL, as evidenced by its significant overexpression and its capacity to modulate BCR signals that are crucial for the growth and survival of the malignant cells.
Previous studies that were performed mainly in T cells established that PTPN22 is a negative regulator of antigen-receptor signaling. Substrate trapping experiments identified LCK, ZAP-70, VAV, CD3ϵ, and TCRζ as targets of PTPN22,27 whereas experiments with knockout mice demonstrated enhanced activation of LCK, ZAP-70, ERK, JNK, and p38 MAPK in PTPN22-deficient T cells.41 Although PTPN22 has also been implicated in BCR signaling,42 its substrates and mechanism of action in B cells have not been fully elucidated. We now show that PTPN22 functions as both a positive and negative regulator of BCR signaling. Down-regulation of PTPN22 in primary CLL cells resulted in enhanced BCR-induced activation of LYN, SYK, BLNK, PKCδ, ERK, JNK, and p38MAPK, analogous to the situation in antigen-stimulated T cells. However, an unexpected finding in these experiments was the reduced activation of AKT, suggesting that PTPN22 operates as a positive regulator of this kinase in CLL cells. The capacity of PTPN22 to act as both a positive and negative regulator of BCR signaling was further confirmed by overexpression experiments, which showed enhanced activation of AKT and reduced activation of LYN, SYK, BLNK, PKCδ, ERK, JNK, and p38MAPK in PTPN22-expressing lymphoma B cells stimulated with anti-IgM. Whether PTPN22 also positively regulates AKT in T cells was not investigated in the current work and remains to be established in future studies.
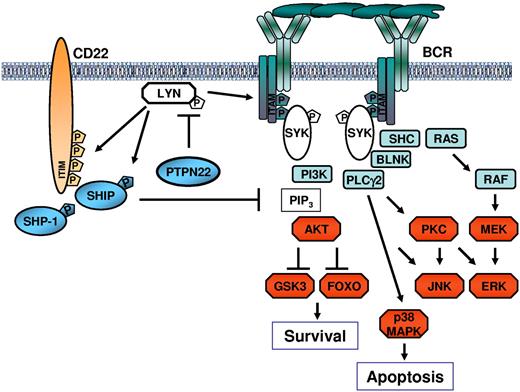
The mechanism through which PTPN22 positively regulates AKT despite having a negative effect on more proximal BCR signaling molecules was shown to rely, at least in part, on reduced recruitment and activation of the phosphatase SHIP (Figure 7). SHIP negatively regulates the PI3K/AKT pathway by dephosphorylating phosphatidylinositol 3,4,5-trisphosphate, which is required for the plasma membrane localization and activation of AKT. Recruitment of SHIP in the vicinity of the activated BCR complex is regulated by LYN, which phosphorylates the CD22 immunoreceptor tyrosine-based inhibitory motifs.43 In addition to regulating SHIP recruitment, LYN directly increases the catalytic activity of SHIP by phosphorylating Y1020 at its C-terminus.36 Thus, by inactivating LYN, PTPN22 could selectively block the pathway that negatively regulates signaling through AKT. Consistent with this mechanism are our findings that the levels of Y822-phosphorylated CD22 and Y1020-phosphorylated SHIP increase after PTPN22 down-regulation and decrease after PTPN22 overexpression, as well as the data from the literature showing that targeted deletion of LYN enhances AKT activation in antigen-stimulated B cells.44 Overall, these data suggest that PTPN22 positively regulates AKT by negatively regulating LYN, although they do not exclude the possibility that PTPN22 may also directly affect the activity of additional molecules that regulate the AKT pathway.
Schematic representation of the mechanism through which PTPN22 blocks proapoptotic and enhances antiapoptotic BCR signals. PTPN22 prevents LYN activation by dephosphorylating the activating Y397 residue. This results in impaired activation of downstream signaling pathways, including pathways that transduce the proapoptotic BCR signal. However, inactivation of LYN by PTPN22 also prevents recruitment and activation of SHIP, which is a key negative regulator of the PI3K/AKT pathway. In the absence of SHIP, AKT continues to be activated by phosphatidylinositol 3,4,5-trisphosphate (PIP3), resulting in a net increase in AKT activity despite the overall attenuation of the BCR signal. Tyrosine residues that are directly phosphorylated by LYN are indicated by “P” in the pentagon.
Schematic representation of the mechanism through which PTPN22 blocks proapoptotic and enhances antiapoptotic BCR signals. PTPN22 prevents LYN activation by dephosphorylating the activating Y397 residue. This results in impaired activation of downstream signaling pathways, including pathways that transduce the proapoptotic BCR signal. However, inactivation of LYN by PTPN22 also prevents recruitment and activation of SHIP, which is a key negative regulator of the PI3K/AKT pathway. In the absence of SHIP, AKT continues to be activated by phosphatidylinositol 3,4,5-trisphosphate (PIP3), resulting in a net increase in AKT activity despite the overall attenuation of the BCR signal. Tyrosine residues that are directly phosphorylated by LYN are indicated by “P” in the pentagon.
The capacity of PTPN22 to positively regulate AKT and negatively regulate p38MAPK signaling may have important implications for the pathogenesis of CLL. Sustained activation of AKT is known to increase the survival of antigen-stimulated CLL and lymphoma B cells,33,44,45 whereas p38MAPK has been shown to induce apoptosis in murine splenocytes and human lymphoma B-cell lines34,35 and was found to display similar activity in primary human CLL cells in the current study. Based on these observations, it would be expected that PTPN22 overexpression would increase the survival of BCR-stimulated CLL cells by enhancing AKT activation and protect them from activation-induced cell death by inhibiting p38MAPK. This possibility was confirmed by our RNA interference experiments, which showed that PTPN22 knockdown significantly reduces the survival of anti-IgM–stimulated CLL cells. Overall, these data suggest that overexpression of PTPN22 could represent a protective mechanism that allows CLL cells to block BCR pathways that induce apoptosis while augmenting the activity of BCR pathways that increase leukemic cell survival. This mechanism could be particularly important for CLL cells that express autoreactive BCRs, which represent the majority of the IGHV-unmutated and a large fraction of the mutated cases.15,–17 Such cells are continuously exposed to autoantigen and would be expected to undergo negative selection. Thus, overexpression of PTPN22 could protect autoreactive CLL cells from negative selection and may also explain why these cells are apparently positively selected and expanded by autoantigen rather than being eliminated by immunologic tolerance mechanisms.
The evidence that antigen drive plays an important role in the pathogenesis of CLL has stimulated efforts to identify the most appropriate therapeutic targets along the BCR signaling pathway. Clinically useful inhibitors of SYK, BTK, and PI3Kδ have already been developed and are showing encouraging activity in initial clinical trials of CLL.10,–12 Our data suggest that PTPN22 could represent another attractive therapeutic target because inhibition or down-regulation of this phosphatase would be expected not only to inhibit BCR survival signals but also convert them into signals that kill the malignant B cells. A prototypic PTPN22 inhibitor40 was tested in our study but was not capable of significantly enhancing apoptosis induction by soluble anti-IgM. This discordance with respect to the results obtained from the RNA interference experiments could be the result of insufficient PTPN22 inhibition but may also indicate that PTPN22 has additional regulatory functions that are unrelated to its catalytic activity. Novel PTPN22 inhibitors with improved potency and selectivity are currently being developed and will hopefully resolve this issue in the near future.46
Another possibility for targeting PTPN22 is to prevent its overexpression. The data presented in this study suggest that PTPN22 overexpression in CLL cells is driven by microenvironmental stimuli that signal through PKC. Although the exact nature of the stimuli that drive PTPN22 overexpression in CLL cells in vivo is currently unknown, several stimuli were shown to be capable of maintaining or inducing PTPN22 expression in vitro, among which the PKC activator TPA and bone marrow stromal cells had a most marked effect. Induction of PTPN22 by bone marrow stromal cells could be efficiently blocked by ruboxistaurin and sotrastaurin, which are 2 oral PKC inhibitors that are currently undergoing phase 2 and phase 3 clinical testing for several nonmalignant diseases, including diabetic retinopathy, psoriasis, and prevention of renal allograft rejection.47,48 Importantly, both compounds significantly enhanced the cytotoxic effect of soluble anti-IgM despite having no direct cytotoxic effect of their own. These data suggest that PKC inhibitors could selectively kill CLL cells in vivo by down-regulating PTPN22, although it remains possible that the enhanced apoptosis induction observed with these agents may also involve additional mechanisms that are independent of PTPN22 expression.
CLL cases with poor prognostic features, such as unmutated IGHV genes, ZAP-70 expression, and CD38 expression showed a trend for higher PTPN22 levels, but the difference was significant only between CD38-positive and CD38-negative cases. The significance of this observation remains unclear at present, although it could suggest that the same microenvironmental stimuli that induce CD38 expression are also responsible for driving PTPN22 overexpression in CLL cells in vivo. Further studies to identify these stimuli are required, as they may lead to the identification of additional approaches for targeting PTPN22 expression.
The PTPN22 R620W polymorphism was initially described as a gain-of-function variant that contributes to the development of autoimmune diseases by decreasing antigen-receptor signaling and thus preventing negative selection of autoreactive lymphocytes.49,50 Studies using T cells derived from carriers of the R620W variant further supported this possibility, demonstrating diminished calcium mobilization, reduced expression of CD25, and diminished IL-10 production on TCR stimulation.42 However, more recent studies have reported enhanced signaling responses in lymphocytes carrying the risk allele.37,39 In light of our finding that PTPN22 functions as both a positive and negative regulator of BCR signaling, it would be interesting to compare the individual activity of the 2 variants on the positively and negatively regulated pathways. Such a study may identify novel functional differences that could help clarify the mechanism through which PTPN22 contributes to the development of multiple autoimmune diseases.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Leukemia & Lymphoma Society (grant R6170-10; D.G.E.), the Italian Association for Cancer Research (grant 5917; D.G.E.), and the National Institutes of Health (grant RO1 CA152194; Z.-Y.Z.).
National Institutes of Health
Authorship
Contribution: R.N. designed and performed research, analyzed and interpreted data, and wrote the manuscript; S.G. and P.G.L. performed research and reviewed the manuscript; Y.H. contributed vital new reagents; Z.-Y.Z. contributed vital new reagents and reviewed the manuscript; L.L. collected, analyzed, and interpreted data and reviewed the manuscript; and D.G.E. designed research, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dimitar G. Efremov, ICGEB, Molecular Hematology Section, Campus Adriano Buzzati-Traverso, Via E Ramarini 32, I-00016 Monterotondo Scalo (Rome), Italy; e-mail: efremov@icgeb.org.




![Figure 6. PKC inhibitors enhance anti-IgM induced apoptosis in CLL cells. (A-B) CLL cells were cocultured with bone marrow stromal cells in the presence and absence of 4μM ruboxistaurin (Rub.) or 4μM sotrastaurin (Sotr.). After 24 hours, an aliquot was taken for Western blot analysis of PTPN22 expression, and the remaining cells were split and cultured for an additional 48 hours with or without 10 μg/mL goat F(ab′)2 anti–human IgM (sol-aIgM). Percentage of viable and dead cells was determined by annexin V/PI staining. (A) One representative experiment. (B) A summary of the results from 18 different experiments. Wilcoxon signed-rank test was used to evaluate significance of differences in leukemic cell viability. (C) The relationship between PTPN22 expression and the capacity of PKC inhibitors (PKCi) to enhance the cytotoxic effect of sol-aIgM was evaluated through Pearson rank correlation. The capacity of PKCi to enhance the cytotoxic effect of sol-aIgM (% PKCi + sol-aIgM cytotoxicity) was defined as the increase in the percentage of apoptosis induced by sol-aIgM + PKCi relative to sol-aIgM alone. The values were normalized against the percentage of viable cells at 48 hours to account for spontaneous apoptosis. The following formula was used: % PKCi + sol-aIgM cytotoxicity = [(%ViablePKCi − %Viablesol-aIgM + PKCi) / %ViablePKCi] × 100 − [(%Viableunst − %Viablesol-aIgM) / %Viableunst] × 100. The exact values are provided in supplemental Table 2. The percentage PKCi + sol-aIgM cytotoxicity was plotted against the amount of PTPN22 at 24 hours in cells without PKCi.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/119/26/10.1182_blood-2012-01-403162/5/m_zh89991292960006.jpeg?Expires=1765889730&Signature=PGAfOfTD6NycVoJqxMPVJdoi4xlNzuo76kCfBKiP6uVc8DRmNjkx~BsMSvRRB9m0JG-vX3Vybhk7sScVbVGYqGzAnkUEmujEDcuL4R1nHGHpIipyNoMug3zx09pS1x5jOfhejdSeCwWy-iYCVGd7dt-wOtv5aSwvQagINn1kzjS9NqmVwiSMTcO3dAlHENH59ar6zamOcPalSMAYtOqKdx-Yrj1WOIZAC3Xm71WvAhRctkUdlFHicJiHiDIVyp1Kfn88TKur0n6MsDR3yj-eQ~ClTqx3SV1P6i8jOx3jbnY7E2CLJCq3xrLjI58BlJSM4pKtf1Pjh5NrgW16-U4EsQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal