Abstract
Targets of curative donor-derived graft-versus-myeloma (GVM) responses after allogeneic hematopoietic stem cell transplantation (HSCT) remain poorly defined, partly because immunity against minor histocompatibility Ags (mHAgs) complicates the elucidation of multiple myeloma (MM)–specific targets. We hypothesized that syngeneic HSCT would facilitate the identification of GVM-associated Ags because donor immune responses in this setting should exclusively target unique tumor Ags in the absence of donor-host genetic disparities. Therefore, in the present study, we investigated the development of tumor immunity in an HLA-A0201+ MM patient who achieved durable remission after myeloablative syngeneic HSCT. Using high-density protein microarrays to screen post-HSCT plasma, we identified 6 Ags that elicited high-titer (1:5000-1:10 000) Abs that correlated with clinical tumor regression. Two Ags (DAPK2 and PIM1) had enriched expression in primary MM tissues. Both elicited Ab responses in other MM patients after chemotherapy or HSCT (11 and 6 of 32 patients for DAPK2 and PIM1, respectively). The index patient also developed specific CD8+ T-cell responses to HLA-A2–restricted peptides derived from DAPK2 and PIM1. Peptide-specific T cells recognized HLA-A2+ MM-derived cell lines and primary MM tumor cells. Coordinated T- and B-cell immunity develops against MM-associated Ags after syngeneic HSCT. DAPK1 and PIM1 are promising target Ags for MM-directed immunotherapy.
Introduction
Clinical studies over the last 2 decades have highlighted the critical contribution of donor-derived immunity against tumors to the long-term curative effects of allogeneic hematopoietic stem cell transplantation (HSCT).1 The potency of this donor-derived graft-versus-tumor (GVT) response is clearly illustrated by the clinical success of therapies such as donor lymphocyte infusion2,3 and reduced intensity HSCT,4,5 which minimize or avoid chemo- and radiotherapy and rely instead on immunity to drive their antitumor effect.2-8 The beneficial GVT effects associated with these responses, however, are typically associated with detrimental GVHD responses.7,9 Preserving the benefits of GVT responses while minimizing toxicity from GVHD thus remains a critical unsolved issue in transplantation medicine.
Defining the target Ags of GVT and GVHD may provide insight into their mechanisms and suggest rational methods for their separation. Minor histocompatibility Ags (mHAgs) make up one major class of Ags against which potent donor-derived T- and B-cell immunity develops after HSCT. mHAgs with broad or nonhematopoietic cell expression are implicated in GVHD,10 whereas those with restricted hematopoietic expression play a well-accepted role in GVT responses.10-12 The extent to which nonpolymorphic tumor-associated Ags are targets is less well understood. Tumors may be distinguished from normal cells by genetic alterations, including chromosomal translocations. Tumors can also overexpress or aberrantly express genes compared with their normal counterparts.13 In support of the existence of immunogenic Ags with tumor-restricted expression, Nishida et al described T-cell immunity against leukemia cells after allogeneic HSCT that was not directed against mHAgs.14 The discovery of such naturally immunogenic tumor-associated Ags (TAAs) could lead to the development of immunotherapeutic strategies to target tumor in a selective fashion and thus avoid GVHD toxicity.
Because allogeneic HSCT can result in durable curative remission, it provides a useful clinical backdrop for identifying Ags that are naturally immunogenic to normal donor cells. However, defining TAAs in the allogeneic setting can be complicated by the presence of alloimmune responses. In the present study, we describe a context in which effective donor-derived tumor immunity occurred in the absence of alloimmunity: myeloablative syngeneic HSCT resulting in durable molecular remission in an individual with multiple myeloma (MM). Dissecting humoral immune responses by serologic screening after immune-mediated therapy15-18 or in premalignant conditions19 has been a successful strategy for identifying TAAs. We therefore examined the B-cell responses developing in this index patient. By screening plasma samples after HSCT against high-density protein microarrays, we identified 2 Ags, DAPK2 and PIM1, which elicited high-titer plasma Ab responses that were coordinated with Ag-specific CD8+ T-cell immunity. Consistent with the notion that these Ags are myeloma-specific targets, we found that peptides derived from these Ags further elicited T-cell responses against HLA-A2+ MM cell lines and primary MM plasma cells. Our results suggest a key role for TAAs that is distinct from mHAgs and that can elicit coordinated T- and B-cell responses to effect GVM immunity.
Methods
Patient samples and cell preparation
Heparinized blood and BM samples were obtained from patients and healthy donors enrolled in clinical research protocols at the Dana-Farber Harvard Cancer Center approved by the Human Subjects Protection Committee. BM mononuclear cells (BMMCs) or PBMCs were isolated by Ficoll/Hypaque density-gradient centrifugation, cryopreserved with 10% DMSO, and stored in vapor-phase liquid nitrogen until the time of analysis. Plasma was isolated by removal of the plasma layer after centrifugation of whole blood and cryopreserved at −80°C until the time of analysis.
Cell lines, plasmids, and peptides
The MM cell lines IM-9, RPMI 8226, MC/CAR, and NCI-H929 were obtained from the American Type Culture Collection and lines OPM1 and OPM2 from Dr Ken Anderson (Dana-Farber Cancer Institute). DNA sequences encoding candidate Ags were acquired from the Dana-Farber Cancer Institute Center for Cancer Systems Biology Human ORFeome Collection (Dr Mark Vidal, Dana-Farber Cancer Institute), or from the PlasmID repository (Dr Joshua LaBaer, Harvard Institute of Proteomics) and cloned into the pDONR221 Gateway vector (Invitrogen). The inserts of all of the acquired plasmids were verified by sequencing and then shuttled into a Gateway-converted mammalian expression vector containing a T7 promoter and a C-terminus GST tag (Wagner Montor, Harvard Institute of Proteomics) using LR Clonase (Invitrogen). DAPK2 and PIM1 were PCR cloned into pcDNA3 using the following primers: PIM1 forward, 5′-AAGG AAGCTT ATGCTCTTGTCCAAAATC-3′, reverse, 5′-CCGG GAATTC CTATTTGCTGGGCCCCG-3′; and DAPK2 forward, 5′-AAGG GGATCC ATGTTCCAGGCCTCAAT-3′, reverse, 5′-CCGG CTCGAG TTAGGAGGTGCTGCTC-3′. Synthetic peptides encoding the influenza M158-66 epitope (FluM1, GILGFVFTL) and predicted HLA-A2+–binding 9-mer peptides derived from DAPK2 and PIM1 were obtained from New England Peptide.
Serologic screening using high-density protein microarrays
Commercial protein microarrays (Human ProtoArray Version 4; Invitrogen) were probed with plasma samples as described previously.20 All samples were screened using arrays from the same printing lot. The microarrays contained approximately 8000 N-terminus GST-fusion human proteins expressed in an insect cell line and spotted on nitrocellulose-coated glass slides. The protein microarrays were processed at 4C according to the manufacturer's recommendations. In brief, after blocking, patient plasma (diluted 1:150) was applied to the microarray surface for 90 minutes, and Ab-Ag interactions were detected with Alexa Fluor 647–conjugated anti–human IgG (H and L chain) Ab (1:2000; Invitrogen). Lot-specific protein spot definitions provided by the microarray manufacturer were manually aligned to the image data. Fluorescence intensities were quantified using GenePix Pro Version 5.0 software (Molecular Devices) at 635 nm, 100% power, and 600 gain. Significant Ab-protein interactions were determined using Prospector Version 5.1.0 analysis software (Invitrogen) and an algorithm20 that incorporates the contribution of protein spot concentration to signal intensity. Signal change between pre-treatment or control plasma and post-treatment plasma was considered significant if the change in both (1) the signal magnitude (Zdelta), defined as Zpost − max(0, Zpre) and (2) the ratio (Zmult), defined as Zpost/max(1,Zpre), were greater than a cutoff value n (n = 5 for significant Ags, called “candidate Ags”). All candidate Ags selected for the final analysis had significantly higher reactivity at the maximum post-HSCT time point compared with 2 normal controls, the patient's donor, and a prechemotherapy patient sample.
Validation and survey of Ag-specific Ab responses to candidate Ags
We first validated the results of serologic screening using an immunoprecipitation-based method.21 In brief, candidate Ags in the pCITE mammalian expression vector were transcribed and translated in vitro with rabbit reticulocyte lysate (TNT T7 Quick Coupled Transcription/Translation; Promega) using biotinylated lysine transfer RNA (Transcend tRNA; Promega). Expressed protein was immunoprecipitated using patient plasma, as described previously.20 Immunoprecipitated proteins were subjected to SDS-PAGE on 10% Tris-HCl polyacrylamide gels (Bio-Rad), transferred to nitrocellulose membranes in Tris-glycine buffer with 20% methanol (blocked overnight in 1× TBS/0.5% Tween-20), and detected using streptavidin-HRP conjugate (1:20 000 dilution; MP Biomedicals).
We further developed ELISA assays in which recombinant DAPK2 and PIM1 proteins, expressed in a baculovirus-infected insect cell line (Ultimate ORF Collection; Invitrogen) were applied at 5 μg/mL to Nunc C96 Maxisorp plates (Fisher Scientific) in carbonate buffer (pH 9.6) overnight at 4°C, then blocked in PBS with 0.05% Tween plus 2% milk. Plasma samples were added to duplicate wells in blocking buffer (1:200 dilution) and incubated for 3 hours at 25°C. Binding of patient Ig to test proteins was detected using goat anti–human IgG-alkaline phosphatase secondary Ab (1:1000 dilution, Jackson ImmunoResearch Laboratories) and visualized with p-nitrophenyl phosphate substrate (Sigma-Aldrich). Absorbance was read at 405 nm. Samples were defined as Ag reactive if the signal was greater than 2 SDs above the mean of 10 normal volunteers.
Quantitative PCR for molecular detection of disease and for gene expression
The clonotypic IgH sequence of patient A was identified using a panel of VH-specific PCR primers, as described previously.22 Based on this sequence, a quantitative TaqMan PCR assay was designed such that a sequence-specific probe was located in the region of junctional diversity (Applied Biosystems). This assay was applied to reverse-transcribed RNA (SuperScript II reverse transcriptase; Invitrogen) isolated from PBMCs, BMMCs, or CD138+-selected (Miltenyi Biotec) BMMCs (RNeasy kit; QIAGEN).
Direct quantification of gene expression of candidate Ags was performed using gene-specific TaqMan Gene Expression Assays (Applied Biosystems), applied to reverse-transcribed RNA (RNeasy; QIAGEN) from total PBMCs immunomagnetically selected CD19+ cells from healthy donors (Miltenyi Biotec) or from BMMCs of MM patients (> 95% malignant plasma cells by immunohistochemistry). All PCR reactions consisted of: 50°C for 1 minute for 1 cycle, 95°C for 10 minutes for 1 cycle, and 40 cycles of 95°C for 15 seconds followed by 60°C for 1 minute (7500 Fast Real-time PCR cycler; Applied Biosystems). Test transcripts were quantified relative to GAPDH transcripts by calculating 2 × (GAPDH CT − target CT). The statistical significance between sample groups was determined by 1-sided exact Wilcoxon rank-sum test.
Generation of cell lysates and Western blotting
Whole-cell lysate was generated from tumor cell lines or from patient samples by lysis with RIPA buffer (1% NP40, 0.5% deoxycholate, 0.1% SDS, 125mM sodium chloride, 50mM HEPES, pH 7.4) in the presence of protease inhibitors. Lysates (20 μg of total protein per lane) were subjected to 4%-15% gradient SDS-PAGE in Tris-glycine buffer, and transferred onto nitrocellulose membranes in Tris-glycine buffer containing 20% methanol. Rabbit polyclonal DAPK2 Ab (1:1000; ProSci) and mouse PIM1 mAb (clone 12H8, 1:1000; Santa Cruz Biotechnology) were used for Western blotting. Ab to β-actin (1:3000; Sigma-Aldrich) was used as a control to ensure equal loading of lanes.
Generation of APCs
To generate dendritic cells, CD14+ cells were isolated from leukapheresis material by immunomagnetic enrichment (Miltenyi Biotec), and cultured in the presence of 50 ng/mL of GM-CSF (Genzyme) and 20 ng/mL of IL-4 (R&D Systems) in medium composed of IMDM (Invitrogen) supplemented with 10% human AB serum (Cellgro), 2mM glutamine (Cellgro), 50 μg/mL of human transferrin (Roche), 5 μg/mL of human insulin (Sigma-Aldrich), and 15 μg/mL of gentamycin (Invitrogen). On days 3 and 6, 50% fresh medium with cytokines was added. On day 7, cells were matured for 48 hours with prostaglandin E2 (1 μg/mL; Sigma-Aldrich), IL-1α (10 ng/mL; BD Biosciences), IL-6 (1000 U/mL; BD Biosciences), and TNFα (10 ng/mL; Genzyme). The TAP-deficient, HLA-A*0201–expressing cell line T2 was obtained from Dr Peter Cresswell (Yale University School of Medicine, New Haven, CT). PIM1 and DAPK2 were expressed in APCs by nucleofecting 20 μg of DNA plasmids encoding these genes into 2 × 106 cells K562 cells, stably expressing HLA-A*0201 in 100 μL of PBS/10% HEPES buffer (Amaxa Nucleofector, Program T-016; Lonza). For some experiments, HLA class I–blocking Ab was used (clone w6/32).
T2-binding assay
The prediction servers IEDB ANN, NetMHC ANN, and MHC-I Multiple Matrix were used to predict HLA-A*0201–binding nonameric peptides derived from DAPK2 and PIM1. Synthesized peptides were tested for their HLA-A2–binding ability using the T2-binding assay.23 In brief, 1 million T2 cells per condition were washed extensively with serum-free IMDM (Invitrogen) and then incubated in serum-free IMDM with or without individual predicted peptides or the influenza M1 peptide at 40μM. Cells were harvested at 0 and 24 hours, stained with FITC-labeled mouse anti–HLA-A*0201 (clone BB7.2; BD Pharmingen), and fixed in 2% formalin overnight. Fixed cells were analyzed by flow cytometry (FACSCanto II; BD Biosciences) to obtain the mean fluorescence intensity (MFI) of 10 000 events. Fluorescence index was calculated as follows: (MFI with peptide − MFI without peptide)/(MFI without peptide).
Detection of DAPK2 and PIM1-specific T-cell responses
Patient reactivity to peptides was tested by ex vivo stimulation using total PBMCs isolated from fresh patient blood collected after HSCT. One to 2 million PBMCs were stimulated with 10μM concentrations of individual peptides in the presence of 10 μg/mL of IL-7 for 7 days, and then tested by ELISpot assay. These peptides included the DAPK2-derived peptide D1156-164, (MLLDKNIPI) and the PIM1-derived peptide P4191-199 (ALLKDTVYT). For some experiments, banked PBMCs were thawed and stimulated with irradiated T2 cells, pulsed with 10 μg/mL of D1 or P4 peptide in RPMI 1640 (Invitrogen) supplemented with 10% human AB serum (Cellgro), 2mM glutamine (Cellgro), 10mM HEPES (Cellgro), and 15 μg/mL of gentamycin (Invitrogen) in the presence of 10 ng/mL of IL-7 (Endogen). ELISpot was performed using autologous matured dendritic cells pulsed with 10μM peptide (20 000 cells/well) co-incubated with the stimulated PBMCs (20 000 cells/well) plated in duplicate on MultiScreen-IP plates (Millipore) for 24 hours. IFN-γ secretion was detected using capture and detection Abs as directed by the manufacturer (Mabtech) and imaged using an ImmunoSpot Series Analyzer (Cellular Technology). Ag-specific reactivity was also detected by staining of cells with PE-conjugated peptide-specific tetramer (provided by the National Institutes of Health Tetramer Core Facility, Emory University), together with anti–CD8-FITC (Beckmann Coulter) and analyzed by flow cytometry (FACSAria II, BD Biosciences).
Generation and testing of D1- and P4-specific T-cell lines
Patient PBMCs were thawed and stimulated weekly for 28 days with irradiated T2 cells (5000 rad) pulsed with 10 μg/mL of peptides D1 or P4. One week after the second stimulation, CD8+ T cells were immunomagnetically selected (Miltenyi Biotec) for reculturing. One week after the fourth restimulation, peptide-specific tetramer-reactive T cells were sorted by flow cytometry and expanded on irradiated (5000 rad) allogeneic PBMCs and EBV-LCL feeder cells in the presence of 10 μL/mL of phytohemagglutinin (Invitrogen) and 100 IU/mL of IL-2 (Novartis). Specificity of the T-cell lines was confirmed by testing with P4- and D1-specific tetramers and by ELISpot. To determine the responses of peptide-specific T cells against tumors, 50 000 cells from the BM of primary MM patients, from healthy donors, or from the MM cell lines were used as T-cell targets for the ELISpot assay. HLA-A2 status was confirmed by flow cytometry using the BB7.2 Ab.
Results
Complete molecular remission of MM after syngeneic HSCT for patient A
We identified a patient (patient A) who achieved complete molecular remission after syngeneic HSCT. As summarized in Table 1, this patient was a 48-year-old HLA-A2+ woman with progressive stage II IgG MM diagnosed 9 years before HSCT. Before HSCT, she underwent induction with dexamethasone, lenalidomide, and bortezomib. At the time of HSCT, she demonstrated persistent residual disease with a serum monoclonal protein level of 0.6 g/dL and detectable monoclonal gammopathy on immunofixation (Figure 1A). After myeloablative conditioning with high-dose melphalan, the patient received CD34+ peripheral blood stem cells (6.85 × 106 cells/kg) from her genotypically identical twin. At approximately 2.6 months after HSCT, neither serum monoclonal protein nor monoclonal gammopathy were detectable. Tumor-specific IgH transcripts remained detectable by quantitative real-time PCR of CD138+ BM RNA, albeit with an almost 4-log reduction from pre-HSCT levels. However, as shown in Figure 1B, repeat analysis of total BMMCs from 24 to > 60 months after HSCT demonstrated continuing molecular remission. Having observed the effective elimination of MM in this patient, we sought to identify the tumor-associated Ags targeted by her syngeneic donor graft.
Clinical characteristics of patient A
| Age/ sex . | Time from diagnosis to HSCT, mo . | Therapies prior to HSCT . | HSCT conditioning regimen . | Stem cell dose, cells/kg body wt . | Stem cell source . | Time from HSCT to complete response, mo* . | Time from HSCT to molecular response, mo . |
|---|---|---|---|---|---|---|---|
| 48/F | 108 | Lenalidomide, bortezomib, dexamethasone | High-dose melphalan | 6.85 × 106 | PBSCs | 2.6 | 24 |
| Age/ sex . | Time from diagnosis to HSCT, mo . | Therapies prior to HSCT . | HSCT conditioning regimen . | Stem cell dose, cells/kg body wt . | Stem cell source . | Time from HSCT to complete response, mo* . | Time from HSCT to molecular response, mo . |
|---|---|---|---|---|---|---|---|
| 48/F | 108 | Lenalidomide, bortezomib, dexamethasone | High-dose melphalan | 6.85 × 106 | PBSCs | 2.6 | 24 |
Defined as negative immunofixation and monoclonal protein of 0 g/dL.
PBSCs indicates peripheral blood stem cells.
Achievement of durable molecular remission after syngeneic HSCT for treatment of MM in patient A. (A) Levels of serum monoclonal protein (g/dL) become undetectable by 1.5 months after HSCT. Arrows indicate the time points screened by protein microarray. Vertical dashed line indicates time of syngeneic stem cell infusion. (B) Expression of transcripts containing the IgH sequence unique to patient A's tumor is detectable by quantitative real-time PCR in BMMCs (black squares and solid line) and PBMCs (gray squares and line) before syngeneic HSCT, but is undetectable beginning at 24 months after HSCT. Values shown are tumor-specific IgH transcripts normalized to GAPDH transcript expression. “Undetectable” indicates samples with acceptable GAPDH values but absent tumor-specific IgH transcript expression.
Achievement of durable molecular remission after syngeneic HSCT for treatment of MM in patient A. (A) Levels of serum monoclonal protein (g/dL) become undetectable by 1.5 months after HSCT. Arrows indicate the time points screened by protein microarray. Vertical dashed line indicates time of syngeneic stem cell infusion. (B) Expression of transcripts containing the IgH sequence unique to patient A's tumor is detectable by quantitative real-time PCR in BMMCs (black squares and solid line) and PBMCs (gray squares and line) before syngeneic HSCT, but is undetectable beginning at 24 months after HSCT. Values shown are tumor-specific IgH transcripts normalized to GAPDH transcript expression. “Undetectable” indicates samples with acceptable GAPDH values but absent tumor-specific IgH transcript expression.
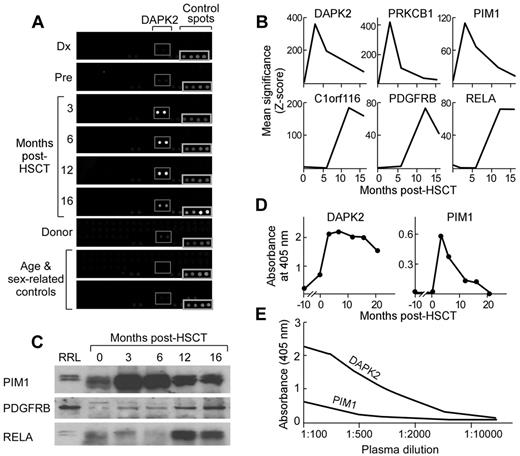
Serologic screening identifies 6 candidate Ags, including DAPK2 and PIM1
We screened plasma samples from our index patient at 9 serial time points before and after HSCT against commercially available protein microarrays consisting of recombinant proteins expressed from approximately 8000 open reading frames in baculovirus-infected insect cell lines and spotted on glass slides. Binding of plasma Ab to spotted proteins was detected with fluorescently labeled anti–human IgG (Figure 2A). Negative controls included plasma from the patient's identical twin donor and from 2 age-, sex-, and parity-matched healthy controls. All 9 samples were screened in 2 replicates against 1 lot of protein microarrays. Significance of Ab binding to protein spots was determined using established analytical methods.20 Candidate Ags were defined as those proteins eliciting significantly more plasma Ab binding from post-HSCT plasma compared with pre-HSCT plasma.
Serologic screening identifies high-titer Ab responses against DAPK2, PDGFRB, PIM1, and PRKCB1 developing after syngeneic HSCT. (A) Plasma samples collected from patient A at serial time points before and after HSCT were screened by ProtoArray protein microarray. Subarrays demonstrating DAPK2 reactivity are highlighted in the gray boxes. Spots seen in the bottom right corner of all subarray images are control spots. (B) Arrays were analyzed using 2 methods described previously.48 Six Ags, DAPK2, PDGFRB, C1orf116, RELA, PIM1, and PRKCB1, elicited significantly greater reactivity after HSCT compared with before HSCT. Significance (Z-scores) scores of Ab reactivity at time points are shown. (C) Ag binding was confirmed by immunoprecipitation of biotinylated target Ags expressed in vitro in rabbit reticulocyte lysate by patient plasma at serial time points. Immunoprecipitated Ags were detected using streptavidin-conjugated secondary Ab. Representative Western blots are shown. The first lane is whole rabbit reticulocyte lysate expressing target Ags that was not subjected to immunoprecipitation. (D) Plasma samples (1:200 dilution) collected from patient A at serial time points before and after HSCT were tested by ELISA (plates coated with 5 μg/mL of recombinant protein). Ab IgG responses against DAPK2 and PIM1 arose and peaked at 3 months after HSCT. (E) Candidate Ags elicit high-titer Ab responses, as indicated by titration studies of serum using ELISA assays. Reactivity remained detectable at dilutions of 1:10 000 (DAPK2) and 1:5000 (PIM1).
Serologic screening identifies high-titer Ab responses against DAPK2, PDGFRB, PIM1, and PRKCB1 developing after syngeneic HSCT. (A) Plasma samples collected from patient A at serial time points before and after HSCT were screened by ProtoArray protein microarray. Subarrays demonstrating DAPK2 reactivity are highlighted in the gray boxes. Spots seen in the bottom right corner of all subarray images are control spots. (B) Arrays were analyzed using 2 methods described previously.48 Six Ags, DAPK2, PDGFRB, C1orf116, RELA, PIM1, and PRKCB1, elicited significantly greater reactivity after HSCT compared with before HSCT. Significance (Z-scores) scores of Ab reactivity at time points are shown. (C) Ag binding was confirmed by immunoprecipitation of biotinylated target Ags expressed in vitro in rabbit reticulocyte lysate by patient plasma at serial time points. Immunoprecipitated Ags were detected using streptavidin-conjugated secondary Ab. Representative Western blots are shown. The first lane is whole rabbit reticulocyte lysate expressing target Ags that was not subjected to immunoprecipitation. (D) Plasma samples (1:200 dilution) collected from patient A at serial time points before and after HSCT were tested by ELISA (plates coated with 5 μg/mL of recombinant protein). Ab IgG responses against DAPK2 and PIM1 arose and peaked at 3 months after HSCT. (E) Candidate Ags elicit high-titer Ab responses, as indicated by titration studies of serum using ELISA assays. Reactivity remained detectable at dilutions of 1:10 000 (DAPK2) and 1:5000 (PIM1).
Six proteins (C1orf116, DAPK2, PDGFRB, PIM1, PRKCB1, and RELA) met the criteria for candidate Ags (Table 2). None elicited significant Ab reactivity from any of the 4 negative control plasma samples based on calculated Z-scores. The increase in significance (Z-score) of Ab binding for post-HSCT plasma was 17- to 10 615-fold higher than pre-HSCT Z-scores (Figure 2B). These significant Ag-specific Ab responses arose in 2 general patterns, early (peaking at 3 months: DAPK2, PIM1, and PRKCB1) or late (peaking at or after 12 months: C1orf116, PDGFRB, and RELA). We validated these statistical findings by confirming the detection of early- and late-peaking patterns of Ag-specific B-cell reactivity by measuring the amount of independently produced Ag immunoprecipitated by post-HSCT plasma–derived Ig from patient A (representative examples are shown in Figure 2C).
Candidate Ags identified by protein microarray screening
| Gene symbol* . | NCBI gene ID . | Chromosome . | Size, aa . | Maximum post-HSCT significance† . | Maximum time point, mo . |
|---|---|---|---|---|---|
| DAPK2 | 79098 | 1q32.1 | 370 | +++ | 3 |
| PIM1 | 23604 | 15q22.31 | 313 | +++ | 3 |
| PRKCB1 | 5159 | 5q31-q32 | 671 | +++ | 3 |
| C1orf116 | 5292 | 6p21.2 | 601 | ++ | 12 |
| PDGFRB | 5579 | 16p11.2 | 1106 | ++ | 12 |
| RELA | 5970 | 11q13 | 551 | + | 15 |
| Gene symbol* . | NCBI gene ID . | Chromosome . | Size, aa . | Maximum post-HSCT significance† . | Maximum time point, mo . |
|---|---|---|---|---|---|
| DAPK2 | 79098 | 1q32.1 | 370 | +++ | 3 |
| PIM1 | 23604 | 15q22.31 | 313 | +++ | 3 |
| PRKCB1 | 5159 | 5q31-q32 | 671 | +++ | 3 |
| C1orf116 | 5292 | 6p21.2 | 601 | ++ | 12 |
| PDGFRB | 5579 | 16p11.2 | 1106 | ++ | 12 |
| RELA | 5970 | 11q13 | 551 | + | 15 |
UniProt identifiers: DAPK2, Q1RMF4; PIM1 isoform 2, P11309–2; PRKCB1, P05771; C1orf116 (SARG), Q9BW04; PDGFRB, P09619; and RELA, Q04206.
+ indicates 10- to 20-fold increase in significance (Z-score) of Ab binding in posttransplantation plasma; ++, 20- to 50-fold increase; and +++, > 50-fold increase.
To more accurately quantify the immune responses against candidate Ags, we performed ELISA assays in which recombinant protein expressed in baculovirus-infected insect cell lines was used as a coating Ag. Using ELISA, the Ab responses against DAPK2 and PIM1 by patient A closely mirrored the reactivity pattern calculated based on significance score, peaking at 3 months after HSCT (Figure 2D), whereas only low or minimal reactivity was detected in pre-HSCT samples. Titration by serial dilution of plasma with maximal reactivity against Ags confirmed the high-titer reactivity against both DAPK2 and PIM1 (1:10 000 and 1:5000, respectively; Figure 2E).
DAPK1 and PIM1 are expressed in primary myeloma cells
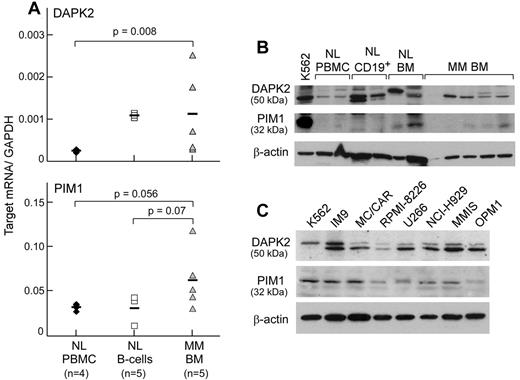
Tumor-associated Ags are commonly overexpressed in tumor tissue.15-18 Using quantitative real-time PCR, DAPK2 and PIM1 were enriched in transcript expression in BM from a primary MM patient containing > 95% malignant plasma cells (Figure 3A). Mean relative transcript levels of DAPK2 (normalized to GAPDH) in 5 primary MM samples were 4.8-fold higher than in 4 healthy donor PBMC samples (P = .008), and comparable transcript levels were observed between primary MM and 5 healthy donor B-cell samples. Mean relative transcript levels of PIM1 in primary MM samples were 2.1- and 2-fold higher than in healthy donor B cells (P = .07) and PBMCs (P = .056), respectively. The RNA expression levels of the 4 other candidate Ags were similar between healthy and malignant hematopoietic tissues.
Expression of DAPK2 and PIM1 is enriched in primary MM cells. (A) DAPK2 and PIM1 have high transcript expression in primary MM BMMCs (gray triangles), but not in normal PBMCs (black diamonds) by gene-specific quantitative real-time PCR. BM specimens contained > 95% tumor by immunohistochemistry. DAPK2, but not PIM1, was also highly expressed in normal CD19+ B cells (white squares). Bars indicate the median value for transcript expression relative to GAPDH. Statistical significance was determined by 1-sided exact Wilcoxon rank-sum test. (B) Western blotting of cell lysates (20 μg of protein per lane) from normal hematopoietic cells and primary MM BMMCs with Ab against either DAPK2 or PIM1 reveals protein expression in 4 (DAPK2) or 3 (PIM1) of 5 primary MM BMMCs. (C) Western blotting of MM cell line lysates (20 μg of protein per lane) showed expression of DAPK2 and PIM1 in all tested MM cell lines.
Expression of DAPK2 and PIM1 is enriched in primary MM cells. (A) DAPK2 and PIM1 have high transcript expression in primary MM BMMCs (gray triangles), but not in normal PBMCs (black diamonds) by gene-specific quantitative real-time PCR. BM specimens contained > 95% tumor by immunohistochemistry. DAPK2, but not PIM1, was also highly expressed in normal CD19+ B cells (white squares). Bars indicate the median value for transcript expression relative to GAPDH. Statistical significance was determined by 1-sided exact Wilcoxon rank-sum test. (B) Western blotting of cell lysates (20 μg of protein per lane) from normal hematopoietic cells and primary MM BMMCs with Ab against either DAPK2 or PIM1 reveals protein expression in 4 (DAPK2) or 3 (PIM1) of 5 primary MM BMMCs. (C) Western blotting of MM cell line lysates (20 μg of protein per lane) showed expression of DAPK2 and PIM1 in all tested MM cell lines.
Western blot analysis of PIM1 and DAPK2 expression (representative examples are shown in Figure 3B) showed that DAPK2 was expressed in 10 of 13 primary MM samples tested, and that PIM1 was expressed in 9 of 13 primary MM samples. DAPK2 was also expressed in healthy donor CD19+ B cells and BMMCs, whereas PIM1 was highly expressed in 1 of 2 healthy donor BMMC samples. Furthermore, PIM1 and DAPK2 were expressed in all 7 MM cell lines (IM9, MC/CAR, MM1S, NCI-H929, OPM1, RPMI 8226, and U266) tested by Western blot analysis (Figure 3C).
Ab responses against DAPK2 and PIM1 are associated with effectively treated MM
To determine whether candidate Ags are commonly immunogenic in other patients with effective antimyeloma immunity, we used ELISA to test serial plasma samples from 6 MM patients with effective GVM responses after allogeneic HSCT. All 6 patients had achieved complete remissions of 2 years or more without clinically evident GVHD. Significant Ab responses to DAPK2 and PIM1 was defined as reactivity greater than 2 SDs above the mean reactivity of 10 normal donors, and was observed in 3 of 6 patients. Two of these 3 patients developed 2-fold or greater increases in Ab reactivity against DAPK2 after allogeneic HSCT, whereas 2 of 3 developed similar increases in reactivity against PIM1 (Figure 4A). Patient 2 showed gradually increasing levels of DAPK2 reactivity from pre-HSCT levels that peaked at 12 months after HSCT (5-fold increase over before HSCT). Patient 3 developed a 2-fold and 4-fold increase in DAPK2 and PIM1 reactivity at 6 months post-HSCT, respectively, that subsequently diminished. Only an early posttransplantation sample was available for patient 5; however, this patient had a striking increase (78-fold) in post-HSCT reactivity against PIM1 compared with pre-HSCT levels.
MM treatment is associated with the development of Ab responses against DAPK2 and PIM1. (A) Plasma samples from 5 MM patients who demonstrated GVM responses (defined as clinical remission > 2 years) without GVHD were tested by ELISA. Three patients (Pt. 2, black bars; Pt. 3, gray bars; and Pt. 5, white bars) had new or markedly increased Ab reactivity to DAPK2 and/or PIM1 after HSCT. The dashed line indicates a cutoff based on 2 SD above the mean of Ab reactivity of 10 normal donors for each Ag. (B) Ab responses against DAPK2 (black bars) and PIM1 (gray bars) are also seen by ELISA in MM patients treated with standard chemotherapy or autologous HSCT, but are minimal in patients with untreated MM or healthy donors.
MM treatment is associated with the development of Ab responses against DAPK2 and PIM1. (A) Plasma samples from 5 MM patients who demonstrated GVM responses (defined as clinical remission > 2 years) without GVHD were tested by ELISA. Three patients (Pt. 2, black bars; Pt. 3, gray bars; and Pt. 5, white bars) had new or markedly increased Ab reactivity to DAPK2 and/or PIM1 after HSCT. The dashed line indicates a cutoff based on 2 SD above the mean of Ab reactivity of 10 normal donors for each Ag. (B) Ab responses against DAPK2 (black bars) and PIM1 (gray bars) are also seen by ELISA in MM patients treated with standard chemotherapy or autologous HSCT, but are minimal in patients with untreated MM or healthy donors.
To further explore the relationship between MM and Ab responses against DAPK2 and PIM1, we performed additional ELISA studies on MM patients before any therapy (“untreated,” n = 10), after standard chemotherapy (n = 16), and after autologous HSCT (n = 10), and in healthy volunteers (n = 10). As summarized in Figure 4B, 6 of 16 MM patients treated with standard chemotherapy (37.5%) and 3 of 10 MM patients treated with autologous HSCT (30%) displayed Ab reactivity against DAPK2. In contrast, reactivity against DAPK2 was seen in only 1 of 10 untreated MM patients (10%) and in 0 of 10 normal volunteers (0%). For PIM1, 3 of 16 chemotherapy-treated patients (18.8%) and 1 of 10 patients receiving autologous HSCT (10%) were reactive against the Ag. One of 10 untreated MM patients (10%) and 1 of 10 normal volunteers (10%) had Ab responses to PIM1. Our results demonstrate that humoral immunity against DAPK2 and PIM1 commonly occurs in the setting of treatment for MM.
DAPK2 and PIM1 peptides elicit T-cell reactivity in patient A
To determine whether the high-titer B-cell responses to PIM1 and DAPK2 were coordinated with cell-mediated Ag-specific immunity, we tested posttransplantation T cells of patient A for their ability to recognize selected DAPK2- and PIM1-derived 9-mers. Because the patient was HLA-A2 positive, we focused our inquiry on DAPK2- and PIM1-derived peptides that were predicted to bind to this HLA allele. We used consensus predictions of the servers IEDB-ANN, NetMHC-ANN, and MHC-I Multiple Matrix to identify a total of 10 DAPK2-derived (D1-D10) and 7 PIM1-derived (P1-P7) nonameric peptides. All 17 peptides were confirmed to bind to HLA-A2 by the T2-binding assay (Table 3). We therefore tested all 17 peptides against fresh patient PBMCs obtained at 2.5 years after HSCT using an ex vivo stimulation assay.
Predicted peptides derived from candidate Ags DAPK2 and PIM1
| Protein . | ID . | Position . | Sequence . | Consensus rank* . | T2 binding . | |
|---|---|---|---|---|---|---|
| FI-0 . | FI-24 . | |||||
| Influenza M1 | M1 | 58 | GILGFVFTL | n/a | 2.34 | 2.17 |
| DAPK2 | D1 | 156 | MLLDKNIPI | 3 | 3.92 | 2.28 |
| D2 | 316 | KLSFSIVSL | 6 | 0.69 | 0.06 | |
| D3 | 220 | LLSGASPFL | 10 | 1.78 | 0.98 | |
| D4 | 129 | FIKQILDGV | 13 | 0.13 | 0 | |
| D5 | 132 | QILDGVNYL | 17 | 1.92 | 0.18 | |
| D6 | 205 | GLEADMWSI | 19 | 1.50 | 0 | |
| D7 | 186 | NIFGTPEFV | 26 | 2.99 | 0.07 | |
| D8 | 81 | VLHHNVITL | 26 | 2.05 | 0.36 | |
| D9 | 211 | WSIGVITYI | 29 | 0.14 | 0 | |
| D10 | 212 | SIGVITYIL | 35 | 0.23 | 0 | |
| PIM1 (isoform 2) | P1 | 1 | MLLSKINSL | 3 | 1.19 | 0 |
| P2 | 183 | KLIDFGSGA | 7 | 0.17 | 0 | |
| P3 | 118 | LILERPEPV | 14 | 1.56 | 2.68 | |
| P4 | 191 | ALLKDTVYT | 16 | 1.03 | 0 | |
| P5 | 147 | FFWQVLEAV | 16 | 0.57 | 0.04 | |
| P6 | 265 | HLIRWCLAL | 21 | 0.85 | 0.14 | |
| P7 | 2 | LLSKINSLA | 21 | 1.8 | 0.39 | |
| Protein . | ID . | Position . | Sequence . | Consensus rank* . | T2 binding . | |
|---|---|---|---|---|---|---|
| FI-0 . | FI-24 . | |||||
| Influenza M1 | M1 | 58 | GILGFVFTL | n/a | 2.34 | 2.17 |
| DAPK2 | D1 | 156 | MLLDKNIPI | 3 | 3.92 | 2.28 |
| D2 | 316 | KLSFSIVSL | 6 | 0.69 | 0.06 | |
| D3 | 220 | LLSGASPFL | 10 | 1.78 | 0.98 | |
| D4 | 129 | FIKQILDGV | 13 | 0.13 | 0 | |
| D5 | 132 | QILDGVNYL | 17 | 1.92 | 0.18 | |
| D6 | 205 | GLEADMWSI | 19 | 1.50 | 0 | |
| D7 | 186 | NIFGTPEFV | 26 | 2.99 | 0.07 | |
| D8 | 81 | VLHHNVITL | 26 | 2.05 | 0.36 | |
| D9 | 211 | WSIGVITYI | 29 | 0.14 | 0 | |
| D10 | 212 | SIGVITYIL | 35 | 0.23 | 0 | |
| PIM1 (isoform 2) | P1 | 1 | MLLSKINSL | 3 | 1.19 | 0 |
| P2 | 183 | KLIDFGSGA | 7 | 0.17 | 0 | |
| P3 | 118 | LILERPEPV | 14 | 1.56 | 2.68 | |
| P4 | 191 | ALLKDTVYT | 16 | 1.03 | 0 | |
| P5 | 147 | FFWQVLEAV | 16 | 0.57 | 0.04 | |
| P6 | 265 | HLIRWCLAL | 21 | 0.85 | 0.14 | |
| P7 | 2 | LLSKINSLA | 21 | 1.8 | 0.39 | |
Consensus rank score represents the composite ranking of peptides predicted by all the 3 servers (IEDB-ANN, NetMHC-ANN, and MHC-I multiple matrix). The lower the composite rank number, the higher the predicted binding affinity.
FI-0 indicates the fluorescence intensity at 0 hours; and FI-24, fluorescence intensity at 24 hours.
After 7 days of expansion to predicted peptides, T-cell reactivity to the peptides was assessed using IFN-γ secretion (ELISpot assay). As shown in Figure 5A, we observed a 3.8-fold increase in IFN-γ secretion in response to stimulation with D1 peptide (8 spot-forming counts [SFCs] per 20 000 cells or 0.038%) compared with the mean SFCs for the other DAPK2-derived peptides (range, 0.6-1.6 SFCs per 20 000 cells [0.003%-0.008%]). Similarly, we observed a striking T-cell response to the PIM1-derived peptide P4 (22 SFCs per 20 000 cells or 0.11%; Figure 5B). Lower levels of reactivity against P1, P2, P5, and P7 were also observed, with frequencies of reactivity in PBMCs of 0.026%, 0.030%, 0.015%, and 0.018%, respectively. The frequency of reactivity for the remainder of the PIM1 peptides was less than 0.005%.
DAPK2- and PIM1-derived nonameric peptides elicit T-cell reactivity in post-HSCT PBMCs from patient A. Fresh post-HSCT PBMCs were stimulated ex vivo with 10μM concentrations of individual peptides in the presence of 10 μg/mL of IL-7 for 7 days, then tested by ELISpot with peptide-pulsed autologous matured dendritic cells (PBMC:APC ratio = 1:1). (A) T cells from patient A 2.5 years after HSCT show reactivity to the DAPK2-derived peptide D1, but not to other DAPK2-derived peptides. (B) T cells from patient A 2.75 years after HSCT are strongly reactive to the PIM1-derived peptide P4 and weakly reactive to P1, P2, P5, and P7.
DAPK2- and PIM1-derived nonameric peptides elicit T-cell reactivity in post-HSCT PBMCs from patient A. Fresh post-HSCT PBMCs were stimulated ex vivo with 10μM concentrations of individual peptides in the presence of 10 μg/mL of IL-7 for 7 days, then tested by ELISpot with peptide-pulsed autologous matured dendritic cells (PBMC:APC ratio = 1:1). (A) T cells from patient A 2.5 years after HSCT show reactivity to the DAPK2-derived peptide D1, but not to other DAPK2-derived peptides. (B) T cells from patient A 2.75 years after HSCT are strongly reactive to the PIM1-derived peptide P4 and weakly reactive to P1, P2, P5, and P7.
Because of the evidence of marked memory responses in patient A to peptides D1 and P4, subsequent experiments were focused on the characterization of T-cell responses against these 2 predicted peptides. D1- and P4-specific T cells were isolated by peptide-specific tetramers and expanded. These T-cell lines were enriched for D1-reactive (6.6%) and P4-reactive (12%) T cells (Figure 6A).
D1- and P4-specific T cells are isolated by tetramer sorting and recognize whole processed and presented Ag. (A) Patient A CD3+ T cells were repeatedly stimulated with either D1 or P4 peptide-pulsed T2 cells before staining with peptide-specific tetramer. Tetramer and CD8 double-positive cells were isolated by FACS (representative FACS plots shown). (B) Sorted D1- or P4-positive T cells were tested by ELISpot against K562 cells stably transfected to express HLA-A2 (K562/A2 cells) that were nucleofected to overexpress either DAPK2 or PIM1. As expected, overexpression of DAPK2 induced apoptosis in nucleofected K562 cells.24 P4-specific T cells recognized processed and presented PIM1, an effect that was blocked by the class I–blocking Ab w6/32. (C) Sorted D1- or P4-positive T cells were also tested against the A2-positive MM cell lines MC/CAR and IM9 and the A2-negative MM cell lines OPM2 and NCI 4929. Both D1- and P4-specific T cells showed reactivity to HLA-A2–positive cell lines, but not to HLA-A2–negative cell lines. IFN-γ secretion was blocked by co-incubation with w6/32 Ab.
D1- and P4-specific T cells are isolated by tetramer sorting and recognize whole processed and presented Ag. (A) Patient A CD3+ T cells were repeatedly stimulated with either D1 or P4 peptide-pulsed T2 cells before staining with peptide-specific tetramer. Tetramer and CD8 double-positive cells were isolated by FACS (representative FACS plots shown). (B) Sorted D1- or P4-positive T cells were tested by ELISpot against K562 cells stably transfected to express HLA-A2 (K562/A2 cells) that were nucleofected to overexpress either DAPK2 or PIM1. As expected, overexpression of DAPK2 induced apoptosis in nucleofected K562 cells.24 P4-specific T cells recognized processed and presented PIM1, an effect that was blocked by the class I–blocking Ab w6/32. (C) Sorted D1- or P4-positive T cells were also tested against the A2-positive MM cell lines MC/CAR and IM9 and the A2-negative MM cell lines OPM2 and NCI 4929. Both D1- and P4-specific T cells showed reactivity to HLA-A2–positive cell lines, but not to HLA-A2–negative cell lines. IFN-γ secretion was blocked by co-incubation with w6/32 Ab.
D1- and P4-specific T cells recognize MM cells in an HLA-A2–restricted manner
We undertook further studies using the D1- and P4-reactive T-cell lines to confirm that D1 and P4 represent processed peptides of DAPK2 and PIM1, respectively. As shown in Figure 6B, P4-specific T cells were reactive to both HLA-A2–expressing K562 cells (K562/A2 cells) that were pulsed with exogenous peptide and K562/A2 cells that were nucleofected with plasmid encoding whole PIM1 protein. IFN-γ secretion in response to peptide or whole protein was blocked by a class I–blocking Ab. Although D1-specific T cells recognized K562/A2 cells pulsed with exogenous D1 peptide (a process abrogated by class I–blocking Ab), we were unable to evaluate reactivity against DAPK2 overexpressed in K562 cells because forced elevated expression of DAPK2 reproducibly caused K562 cells to undergo apoptosis, as described previously for this molecule.24
To further confirm that D1 and P4 peptides are naturally processed and presented by myeloma cells in the context of HLA-A2, we tested 2 other classes of target cells. First, we tested the T-cell lines against 2 HLA-A2+ (MC/CAR and IM-9) and 2 HLA-A2–negative (OPM-2 and NCI-H929) MM cell lines. Consistent with the HLA-A2–restricted presentation of P4 and D1 by these MM cell lines, we observed reactivity by both D1- and P4-specific T-cell lines against 2 of 2 HLA-A2+ myeloma cell lines (with reactivity that was blocked by class I–blocking Ab), and against neither of the HLA-A2–negative cell lines (Figure 6C).
Second, we further tested D1- and P4-reactive T cells against CD138-positive cells isolated from BM from primary MM patients (Figure 7). Both D1- and P4-specific T cells were reactive against 4 of 5 HLA-A2–positive BMMC samples from MM patients (average 35 SFCs per 20 000 cells). In contrast, D1- and P4-specific T cells were not reactive against any of the 4 HLA-A2–negative patient samples (average 3.5 SFCs per 20 000 cells) nor the 3 HLA-A2–positive healthy donor BMMC samples. These results demonstrate that the D1 and P4 immunogenic epitopes are commonly presented on myeloma cells by HLA-A2 molecules.
D1- and P4-specific CD8 T cells recognize primary MM tissue from HLA-A2–positive patients. Sorted D1- or P4-positive CD8 T cells were tested by ELISpot against MM BMMCs isolated from fresh BM from HLA-A2–positive donors. D1-specific (A) and P4-specific (B) T cells recognized primary MM BMMCs from HLA-A2–positive donors (n = 5), but not healthy HLA-A2–positive BMMCs (n = 2) or HLA-A2–negative MM BMMCs (n = 4).
D1- and P4-specific CD8 T cells recognize primary MM tissue from HLA-A2–positive patients. Sorted D1- or P4-positive CD8 T cells were tested by ELISpot against MM BMMCs isolated from fresh BM from HLA-A2–positive donors. D1-specific (A) and P4-specific (B) T cells recognized primary MM BMMCs from HLA-A2–positive donors (n = 5), but not healthy HLA-A2–positive BMMCs (n = 2) or HLA-A2–negative MM BMMCs (n = 4).
Discussion
Syngeneic HSCT for patients with MM produces higher complete remission rates, decreased relapse rates, and improved survival compared with autologous HSCT.25-27 The clinical superiority of syngeneic over autologous HSCT has been attributed to the absence of tumor contamination28,29 in the donor graft, and has been hypothesized to also involve the ability to reconstitute normal donor immunity in a MM Ag-rich environment. The results of the present study demonstrate the development of bona fide GVM immunity in the syngeneic setting, providing evidence that immunologic factors contribute to improved outcomes after syngeneic HSCT. Our findings are supported by previous studies of MM patients successfully treated with donor lymphocyte infusion, an established therapy with well-characterized GVM activity3,4,7,8 in which potent B- and T-cell responses against TAAs developed in temporal association with effective MM elimination.15,30 In the present study, disease regression after syngeneic HSCT was likewise associated with potent coordinated B- and T-cell responses against 2 overexpressed MM-associated Ags (DAPK2 and PIM1). Through this system, we identified DAPK2 and PIM1 as potentially promising targets for myeloma-specific immunotherapy, because Ag-specific T cells against both Ags were consistently reactive against HLA-A2+ primary tumor cells.
Both PIM1 and DAPK2 have been shown previously to play functional roles in lymphoid malignancies, including MM. PIM1 is a known proto-oncogene that participates in the c-myc–signaling pathway31 and has established associations with B-cell lymphomas and other malignancies.32 Epigenetic silencing of DAPK2, a proapoptotic serine/threonine protein kinase33 with a tumor-suppressor function, and of related death-associated protein kinases has been observed in multiple neoplasms,34 including MM,35 in which loss of expression appears to worsen clinical outcomes. Based on criteria reported recently by Cheever et al for the selection of optimal Ags for immunotherapy based on function and expression,36 PIM1 and DAPK2 are both promising targets (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). One criterion not included in this schema, however, is the ability of Ags to elicit immunity in the setting of therapeutic response. This may be an important and desirable characteristic, because the limitations in the ability of T cells with specificities against existing tumor-specific Ags to expand in the posttransplantation setting has been reported, suggesting that chronic antigenic exposure induces replicative senescence.37
Whereas the precise mechanism of immunogenicity for DAPK2 and PIM1 remains unclear, the characteristics of these Ags provide some clues. One possible mechanism is the expression of tumor-specific genetic alterations. Malignant plasma cells of MM have been documented to harbor translocations and somatic mutations, albeit at low frequency,38,39 and these may produce new epitopes that may be recognized by donor T and B cells. We were unable to perform tumor-mutation analysis in our index patient because of no available tumor sample, but mutations in DAPK2 and PIM1 were not noted among a large series of MM samples analyzed by whole-tumor sequencing.39 Moreover, mutation is unlikely to be the mechanism underlying the immunogenicity of PIM1 and DAPK2, because the immunogenic D1 and P4 A2-binding peptides were derived from wild-type DAPK2 and PIM1 sequences. Conversely, aberrant expression or overexpression of Ags in malignant tissues (which we observed for DAPK2 and PIM1) has been proposed as a factor influencing immunogenicity and other Ab-defined targets.15-18 Cellular localization may also function to enhance immunogenicity. Recent studies suggest that proteins complexed with nucleic acid are immunostimulatory,40 providing a plausible mechanism of immunogenicity for the nuclear protein PIM1. Expression of DAPK2 generates apoptosis,24 and apoptotic blebs have been reported to possess enriched expression of immunogenic Ags.41
Mounting evidence suggests that tumor Ags that elicit coordinated T- and B-cell immunity are viable targets for tumor-specific immunotherapy.42,43 The phenomenon of cross-presentation, in which immune complexes of Ig and exogenous Ag bind FcγR on dendritic cells and in which internalized Ag is presented on MHC class I molecules,6 provides a link between B-cell and CD8+ T-cell responses. Both post-vaccination patient serum44 and mAbs45-47 have been shown to enhance cell-mediated tumor immunity, and these mechanisms are potentially relevant for DAPK2 and PIM1.
Our tool kit for generating effective vaccines has increased greatly in recent years, and includes the development of novel vaccine-delivery methods, more potent adjuvants, and highly active checkpoint blockade inhibitors.48 In this context, defining true tumor-specific Ags is of utmost importance to ensure that potent and focused immune responses leading to effective destruction of tumor cells can be implemented without eliciting autoimmunity. DAPK2 and PIM1, as targets of coordinated humoral and cellular immunity identified in the context of an effective GVM response, are promising targets for future MM-specific immunotherapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Maris Handley, Suzan Lazo-Kallanian, and John Daley for their assistance with flow cytometry; Dr John Donovan for his vital help in designing the tumor-specific IgH-quantitative PCR assay; and the clinical transplantation team at the Dana-Farber Cancer Institute for their assistance with patient samples.
This work was supported by the Multiple Myeloma Research Foundation and the Pasquarello Tissue Bank. C.J.W. also received a Miles and Eleanor Shore Award, a grant from the National Cancer Institute (5R21CA115043-2), the Early Career Physician-Scientist Award of the Howard Hughes Medical Institute, and a clinical investigator award supported by the Damon-Runyon Cancer Research Foundation (CI-38-07).
National Institutes of Health
Authorship
Contribution: M.A.B. and C.J.W. designed the study, performed the experiments, analyzed the data, and wrote the manuscript; A.A., W.Z., and L.Z. performed the experiments; G.L.Z., D.N., and V.B. analyzed the data; and Y.-t.T., R.P., N.M., R.J.S., J.R., E.P.A., and K.C.A. provided vital clinical samples.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Catherine J. Wu, MD, Dana-Farber Cancer Institute, Harvard Institutes of Medicine, Rm 416B, 77 Avenue Louis Pasteur, Boston MA 02115; e-mail: cwu@partners.org.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal