Abstract
Specific imatinib-resistant BCR-ABL1 mutations (Y253H, E255K/V, T315I, F317L, and F359V/C) predict failure of second-line nilotinib or dasatinib therapy in patients with chronic myeloid leukemia; however, such therapy also fails in approximately 40% of patients in the chronic phase of this disease who do not have these resistant mutations. We investigated whether sensitive mutation analysis could identify other poor-risk subgroups. Analysis was performed by direct sequencing and sensitive mass spectrometry on 220 imatinib-resistant patients before they began nilotinib or dasatinib therapy. Patients with resistant mutations by either method (n = 45) were excluded because inferior response was known. Of the remaining 175 patients, 19% had multiple mutations by mass spectrometry versus 9% by sequencing. Compared with 0 or 1 mutation, the presence of multiple mutations was associated with lower rates of complete cytogenetic response (50% vs 21%, P = .003) and major molecular response (31% vs 6%, P = .005) and a higher rate of new resistant mutations (25% vs 56%, P = .0009). Sensitive mutation analysis identified a poor-risk subgroup (15.5% of all patients) with multiple mutations not identified by standard screening.
Introduction
Mutation analysis is required to aid subsequent therapy selection for patients with chronic myeloid leukemia (CML) for whom imatinib therapy fails,1,2 because certain BCR-ABL1 kinase domain mutations confer clinical resistance to nilotinib (Y253H, E255K/V, T315I, and F359V/C)3 or dasatinib (V299L, T315I/A, and F317L/I/V/C)4-7 and are associated with poor outcome.3,7 However, second-line nilotinib or dasatinib therapy also fails in approximately 40% of patients with imatinib-resistant chronic phase (CP) CML without these resistant mutations.3,7 We examined the BCR-ABL1 kinase domain of imatinib-resistant CML patients before commencing nilotinib/dasatinib therapy using sensitive multiplex mass spectrometry,8 to determine whether the number of mutations harbored by individual patients impacted subsequent response. Multiple sensitive mutations were associated with poor response and a high rate of new resistant mutations.
Methods
The patients analyzed were a subset of patients from previously reported studies3,7,9-11 and have been described formerly.8 The trials were run in accordance with the Declaration of Helsinki, and approvals were obtained from the relevant institutional review boards of all participating institutions. Peripheral blood samples of 220 imatinib-resistant CML patients (CP n = 100, accelerated phase n = 64, blast crisis n = 56) collected before subsequent treatment with nilotinib (n = 89) or dasatinib (n = 131) were investigated. The median follow-up during nilotinib/dasatinib treatment for patients in CP, accelerated phase, and blast crisis was 18 (range 2-33), 12 (range 1-36), and 3 (range 1-27) months, respectively. BCR-ABL1 mutation analysis was performed by direct sequencing (detection limit 10%-20%)12 and retrospectively by mass spectrometry (Sequenom MassARRAY; detection limit 0.05%-0.5%),8 which detects 31 common mutations, including those clinically resistant to nilotinib3 or dasatinib.4-7 Mutations that were not clinically resistant to the second-line inhibitor received by the patient were classified as sensitive. Frequencies were compared with the χ2 and Fisher exact tests.
Results and discussion
BCR-ABL1 mutations (n = 301) were detected before nilotinib/dasatinib therapy was begun (switchover) in 139 of 220 imatinib-resistant patients (supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Of these, 44% (132/301) were low-level mutations that were detected by mass spectrometry alone (detected in 64 patients). We previously demonstrated that at switchover, mass spectrometry could detect mutations that conferred clinical resistance to the inhibitor received in 9% more patients than sequencing and that these low-level resistant mutations rapidly expanded and were associated with subsequent treatment failure.8 Therefore, when we assessed the impact of multiple mutations on response to subsequent nilotinib/dasatinib therapy, patients with these mutations detected by either method (n = 45) were excluded, because poor response was known (3 of 45 achieved complete cytogenetic response [CCyR]).8
In the switchover samples of the remaining 175 patients, 159 mutations were detected in 86 patients by mass spectrometry, but just 108 mutations were detected in 89 patients by sequencing. Thirteen rare mutations detected by sequencing were not included in the mass spectrometry assay design (the response of patients with these mutations has been detailed previously8 ). Mass spectrometry detected all other mutations detected by sequencing, plus 64 low-level mutations not detected by sequencing. Multiple (≥ 2) sensitive mutations were detected in more of the 175 patients by mass spectrometry (34/175, 19%; 2-9 mutations per patient) than by sequencing (16/175, 9%; 2-3 mutations per patient; P = .009).
During nilotinib/dasatinib therapy, 78 (45%) of 175 patients achieved CCyR and 46 (26%) achieved major molecular response. Sequencing was used to monitor patients during nilotinib/dasatinib therapy for emerging new mutations. New mutations were detected in 65 (37%) of 175 patients (n = 87 mutations). Sixty-six of these new mutations (76%) were considered clinically resistant to the inhibitor received (T315I; n = 23).
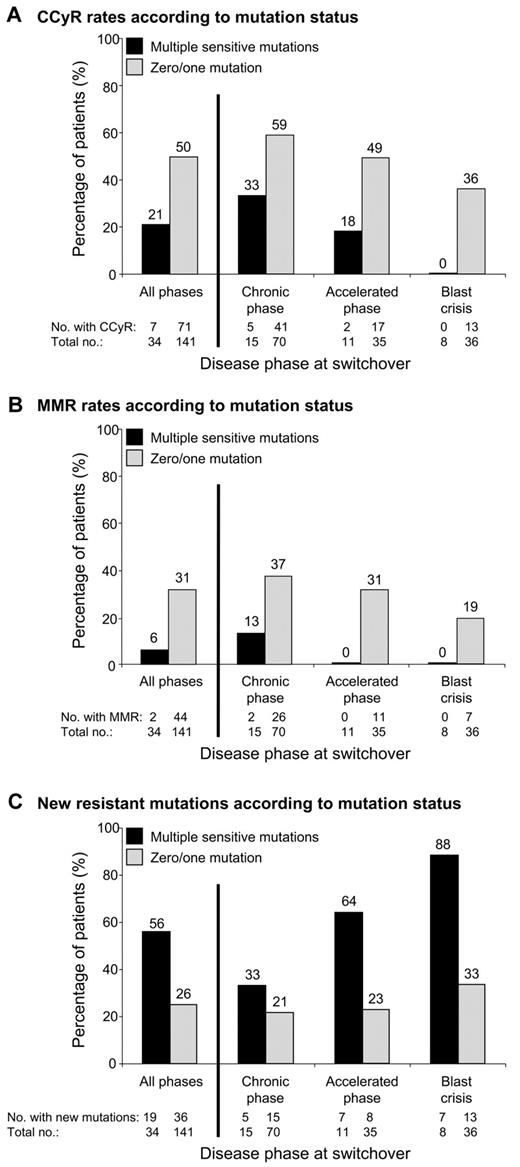
We investigated whether the number of sensitive mutations detected by mass spectrometry at switchover impacted response to subsequent nilotinib/dasatinib therapy. Consistent with previous reports,3,6 patients with at least 1 mutation (n = 86) were more likely to develop new resistant mutations than patients without mutations (n = 89; 42% vs 21%, P = .006). However, no differences in the CCyR (40% vs 49%, P = .24) or major molecular response (27% vs 26%, P = .97) rates were observed between the 2 groups. When we analyzed the subgroup of patients with multiple mutations (n = 34), we could segregate a group with poorer response to nilotinib/dasatinib therapy. The frequency of multiple mutations for patients in CP, accelerated phase, and blast crisis was 18% (15/85), 24% (11/46), and 18% (8/44), respectively. Patients with 0 or 1 mutation, and similarly patients with 2 or > 2 mutations, were grouped together, because no difference in response was observed within these subgroups (data not shown). Likewise, similar responses were observed for nilotinib-treated patients and dasatinib-treated patients. Multiple mutations at switchover were associated with lower rates of CCyR (21% vs 50%, P = .003; Figure 1A) and major molecular response (6% vs 31%, P = .005; Figure 1B) and higher rates of new resistant mutations (56% vs 26%, P = .0013; Figure 1C; Table 1). Supplemental Figure 2 shows the failure-free survival1 rates for the subgroup of CP patients. We were unable to assess whether other factors at switchover, such as prior response to imatinib, were associated with multiple sensitive mutations because this information was not available.
The association between multiple mutations and response to second-line kinase inhibitor therapy. Among the 220 imatinib-resistant CML patients analyzed for BCR-ABL1 kinase domain mutations by sequencing and mass spectrometry before commencing nilotinib/dasatinib therapy (switchover), 175 did not have detectable mutations known to confer clinical resistance to the inhibitor they received. Patients were categorized on the basis of the presence (n = 34) or absence (n = 141) of multiple sensitive mutations detected by mass spectrometry at switchover. (A) Cumulative incidence of CCyR and/or its equivalent of ≤ 1% BCR-ABL1 IS (International Scale),26 according to disease phase at switchover. (B) Cumulative incidence of major molecular response (MMR; ≤ 0.1% BCR-ABL1 IS), according to disease phase at switchover. (C) Rate of new resistant mutations detected by direct sequencing during nilotinib/dasatinib therapy, according to disease phase at switchover.
The association between multiple mutations and response to second-line kinase inhibitor therapy. Among the 220 imatinib-resistant CML patients analyzed for BCR-ABL1 kinase domain mutations by sequencing and mass spectrometry before commencing nilotinib/dasatinib therapy (switchover), 175 did not have detectable mutations known to confer clinical resistance to the inhibitor they received. Patients were categorized on the basis of the presence (n = 34) or absence (n = 141) of multiple sensitive mutations detected by mass spectrometry at switchover. (A) Cumulative incidence of CCyR and/or its equivalent of ≤ 1% BCR-ABL1 IS (International Scale),26 according to disease phase at switchover. (B) Cumulative incidence of major molecular response (MMR; ≤ 0.1% BCR-ABL1 IS), according to disease phase at switchover. (C) Rate of new resistant mutations detected by direct sequencing during nilotinib/dasatinib therapy, according to disease phase at switchover.
Details on response during nilotinib or dasatinib therapy of imatinib-resistant patients with multiple mutations detected by sensitive multiplex mass spectrometry at switchover (mutations were considered not clinically resistant to the inhibitor received by the patient)
| Disease phase . | No. of mutations by mass spectrometry . | Mutations by mass spectrometry . | Therapy received . | New mutations by sequencing during nilotinib/dasatinib* . | Months to CCyR . | Months to MMR . | Follow-up, mo . |
|---|---|---|---|---|---|---|---|
| CP | 2 | M244V, H396R | Dasatinib | M244V | No CCyR | No MMR | 7 |
| CP | 2 | M351T, H396R | Dasatinib | 24 | No MMR | 30 | |
| CP | 2 | M244V, G250E | Dasatinib | 3 | No MMR | 18 | |
| CP | 2 | G250E, E255K | Dasatinib | 3 | 11 | 24 | |
| CP | 2 | G250E, M351T | Dasatinib | T315I | No CCyR | No MMR | 18 |
| CP | 3 | Q252H, E255K, H396R | Dasatinib | T315I | No CCyR | No MMR | 5 |
| CP | 3 | G250E, E355G, E459K | Dasatinib | G250E, F317L | No CCyR | No MMR | 22 |
| CP | 3 | M244V, L248V, F359I | Dasatinib | V299L | No CCyR | No MMR | 20 |
| CP | 6 | M244V, L248V, G250E, Y253H, M351T, E355G | Dasatinib | 6 | 9 | 12 | |
| CP | 8 | L248V, G250E, E255V, M351T, F359V, H396R, E459K, F486S | Dasatinib | F317L × 3† | No CCyR | No MMR | 10 |
| CP | 2 | M244V, F359I | Nilotinib | F359I | No CCyR | No MMR | 12 |
| CP | 2 | V299L, F317L | Nilotinib | Q252H | No CCyR | No MMR | 4 |
| CP | 2 | M244V, G250E | Nilotinib | No CCyR | No MMR | 15 | |
| CP | 3 | M351T, F486S, F317L | Nilotinib | F317L | No CCyR | No MMR | 18 |
| CP | 5 | G250E, M351T, E355G, F359I, H396R | Nilotinib | G250E, F359I, H396R | No CCyR | No MMR | 3 |
| AP | 2 | M244V, H396R | Dasatinib | 21 | No MMR | 21 | |
| AP | 2 | G250E, H396R | Dasatinib | T315A | No CCyR | No MMR | 16 |
| AP | 2 | H396R, E459K | Dasatinib | No CCyR | No MMR | 12 | |
| AP | 2 | Y253H, E255K | Dasatinib | T315I | No CCyR | No MMR | 4 |
| AP | 3 | M244V, E355G, F359V | Dasatinib | M244V, T315I | 3 | No MMR | 35 |
| AP | 3 | G250E, M351T, F359V | Dasatinib | F317L | No CCyR | No MMR | 36 |
| AP | 3 | M244V, M351T, H396R | Dasatinib | No CCyR | No MMR | 33 | |
| AP | 4 | G250E, Y253F, M351T, H396R | Dasatinib | T240A‡, G250E | No CCyR | No MMR | 5 |
| AP | 4 | Y253F, F359I, F359V, H396R | Dasatinib | T315I | No CCyR | No MMR | 18 |
| AP | 8 | M244V, L248V, E255V, E279K, M351T, E355A, E459K, F486S | Dasatinib | L248V, T315I,E355A | No CCyR | No MMR | 24 |
| AP | 2 | M351T, H396R | Nilotinib | Y253H, E255V | No CCyR | No MMR | 18 |
| BC | 2 | F359V, H396R | Dasatinib | F317L | No CCyR | No MMR | 8 |
| BC | 2 | Y253H, E459K | Dasatinib | T315I | No CCyR | No MMR | 3 |
| BC | 4 | G250E, E279K, H396R, F486S | Dasatinib | F317L × 2§ | No CCyR | No MMR | 8 |
| BC | 9 | M244V, L248V, G250E, Y253H, M351T, F359V, H396R, E459K, F486S | Dasatinib | F317L | No CCyR | No MMR | 4 |
| BC | 2 | M244V, Q252H | Nilotinib | Y253H, E255K, F359V | No CCyR | No MMR | 2 |
| BC | 2 | Y253F, F359I | Nilotinib | No CCyR | No MMR | 3 | |
| BC | 2 | V299L, F317I | Nilotinib | E255V | No CCyR | No MMR | 2 |
| BC | 2 | M244V, D276G | Nilotinib | D276G, E255V | No CCyR | No MMR | 5 |
| Disease phase . | No. of mutations by mass spectrometry . | Mutations by mass spectrometry . | Therapy received . | New mutations by sequencing during nilotinib/dasatinib* . | Months to CCyR . | Months to MMR . | Follow-up, mo . |
|---|---|---|---|---|---|---|---|
| CP | 2 | M244V, H396R | Dasatinib | M244V | No CCyR | No MMR | 7 |
| CP | 2 | M351T, H396R | Dasatinib | 24 | No MMR | 30 | |
| CP | 2 | M244V, G250E | Dasatinib | 3 | No MMR | 18 | |
| CP | 2 | G250E, E255K | Dasatinib | 3 | 11 | 24 | |
| CP | 2 | G250E, M351T | Dasatinib | T315I | No CCyR | No MMR | 18 |
| CP | 3 | Q252H, E255K, H396R | Dasatinib | T315I | No CCyR | No MMR | 5 |
| CP | 3 | G250E, E355G, E459K | Dasatinib | G250E, F317L | No CCyR | No MMR | 22 |
| CP | 3 | M244V, L248V, F359I | Dasatinib | V299L | No CCyR | No MMR | 20 |
| CP | 6 | M244V, L248V, G250E, Y253H, M351T, E355G | Dasatinib | 6 | 9 | 12 | |
| CP | 8 | L248V, G250E, E255V, M351T, F359V, H396R, E459K, F486S | Dasatinib | F317L × 3† | No CCyR | No MMR | 10 |
| CP | 2 | M244V, F359I | Nilotinib | F359I | No CCyR | No MMR | 12 |
| CP | 2 | V299L, F317L | Nilotinib | Q252H | No CCyR | No MMR | 4 |
| CP | 2 | M244V, G250E | Nilotinib | No CCyR | No MMR | 15 | |
| CP | 3 | M351T, F486S, F317L | Nilotinib | F317L | No CCyR | No MMR | 18 |
| CP | 5 | G250E, M351T, E355G, F359I, H396R | Nilotinib | G250E, F359I, H396R | No CCyR | No MMR | 3 |
| AP | 2 | M244V, H396R | Dasatinib | 21 | No MMR | 21 | |
| AP | 2 | G250E, H396R | Dasatinib | T315A | No CCyR | No MMR | 16 |
| AP | 2 | H396R, E459K | Dasatinib | No CCyR | No MMR | 12 | |
| AP | 2 | Y253H, E255K | Dasatinib | T315I | No CCyR | No MMR | 4 |
| AP | 3 | M244V, E355G, F359V | Dasatinib | M244V, T315I | 3 | No MMR | 35 |
| AP | 3 | G250E, M351T, F359V | Dasatinib | F317L | No CCyR | No MMR | 36 |
| AP | 3 | M244V, M351T, H396R | Dasatinib | No CCyR | No MMR | 33 | |
| AP | 4 | G250E, Y253F, M351T, H396R | Dasatinib | T240A‡, G250E | No CCyR | No MMR | 5 |
| AP | 4 | Y253F, F359I, F359V, H396R | Dasatinib | T315I | No CCyR | No MMR | 18 |
| AP | 8 | M244V, L248V, E255V, E279K, M351T, E355A, E459K, F486S | Dasatinib | L248V, T315I,E355A | No CCyR | No MMR | 24 |
| AP | 2 | M351T, H396R | Nilotinib | Y253H, E255V | No CCyR | No MMR | 18 |
| BC | 2 | F359V, H396R | Dasatinib | F317L | No CCyR | No MMR | 8 |
| BC | 2 | Y253H, E459K | Dasatinib | T315I | No CCyR | No MMR | 3 |
| BC | 4 | G250E, E279K, H396R, F486S | Dasatinib | F317L × 2§ | No CCyR | No MMR | 8 |
| BC | 9 | M244V, L248V, G250E, Y253H, M351T, F359V, H396R, E459K, F486S | Dasatinib | F317L | No CCyR | No MMR | 4 |
| BC | 2 | M244V, Q252H | Nilotinib | Y253H, E255K, F359V | No CCyR | No MMR | 2 |
| BC | 2 | Y253F, F359I | Nilotinib | No CCyR | No MMR | 3 | |
| BC | 2 | V299L, F317I | Nilotinib | E255V | No CCyR | No MMR | 2 |
| BC | 2 | M244V, D276G | Nilotinib | D276G, E255V | No CCyR | No MMR | 5 |
CCyR indicates complete cytogenetic response; MMR, major molecular response; CP, chronic phase; AP, accelerated phase; and BC, blast crisis.
Mutations became detectable by sequencing during nilotinib or dasatinib therapy. Mutations are coded according to the inhibitor received. Underlined mutations do not confer clinical resistance to the inhibitor received and were detected by mass spectrometry at switchover. Mutations in bold confer clinical resistance to the inhibitor received and were not detected at switchover by mass spectrometry.
Three different F317L mutations were acquired in this patient (c.949T > C, c.951C > G, and c.951C > A).
Mutation not included in the design of the mass spectrometry assay.
Two different F317L mutations were detected in this patient (c.949T > C and c.951C > A).
Interestingly, of the 64 low-level mutations detected at switchover, 12 (19%) expanded during nilotinib/dasatinib therapy and were detected by sequencing at a median of 3 months after switchover (Table 1). These mutant clones may have expanded because of differential inhibitor sensitivity. Indeed, most had lower in vitro sensitivity to the inhibitor received than did unmutated BCR-ABL1.13,14
The present study suggests that the detection of multiple mutations at switchover, in the absence of resistant mutations, could classify a group of imatinib-resistant CML patients with poorer response to nilotinib/dasatinib therapy. This subgroup represented 15.5% of all patients in the present study cohort (34/220). Multiple mutations after imatinib resistance may be a marker of an increased propensity for subsequent selection of resistant mutations. Clonal diversity, the number of clones in a tumor, has been associated with cancer progression.15 Genetic instability, a hallmark of cancer and a feature of CML progression,16-19 drives clonal diversity if viable mutants can expand into detectable clones.15 Clonal diversity allows for interclonal cooperativity, in which clones with different mutations complement each other to drive progression, which has been observed in solid tumors.20 Paracrine protection of imatinib-sensitive leukemic cells by low levels of imatinib-resistant cells with BCR-ABL1 mutations has also been reported.21
A recent study found no correlation between the number of low-level mutations (detection limit 1%) in tyrosine kinase inhibitor–naïve, Philadelphia chromosome–positive acute lymphoblastic leukemia patients and the likelihood of relapse when taking dasatinib.22 However, only 15 patients were studied, and all had ≥ 2 mutations, which may signify high clonal diversity in these patients. Another study reported that multiple mutations by sequencing at switchover in imatinib-resistant CML patients were associated with low response and progression-free survival rates on second-line therapy.23 However, only 7 patients were identified with multiple mutations (3% of patients screened), and 5 of 7 with multiple mutations already had mutations that the authors classified as highly resistant to the second-line therapy received, which may be associated with the poor response.
Direct sequencing is the method currently recommended to assess BCR-ABL1 mutation status.2 However, we8 and others24 have demonstrated that low-level mutations are clinically significant for imatinib-resistant patients. Therefore, we believe that low-level mutation detection should be incorporated into clinical practice to aid therapy selection after imatinib failure. Among the 220 patients analyzed in the present study, the number of mutations detected per patient by mass spectrometry (maximum of 10; 18 [13%] of 139 patients with mutations had ≥ 4) far exceeded the number concurrently detected by sequencing (maximum of 4; 2 [1%] of 139 patients had 4) and the numbers previously reported by use of sequencing alone.25 This suggests that mass spectrometry detected a pool of subclonal mutants, each with a small survival advantage after imatinib therapy that was insufficient for their clonal predominance. The study demonstrates the advantage of sensitive multiplex mutation analysis, as opposed to current sensitive singleplex techniques such as allele-specific oligonucleotide PCR. Ultra-deep next-generation sequencing, however, may allow simultaneous detection of all mutations.
In conclusion, sensitive BCR-ABL1 mutation analysis identified a poor-risk subgroup of imatinib-resistant CML patients with multiple mutations who were not identified by sequencing. This subgroup represented 15.5% of all patients, who would not otherwise be classified as being at risk of poor response on the basis of their mutation status determined by conventional sequencing. This poor-risk subgroup may warrant closer monitoring, experimental approaches, or stem cell transplantation to reduce the high risk of second-line kinase inhibitor failure.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to thank the patients, clinicians, and study center coordinators who contributed samples and follow-up data for this study and the staff of the Leukemia Unit, Genetics and Molecular Pathology, SA Pathology, for their excellent technical support.
This work was supported by a grant from the National Health and Medical Research Council (565170).
Authorship
Contribution: W.T.P. contributed to experimental design, performed research, analyzed data, and wrote the manuscript; M.H. performed research and contributed to manuscript preparation; H.S.S. contributed to experimental design and manuscript preparation; T.P.H. contributed to experimental design and contributed significantly to manuscript preparation; and S.B. contributed to experimental design, analyzed data, and contributed significantly to manuscript preparation.
Conflict-of-interest disclosure: S.B. and T.H. have received research funding and honoraria from Novartis Pharmaceuticals, Bristol-Myers Squibb, and Ariad Pharmaceuticals. The remaining authors declare no competing financial interests.
Correspondence: Wendy T. Parker, Department of Genetics and Molecular Pathology, SA Pathology (RAH Campus), PO Box 14, Rundle Mall, Adelaide, SA, 5000, Australia; e-mail: wendy.parker@health.sa.gov.au.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal