Abstract
Pim kinases are Ser/Thr kinases with multiple substrates that affect survival pathways. These proteins are overexpressed in acute myeloid leukemia (AML) blasts and we hypothesized that Pim kinase inhibition would affect AML cell survival. Imidazo[1,2-b]pyridazine compound, SGI-1776 inhibits Pim-1, Pim-2 and Pim-3, and was evaluated in AML-cell line, -xenograft model, and -primary blasts. Treatment of AML cells with SGI-1776 results in a concentration-dependent induction of apoptosis and we investigated its effect on Pim kinase functions. Phosphorylation of traditional Pim kinase targets, c-Myc(Ser62) and 4E-BP1 (Thr36/Thr47), were both decreased in actively cycling AML cell lines MV-4-11, MOLM-13 and OCI-AML-3. Levels of antiapoptotic proteins Bcl-2, Bcl-xL, XIAP, and proapoptotic Bak and Bax were unchanged; however, a significant reduction in Mcl-1 was observed. This was correlated with inhibition of global RNA and protein synthesis and MCL-1 transcript decline after SGI-1776 treatment. These data suggest that SGI-1776 mechanism in AML involves Mcl-1 protein reduction. Consistent with cell line data, xenograft model studies with mice bearing MV-4-11 tumors showed efficacy with SGI-1776. Importantly, SGI-1776 was also cytotoxic in AML primary cells, irrespective of FLT3 mutation status and resulted in Mcl-1 protein decline. Pim kinase inhibition may be a new strategy for AML treatment.
Introduction
Pim (provirus integration site for Moloney murine leukemia virus) family proteins are highly conserved serine/threonine kinases that have been implicated in cancer progression and the development of resistance to chemotherapeutic agents (for a review, see Shah et al1 ). Three Pim kinases (Pim-1, -2 and -3) have been identified, each with variant isoforms of the expressed protein because of alternate start sites. In humans, PIM1, PIM2, and PIM3 genes are located on chromosome 6p21, Xp11.23, and 22q13, respectively.2,3 At the amino acid level, there is substantial homology between Pim-1 and Pim-2 (53%)4 and Pim-3 (69%).5 Pim kinases have overlapping functions and compensate for one another, and their numerous targets include regulators of transcription, translation, cell cycle, survival, and drug resistance (Figure 1A).
Pim kinases in cancer. (A) Pim kinase pathways (adapted from Chen et al26). (B) Chemical structure of imidazo[1,2-b]pyridazine compound SGI-1776.
Pim kinases in cancer. (A) Pim kinase pathways (adapted from Chen et al26). (B) Chemical structure of imidazo[1,2-b]pyridazine compound SGI-1776.
Pertaining to cancer biology, increased levels of Pim kinase proteins have been strongly implicated in cell survival and tumorigenesis. Pim kinases are overexpressed in both solid tumors such as colon,6 prostate cancer7 and hematologic malignancies including lymphomas,8,9 chronic lymphocytic leukemia (CLL)8,10 and acute leukemias.11 Specifically in AML, up-regulation of Pim may be because of overexpression of HOXA912-14 and STAT activation,15 which can act as transcription factors16,17 for Pim. These oncogenic kinases appear to play critical roles in leukemogenesis, and resistance to chemotherapy18 and radiotherapy.19 Consistent with these reports, knockdown of Pim kinases was shown to impair the survival of resistant forms of FLT3- and BCR-ABL–transformed leukemia cells.20
FLT3-ITD is one of the most prevalent activating mutations identified in AML (15%-30%; reviewed in Meshinchi et al21 ), and is associated with inferior disease-free survival and increased relapse rate.22 Allele analysis revealed that homozygous FLT3-ITD is a strong adverse prognostic factor in de novo AML in patients with normal cytogenetics.23 One of the challenges of FLT3 inhibition therapy is the development of resistance and Pim-1 has been shown to contribute to this increased resistance.24 Pim itself can phosphorylate FLT3 in a feedback loop (Figure 1A),25 and consequently Pim kinase inhibition may be an alternative strategy to target AML with FLT3 activating mutations.
Given the oncogenic nature of Pim kinases, there has been increasing interest in developing Pim kinase inhibitors for the treatment of cancer.26 SGI-1776 is an imidazo[1,2-b]pyridazine (Figure 1B) small molecule that is inhibitory to all 3 Pim kinases: the IC50 values are 7nM, 363nM, and 69nM for Pim-1, -2 and -3, respectively.10 In addition to Pim, SGI-1776 also potently targets FLT3 (IC50 = 44nM). Using established AML cell lines with wild-type and mutated FLT3, AML mouse model system, and primary AML leukemic blasts with variety of FLT3 mutation status, we elucidate the mechanism of action of SGI-1776 and its cytotoxicity in AML.
Methods
Drugs
SGI-1776 was obtained from SuperGen and was dissolved in DMSO and stored at −20°C. All experiments including a vehicle control were conducted using 0.1% DMSO.
Cell lines
MV-4-11, MOLM-13, and OCI-AML-3 cell lines were obtained from ATCC. The cells were cultured in IMDM (ATCC) supplemented with 10% FBS and grown in a 37°C incubator with 5% CO2. Cells were routinely tested for Mycoplasma infection using a commercially available kit (Cambrex).
Patient samples
The present in vitro studies were carried out in freshly isolated primary blasts obtained from peripheral blood of patients with AML (n = 6). For all investigations, freshly isolated AML blasts were used. All patients signed a written informed consent in accordance with the Declaration of Helsinki to participate in the laboratory protocol, which was approved by the Institutional Review Board of the University of Texas M. D. Anderson Cancer Center.
Clinical laboratory end points
Determination of FLT3 mutation status analysis for the patients in our study were conducted by fluorescent multiplex PCR and restriction digestion method followed by capillary electrophoresis performed by the molecular diagnostic laboratory at M. D. Anderson Cancer Center. FISH analysis data were provided by the clinical cytogenetics laboratory, Department of Hematopathology at M. D. Anderson Cancer Center.
Isolation of primary leukemia cells
Whole blood was collected in heparinized tubes, then the leukemia cells were isolated using Ficoll-Hypaque (specific gravity, 1.086; Life Technologies) density gradient separation as previously described.10 The blasts were washed twice with cold PBS, resuspended in 10 mL of RPMI 1640, supplemented with 10% autologous plasma, and maintained at 1 × 107 cells/mL. The cell number and mean cell volume were determined using a Coulter channelyzer (Coulter Electronics).
Cell death/apoptosis assay
AML cell line and primary cells were treated with DMSO or various concentrations of SGI-1776 for 24 hours. Cells (1 × 106) were washed, then resuspended in 100 μL of annexin binding buffer (Roche), mixed with 5 μL of -FITC solution (BD Pharmingen) and 5 μL of propidium iodide (PI; 50 μg/mL, Sigma-Aldrich) solution. For each sample, 10 000 cells were measured using a Becton Dickinson FACSCalibur flow cytometer.
Protein expression analysis
AML cell line and primary cells were treated with DMSO or SGI-1776, then cell lysates were prepared and protein content was measured as described.10 Aliquots (30 μg) of total protein were loaded onto SDS-polyacrylamide gels (12% or 4%-12%), transferred to nitrocellulose membranes (GE Osmonics Labstore), then blocked and probed with antibody in Odyssey blocking buffer (LI-COR Inc), as previously described.10 Antibodies: Bad (Cell Signaling Technology), phospho-Bad (Ser112; Cell Signaling Technology), Bak (Millipore), Bax (BD Pharmingen), Bcl-2 (Dako), Bcl-xL (BD Transduction Laboratories), c-Myc (clone C33; Santa Cruz Biotechnology), c-Myc (Ser62; Abcam), GAPDH (Cell Signaling Technology), total histone H3 (Cell Signaling Technology), histone H3 (Ser10; Millipore), Mcl-1 (Santa Cruz Biotechnology), PARP (Enzo Life Sciences International), 4E-BP1 (Santa Cruz Biotechnology), phospho-4E-BP1 (Thr37/46; Cell Signaling Technology) were used. Membranes were probed with infrared-labeled secondary antibodies and visualized using a LI-COR Odyssey Infrared Imager as described.10
RNA/protein synthesis assay
AML cells were incubated with SGI-1776 for 24 hours. One hour before removal of each aliquot, either 1 μCi [3H]uridine (Moravek Biochemicals) for RNA synthesis measurement, or 5 μCi [3H]leucine (Moravek) for protein synthesis assay, was added to the cell cultures, then analyzed using a multiscreen assay system (Millipore Corp) as previously described.10 Radioactivity was measured in treated and untreated (control) cells and expressed as percent of control.
Gene expression analysis
RNA was isolated with the RNeasy Mini kit (QIAGEN) and the gene expression levels were measured on an ABI prism 7900 Sequence Detection System (Applied Biosystems) using 1-step real-time TaqMan RT-PCR. Taqman primers and probes for MCL-1 and 18S were purchased from Applied Biosystems. Each RNA sample was assayed in triplicate and the relative gene expression levels were normalized with 18S.
Clonogenic assay
MV-4-11 cells were untreated or treated with SGI-1776 for 24 hours, then washed with PBS to remove drug. Cells then were added to MethoCult H4230 (StemCell Technologies) at 500-1000 cells/mL and dispensed to 6-well plates at 1 mL per well in triplicates. Colonies > 50 cells were counted after 6-10 days of culture at 37°C.
In vivo efficacy studies
All animal experiments were performed in accordance with the regulations of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Female NOD-SCID mice were allowed to acclimate for a period of approximately one week between animal receipt and cell inoculation. The conditions for animal room environment and photoperiod were 20-25°C, 40%-70% humidity, and 12 hours of light/12 hours of dark cycle. Each mouse was inoculated subcutaneously at the right flank with MV-4-11 tumor cells (5 × 106). The treatments started when the tumor size reached 80-150 mm3. Mice were randomized to treatment groups based on their tumor sizes; tumor size was measured in 2 dimensions using a caliper, and the volume was expressed in mm3 using the formula: V = 0.5 a × b2 where a and b are the long and short diameters of the tumor, respectively. Pretreatment randomization ensures that each group has approximately the same mean tumor size. Mice were treated with vehicle (5% dextrose), SGI-1776 or cytarabine (ara-C; Sigma-Aldrich). SGI-1776 and ara-C were formulated in 5% dextrose. SGI-1776 was administered by oral gavage (PO) on a daily × 5/week or twice/week schedule; ara-C was administered by intraperitoneal injection 3 times/week for 3 consecutive weeks. Animals were euthanized when their measured tumor size was greater than 3000 mm3 or when they lost ≥ 20% initial body weight; if the body weight loss ≥ 15%, treatment was stopped at first until mice regained body weight. Mice were euthanized when body weight loss was still ≥ 20% even after stopping treatment. T/C value (in %) is an indication of antitumor efficacy, where T and C are the mean tumor volume of the treated and control groups, respectively, on a given day. The differences between the mean tumor sizes for comparing groups was analyzed using the ANOVA test, where P ≤ .05 was considered to be statistically significant.
Results
SGI-1776 in AML cell lines
Pim kinase inhibition in MV-4-11, MOLM-13 and OCI-AML-3.
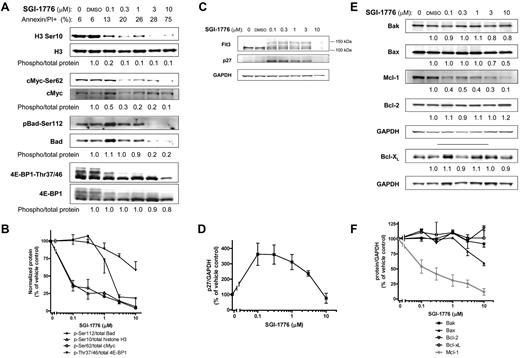
AML cell lines MV-4-11 (homozygous FLT3-ITD), MOLM-13 (heterozygous FLT3-ITD) and OCI-AML-3 (wild-type FLT3) with various FLT3 mutations were used. First, we determined the effect of SGI-1776 on Pim kinase function by evaluating changes in traditional Pim substrate proteins histone H3, c-Myc and Bad in MV-4-11 cell line (Figure 2A). Even at 0.1μM SGI-1776, phosphorylation of histone H3 at Ser10 was reduced to only ∼ 20% of vehicle control. c-Myc(Ser62) phosphorylation was decreased by ∼ 50% relative to control at 0.1μM SGI-1776, which was further reduced by 80% with 1μM (Figure 2A-B). Similarly, in MOLM-13 and OCI-AML-3 cell lines, and there was a significant reduction in c-Myc phosphorylation after treatment with SGI-1776 (supplemental Figure 3, available on Blood Web site; see the Supplemental Materials link at the top of the online article). The levels of both total and phospho-Bad (Ser112) protein levels decreased after SGI-1776 treatment (Figure 2A-B). Phosphorylation of translation regulator 4E-BP1 (Thr37/Thr46) was also decreased, and there is a clear reduction of hyperphosphorylated 4E-BP1 even at 0.1μM SGI-1776 treatment in MV-4-11 (Figure 2A), MOLM-13 and OCI-AML-3 (supplemental Figure 3).
Immunoblot analysis of Pim target proteins in AML cell line treated with SGI-1776. MV-4-11 cells were either untreated, treated with 0.1% vehicle DMSO alone, 0.1, 0.3, 1, 3, or 10μM SGI-1776 and cells were harvested after 24 hours and lysed. (A) The phospho-protein levels of histone H3 (Ser10), c-Myc (Ser62), Bad(Ser112), 4E-BP1 (Thr37/46), were analyzed using immunoblot and normalized to each respective total protein levels. (B) Phospho-protein levels relative to total protein levels. Data are the average of 3 independent experiments ± SEM. (C) Protein levels of FLT3 and p27 were analyzed using immunoblot and normalized to GAPDH as a loading control. (D) p27 protein levels normalized to GAPDH protein. Data are the average of 3 independent experiments ± SEM. (E) Protein levels of proapoptotic Bak, Bax, and antiapoptotic Mcl-1, Bcl-2, and Bcl-xL were analyzed using immunoblot and normalized to GAPDH as a loading control. (F) Pro- and antiapoptotic protein levels normalized to GAPDH protein with Mcl-1 shown in gray. Data are the average of 3 independent experiments ± SEM.
Immunoblot analysis of Pim target proteins in AML cell line treated with SGI-1776. MV-4-11 cells were either untreated, treated with 0.1% vehicle DMSO alone, 0.1, 0.3, 1, 3, or 10μM SGI-1776 and cells were harvested after 24 hours and lysed. (A) The phospho-protein levels of histone H3 (Ser10), c-Myc (Ser62), Bad(Ser112), 4E-BP1 (Thr37/46), were analyzed using immunoblot and normalized to each respective total protein levels. (B) Phospho-protein levels relative to total protein levels. Data are the average of 3 independent experiments ± SEM. (C) Protein levels of FLT3 and p27 were analyzed using immunoblot and normalized to GAPDH as a loading control. (D) p27 protein levels normalized to GAPDH protein. Data are the average of 3 independent experiments ± SEM. (E) Protein levels of proapoptotic Bak, Bax, and antiapoptotic Mcl-1, Bcl-2, and Bcl-xL were analyzed using immunoblot and normalized to GAPDH as a loading control. (F) Pro- and antiapoptotic protein levels normalized to GAPDH protein with Mcl-1 shown in gray. Data are the average of 3 independent experiments ± SEM.
In addition to these traditional substrates, SGI-1776 induced changes in Pim targets p27 and FLT3, which are proteins important for AML cell growth and survival. Pim-1 has been reported to phosphorylate p27, which results in p27 binding to 14-3-3 protein and subsequent nuclear export and proteasome-dependent degradation.27,28 Consistent with these findings, treatment of MV-4-11 cells with 0.1-0.3μM SGI-1776 resulted in a 3- to 4-fold increase in p27 levels (Figure 2C-D).
Pim-1 has been reported to be involved with FLT3 posttranslational modification and signaling. Two forms of human FLT3 have been reported; an unglycosylated form (∼ 130 kDa) and a membrane-bound N-linked glycosylated form (∼ 150 kDa),29,30 and Pim-1 can stabilize the 130 kDa isoform.25 Treatment of MV-4-11 cells with SGI-1776 resulted in changes in FLT3 protein consistent with Pim kinase inhibition. There were lower levels of the 130 kDa and higher levels of the 150 kDa FLT3 isoform in cells treated with 0.1-3μM SGI-1776 (Figure 2C). Taken together, our results indicate that SGI-1776 effectively inhibits Pim kinase function in AML cell lines.
Changes in Mcl-1 and other protein levels.
To determine whether the reduction in Pim kinase target phosphorylation correlated with changes in pro- and antiapoptotic protein levels, we evaluated the levels of various cell survival proteins. Consistent with the reduction in phosphorylation of Pim kinase targets, there was a significant dose-dependent reduction of Mcl-1 protein in AML cell lines treated with SGI-1776. At low concentrations (0.1-1μM), there was an approximately ∼ 50% decrease in Mcl-1 protein levels relative to vehicle DMSO alone after 24 hours in MV-4-11 (Figure 2C) and MOLM-13 (supplemental Figure 3). At 3 and 10μM, there was a further reduction of Mcl-1 protein levels to only 30% and ∼ 10% of control, respectively. A decline in Mcl-1 protein levels was observed only at higher concentrations in OCI-AML-3, and there was only ∼ 30% of control remaining at 10μM (supplemental Figure 3). A decrease was neither observed in related antiapoptotic proteins Bcl-2 or Bcl-xL (Figure 2C,E), nor were there significant changes in the protein levels of proapoptotic Bak and Bax in MV-4-11 (Figure 2D-E). Stabilization of p53 was not observed in SGI-1776–treated MV-4-11 cells (supplemental Figure 2).
Effect on RNA and protein synthesis.
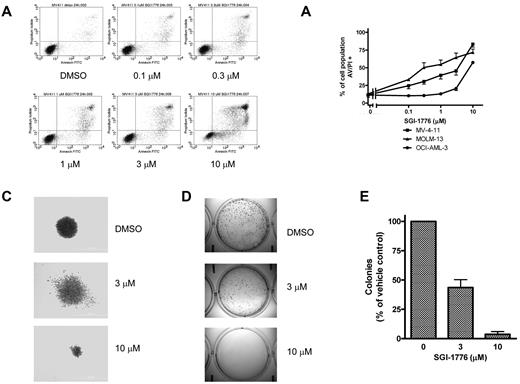
To investigate the mechanism of Mcl-1 protein level decline, we examined the effects of SGI-1776 on gene expression. Because there is cooperation between Pim kinases and c-Myc–driven transcription, we evaluated the effects of Pim inhibition on new transcript synthesis. AML cell lines were treated with increasing concentrations of SGI-1776 for 24 hours, then pulsed with [3H]-labeled uridine to measure RNA synthesis. There was a dose-dependent decrease in RNA synthesis in all 3 cell lines, with MOLM-13 and MV-4-11 as the most sensitive to SGI-1776. In both cell lines, 0.1μM SGI-1776 treatment resulted in the inhibition of global RNA synthesis to < 50% of control, which was further reduced to ∼ 25% of control at 1μM SGI-1776 (Figure 3A). In OCI-AML-3, RNA synthesis was inhibited to ∼ 80% and 50% of control at 1 and 10μM SGI-1776, respectively.
Effect of SGI-1776 in AML cell lines. (A) Inhibition of RNA synthesis in AML cell lines treated with SGI-1776. MV-4-11 (squares), MOLM-13 (triangles), and OCI-AML-3 (circles) lines were incubated with 0.1% DMSO alone, or 0.1, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours, then 1 hour before harvesting the cells, [3H]uridine was added to the cell culture as described in “RNA protein synthesis assay.” The results represent an average of triplicate experiments ± SEM. (B) Dose-dependent reduction of MCL-1 transcript levels in AML cell line treated with SGI-1776. MV-4-11 cells were treated with 0.1% vehicle DMSO alone, 0.1, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours, and the RNA was isolated. MCL-1 transcript levels were measured using real-time RT-PCR and normalized using 18S transcripts. Each RNA sample was assayed in triplicate and the results are expressed as a percentage of the MCL-1 expression level in cells treated with vehicle DMSO alone. The results represent an average of triplicate experiments ± SEM (C) Inhibition of protein synthesis in AML cell line treated with SGI-1776. MOLM-13 cells were incubated with 0.1% DMSO alone, or 0.1, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours, then 1 hour before harvesting the cells, [3H]leucine was added to the cell culture as described in “RNA protein synthesis assay.” The results represent an average of triplicate experiments ± SEM.
Effect of SGI-1776 in AML cell lines. (A) Inhibition of RNA synthesis in AML cell lines treated with SGI-1776. MV-4-11 (squares), MOLM-13 (triangles), and OCI-AML-3 (circles) lines were incubated with 0.1% DMSO alone, or 0.1, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours, then 1 hour before harvesting the cells, [3H]uridine was added to the cell culture as described in “RNA protein synthesis assay.” The results represent an average of triplicate experiments ± SEM. (B) Dose-dependent reduction of MCL-1 transcript levels in AML cell line treated with SGI-1776. MV-4-11 cells were treated with 0.1% vehicle DMSO alone, 0.1, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours, and the RNA was isolated. MCL-1 transcript levels were measured using real-time RT-PCR and normalized using 18S transcripts. Each RNA sample was assayed in triplicate and the results are expressed as a percentage of the MCL-1 expression level in cells treated with vehicle DMSO alone. The results represent an average of triplicate experiments ± SEM (C) Inhibition of protein synthesis in AML cell line treated with SGI-1776. MOLM-13 cells were incubated with 0.1% DMSO alone, or 0.1, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours, then 1 hour before harvesting the cells, [3H]leucine was added to the cell culture as described in “RNA protein synthesis assay.” The results represent an average of triplicate experiments ± SEM.
To evaluate whether RNA synthesis inhibition correlated with decreased MCL-1 transcripts levels, MV-4-11 cells were treated with SGI-1776 for 24 hours, then the cellular RNA was isolated. MCL-1 transcripts were quantified and normalized with 18S as an internal standard. At 1 and 10μM SGI-1776 there was a reduction to ∼ 40% and ∼ 15% of control (Figure 3B). In addition to down-regulation of MCL-1 transcription, decreased Mcl-1 protein levels may arise via impaired protein translation because treatment of AML cells with SGI-1776 resulted in decreased 4E-BP1 (Thr36/Thr47) phosphorylation (Figure 2A, supplemental Figure 3). To evaluate the effect of SGI-1776 on new protein synthesis, MOLM-13 was treated with SGI-1776 for 24 hours, then pulsed with [3H]-labeled leucine. At 0.3μM SGI-1776, protein synthesis decreased to ∼ 50% of control, which further decreased to ∼ 25% of control with 3μM SGI-1776 (Figure 3C). Because both MCL-1 transcript and protein have short half-lives, these results correlate with the decline in Mcl-1 protein levels.
Apoptosis induction and loss of clonogenic survival.
Based on the decline in antiapoptotic Mcl-1 protein level, we further investigated the consequences of Pim kinase inhibition on cell viability. MV-4-11, MOLM-13, and OCI-AML-3 cells were treated with SGI-1776 then stained with either annexin/PI or with DiOC6/PI to assess cell survival. There was a dose dependent increase in annexin/PI positive cells in all 3 lines (Figure 4A-B), however in DiOC6/PI-stained MV-4-11 cells there was increase in PI-positive cells without loss of DiOC6 (supplemental Figure 1). These results indicate that apoptosis induction by SGI-1776 occurs without loss of mitochondrial membrane potential. Induction of apoptosis was also confirmed via immunoblot analysis of PARP cleavage (supplemental Figure 2). There were higher levels of apoptosis induction in MV-4-11 and MOLM-13, which are FLT3-ITD homozygous and heterozygous, respectively. Increased cytotoxicity from SGI-1776 is expected in FLT3-ITD because of increased expression of Pim kinases in FLT3-ITD24 and also because Pim-1 phosphorylates FLT3 (Tyr591), which is the docking site for STAT5 with FLT3-ITD but not FLT3 wild-type.25 Finally, SGI-1776 also directly inhibits FLT3 kinase, which is constitutively active in FLT3-ITD.
Induction of cell death and reduction of clonogenic survival by SGI-1776 in AML cell lines. (A) Flow cytometry analysis of annexin-FITC/propidium iodide staining of MV-4-11 cells that were either untreated, treated with 0.1% DMSO vehicle alone, 0.1, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours. (B) Graphical representation of annexin/PI positivity in MV-4-11 (squares), MOLM-13 (triangles), and OCI-AML-3 (circles) cells treated with 0.1% DMSO vehicle alone, 0.1, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours. The results represent an average of triplicate experiments ± SEM (C-E) Suppression of clonogenic survival of MV-4-11 cells by SGI-1776. Clonogenic assay were performed as described in “Clonogenic assay.” (C) Representative wells of MV-4-11 colonies cultured in drug-free methycellulose after SGI-1776 washout. (D) Images of formed colonies at 10× magnification. Note that the colonies are more densely populated in vehicle DMSO alone than with SGI-1776 treatment. (E) The average percentage of colonies formed relative to DMSO of 3 independent experiments ± SEM.
Induction of cell death and reduction of clonogenic survival by SGI-1776 in AML cell lines. (A) Flow cytometry analysis of annexin-FITC/propidium iodide staining of MV-4-11 cells that were either untreated, treated with 0.1% DMSO vehicle alone, 0.1, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours. (B) Graphical representation of annexin/PI positivity in MV-4-11 (squares), MOLM-13 (triangles), and OCI-AML-3 (circles) cells treated with 0.1% DMSO vehicle alone, 0.1, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours. The results represent an average of triplicate experiments ± SEM (C-E) Suppression of clonogenic survival of MV-4-11 cells by SGI-1776. Clonogenic assay were performed as described in “Clonogenic assay.” (C) Representative wells of MV-4-11 colonies cultured in drug-free methycellulose after SGI-1776 washout. (D) Images of formed colonies at 10× magnification. Note that the colonies are more densely populated in vehicle DMSO alone than with SGI-1776 treatment. (E) The average percentage of colonies formed relative to DMSO of 3 independent experiments ± SEM.
Clonogenic survival of MV-4-11 was suppressed in cells treated with 3 or 10μM SGI-1776. Pretreatment with SGI-1776 resulted in a reduction in the total number of colonies as well as alteration in the morphology of the formed colonies (Figure 4C-E). Qualitatively, there is clear reduction in colony size after pretreatment with SGI-1776; the colonies shown in Figure 4C are at equal magnification. Quantitatively, compared with vehicle DMSO alone, at 3 and 10μM SGI-1776, there were < 50% and 5% of colonies, respectively (Figure 4E).
AML xenograft model
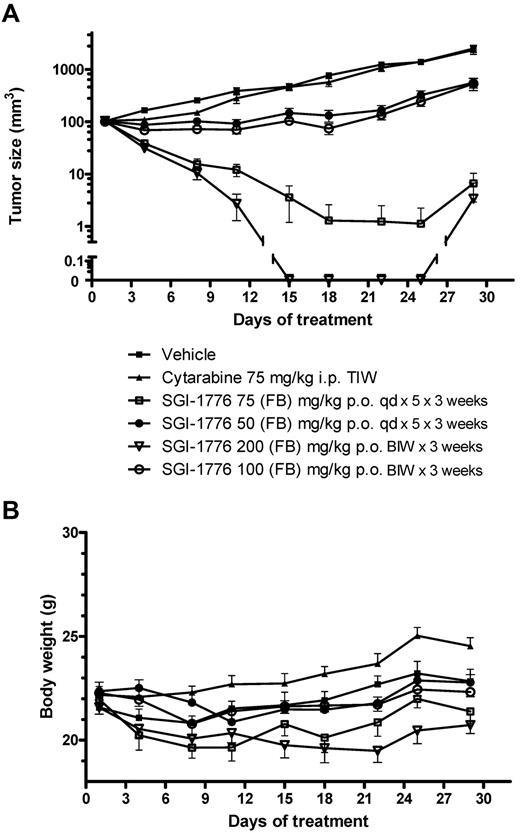
In vivo antitumor activity by SGI-1776.
Potent and sustained antitumor activity was seen in MV-4-11 xenografts with oral administration of SGI-1776 (Figure 5A), with tumors in the 75 mg/kg and 200 mg/kg treatment groups disappearing or became almost impalpable within 1 week after treatment. On day 22 of the experiment, 8 of 9 mice treated with 75 mg/kg SGI-1776 and all 10 mice treated with 200 mg/kg experienced a complete regression of their tumors; only 3 mice in 75 mg/kg group and 1 mouse in the 200 mg/kg group experienced a minor regrowth of their tumors at the completion of observation period. Treatment with 50 mg/kg SGI-1776 on a daily schedule or 100 mg/kg on a biweekly schedule resulted in some antitumor effect; no regressions were observed on day 18 but a significant 13% T/C after 50 mg/kg and 11% after 100 mg/kg was observed on day 22. Of note was the observation that potent efficacy was achieved with an intermittent treatment schedule (2 oral doses/wk) at 100 or 200 mg/kg. The MV-4-11 tumors were completely resistant to standard chemotherapy agent cytarabine (ara-C) administered intraperitoneally at a schedule of 75 mg/kg 3 times weekly. Mouse body weights were generally stable during the dosing period (Figure 5B). However, body weight losses (≤ 10%) were evident in mice treated with SGI-1776 75 mg/kg, qd × 5 × 3 and with 200 mg/kg biweekly; mice could regain their body weight after dosing holidays.
Effect of SGI-1776 in vivo in MV-4-11 AML xenografts. SGI-1776 was tested in MV-4-11 tumors on a daily for 5 days (qd) and twice weekly (BIW) treatment schedules. Each treatment group included 10 mice. (A) Average tumor size for the different treatment groups ± SEM. (B) Average % body weight change compared with average body weight on day 1.
Effect of SGI-1776 in vivo in MV-4-11 AML xenografts. SGI-1776 was tested in MV-4-11 tumors on a daily for 5 days (qd) and twice weekly (BIW) treatment schedules. Each treatment group included 10 mice. (A) Average tumor size for the different treatment groups ± SEM. (B) Average % body weight change compared with average body weight on day 1.
SGI-1776 in AML primary cells
Pim kinase inhibition in primary cells.
We investigated SGI-1776 in primary cells from patients with AML and results compared with those in MV-4-11. Myeloid cells were isolated and cultured with various concentrations of SGI-1776 for 24 hours then the relative protein levels were assessed via immunoblot. Although there were no significant changes in histone H3 (Ser10) phosphorylation there was a decrease in c-Myc (Ser62) phosphorylation in treated cells (Figure 6A), which is consistent with our report in CLL.10 In contrast to our studies in CLL primary cells, where there was neither a reduction in phospho-Bad (Ser112) nor in total Bad protein levels, in AML primary cells treated with SGI-1776 levels of both proteins decline (Figure 6A). Interestingly, this decline in both phospho- and total Bad protein was seen in both AML cell line and primary cells (Figure 2A-B). Consistent with our studies in AML cell line, there was also a substantial reduction in 4E-BP1 phosphorylation after SGI-1776 treatment in all AML primary samples evaluated (Figure 6B-C). The reduction in 4E-BP1 phosphorylation was in a dose-dependent manner and was observed in cells from both FLT3 wild-type and FLT3-ITD mutated AML patients. The decline in Mcl-1 protein observed in MV-4-11 was also mirrored in the AML primary cells (Figure 6D-E). Mcl-1 protein decline was observed in AML blasts from patients with wild-type FLT3 (n = 3) as well as FLT3-ITD mutation (n = 2), which was correlated with RNA synthesis inhibition (Figure 6F). Despite the decline in Mcl-1 protein levels, there were no significant changes in Bcl-2, Bcl-xL, Bak, or Bax (Figure 6G).
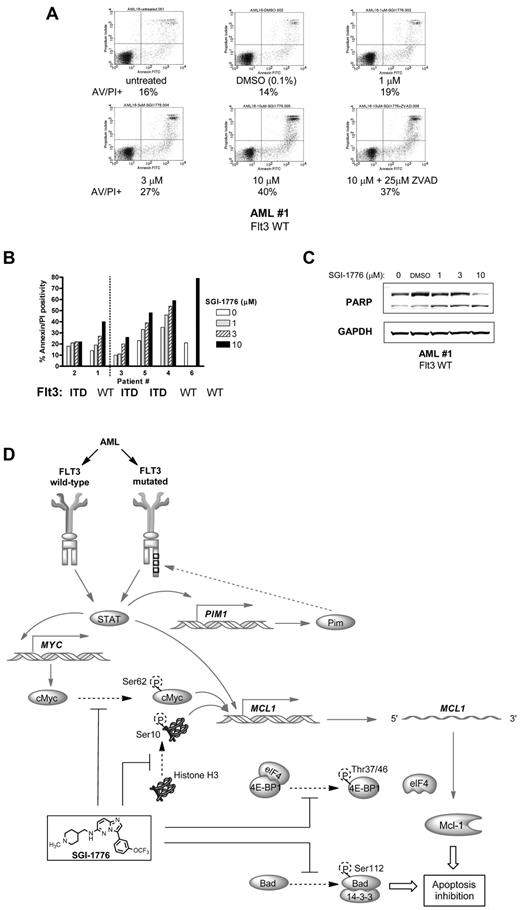
Molecular mechanism of action of SGI-1776 in primary AML cells. (A) Immunoblot analysis of Pim kinase targets in AML primary cells treated with SGI-1776. AML blasts were treated with 0.1% vehicle DMSO alone, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours, then harvested and lysed. Immunoblot analysis of the total and phospho-protein levels of (A) histone H3 (Ser10), c-Myc(Ser62), and Bad(Ser112) from lysates (patient 1). (B) Immunoblot analysis of the total and phospho-protein levels of 4E-BP1 (Thr37/46) from AML primary cells (patients 4 and 5). (C) Quantitation of phospho-4E–BP1 protein levels normalized to total 4E-BP1 levels in AML primary cells (patients 1, 2, 3, 4, and 5). (D) Immunoblot analysis of Mcl-1 protein using GAPDH as a loading control in AML primary cells (patients 4, 5). (E) Quantification of Mcl-1 protein levels normalized to GAPDH levels in 5 AML primary samples (patients 1, 2, 3, 4, and 5) treated with SGI-1776. (F) Inhibition of RNA synthesis in AML primary cells treated with SGI-1776. AML primary cells (patients 1, 2, 3, 4, and 6) were incubated with 0.1% DMSO alone, 1, 3, or 10μM SGI-1776 for 24 hours, then 1 hour before harvesting the cells, [3H]uridine was added to the cell culture as described in “RNA/protein synthesis assay.” (G) Immunoblot of anti- and proapoptotic proteins Bcl-xL, Bcl-2, Bak, and Bax in AML primary cells (patients 4 and 5).
Molecular mechanism of action of SGI-1776 in primary AML cells. (A) Immunoblot analysis of Pim kinase targets in AML primary cells treated with SGI-1776. AML blasts were treated with 0.1% vehicle DMSO alone, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours, then harvested and lysed. Immunoblot analysis of the total and phospho-protein levels of (A) histone H3 (Ser10), c-Myc(Ser62), and Bad(Ser112) from lysates (patient 1). (B) Immunoblot analysis of the total and phospho-protein levels of 4E-BP1 (Thr37/46) from AML primary cells (patients 4 and 5). (C) Quantitation of phospho-4E–BP1 protein levels normalized to total 4E-BP1 levels in AML primary cells (patients 1, 2, 3, 4, and 5). (D) Immunoblot analysis of Mcl-1 protein using GAPDH as a loading control in AML primary cells (patients 4, 5). (E) Quantification of Mcl-1 protein levels normalized to GAPDH levels in 5 AML primary samples (patients 1, 2, 3, 4, and 5) treated with SGI-1776. (F) Inhibition of RNA synthesis in AML primary cells treated with SGI-1776. AML primary cells (patients 1, 2, 3, 4, and 6) were incubated with 0.1% DMSO alone, 1, 3, or 10μM SGI-1776 for 24 hours, then 1 hour before harvesting the cells, [3H]uridine was added to the cell culture as described in “RNA/protein synthesis assay.” (G) Immunoblot of anti- and proapoptotic proteins Bcl-xL, Bcl-2, Bak, and Bax in AML primary cells (patients 4 and 5).
Apoptosis induction by SGI-1776 in AML primary cells.
There was a dose-dependent increase in apoptotic cells after treatment with SGI-1776, and there was ∼ 25% increase in apoptosis with 10μM SGI-1776 (Figure 7A-B). Inclusion of caspase inhibitor ZVAD did not significantly alter the level of apoptosis induced by SGI-1776, and the percentage of apoptotic cells was 40% with 10μM SGI-1776 alone and 37% with the coincubation with ZVAD (Figure 7A). Further analysis of cell death was evaluated using cells from multiple AML patients of various FLT3 mutation status. These experiments were conducted with cells cultured in media supplemented with autologous plasma (Figure 7B left of dotted line) and similar results were obtained when cells were cultured with FBS-supplemented media (Figure 7B right of dotted line), with even higher cytotoxicity toward AML cells. Because > 95% of the SGI-1776 is bound to human plasma protein, at 10μM SGI-1776, the free level of the drug is < 500nM.10 The average increase in apoptosis in the 4 treated AML samples cultured in FBS-supplemented media was 31% (Figure 7B). Apoptosis induction was further confirmed by PARP cleavage (Figure 7C). Overall cytotoxicity was lower in primary cells than in AML cell lines, which may be because primary cells in the peripheral blood are not actively cycling. In addition, AML primary cells originated from patients with heterogeneous cytogenetic profiles and prior treatment history. Actively cycling cell lines may be more susceptible to Pim kinase inhibition, because several Pim targets such as p27 are involved with cell cycle regulation. In contrast with primary AML blasts, little to no cytotoxicity was observed with SGI-1776 in lymphocytes from healthy donors where the increase in apoptosis averaged only 5%.10 These data elucidate that SGI-1776 is preferentially cytotoxic to primary AML cells with little toxicity to normal lymphocytes.
Induction of apoptosis by SGI-1776 in AML primary cells. (A) Flow cytometry analysis of annexin-FITC/propidium iodide staining of AML primary cells (patient 1) that were either untreated, treated with 0.1% DMSO vehicle alone, 1, 3, 10μM SGI-1776 or 10μM SGI-1776 with 25μM ZVAD for 24 hours. (B) Graphical representation of annexin/PI positive cells in AML cells cultured with increasing concentrations of SGI-1776 in media supplemented with autologous plasma (patients 1 and 2 left of dotted line) or with FBS (patients 3, 4, 5, and 6 right of dotted line). (C) Immunoblot analysis of PARP protein using GAPDH as a loading control in AML primary cells (patient 1). (D) Summary of relevant AML pathway signaling pathways (solid arrows) and Pim kinase phosphorylation targets (dashed arrows). In all 3 AML cell lines and primary AML cells, inhibition of Pim kinase function by SGI-1776 results in the decrease of downstream target protein Mcl-1 and induction of apoptosis.
Induction of apoptosis by SGI-1776 in AML primary cells. (A) Flow cytometry analysis of annexin-FITC/propidium iodide staining of AML primary cells (patient 1) that were either untreated, treated with 0.1% DMSO vehicle alone, 1, 3, 10μM SGI-1776 or 10μM SGI-1776 with 25μM ZVAD for 24 hours. (B) Graphical representation of annexin/PI positive cells in AML cells cultured with increasing concentrations of SGI-1776 in media supplemented with autologous plasma (patients 1 and 2 left of dotted line) or with FBS (patients 3, 4, 5, and 6 right of dotted line). (C) Immunoblot analysis of PARP protein using GAPDH as a loading control in AML primary cells (patient 1). (D) Summary of relevant AML pathway signaling pathways (solid arrows) and Pim kinase phosphorylation targets (dashed arrows). In all 3 AML cell lines and primary AML cells, inhibition of Pim kinase function by SGI-1776 results in the decrease of downstream target protein Mcl-1 and induction of apoptosis.
Discussion
Pim kinase signaling involves numerous pathways, and thus the biologic effects of SGI-1776 in AML cells may be multifaceted (Figure 7D). Significant transcription inhibition was observed in AML cell line and primary cells, which is in agreement with our hypothesis that c-Myc driven transcription is disrupted by Pim kinase inhibition. This may be because of decreased c-Myc(Ser62) phosphorylation (Figures 2A-B, 6A, supplemental Figure 3), which is needed for c-Myc protein stability and decreased histone H3(Ser10) phosphorylation (Figure 2A-B) that is required for binding of Myc/Max dimers to the E-box31 and initiation of Myc-driven transcription. In addition to c-Myc, other transcription factors associated with AML may also be affected by Pim kinase inhibition. AML patients harboring FLT3 mutations have been found to have increased levels of phosphorylation of forkhead transcription factor FoxO3a, which is correlated with higher rates of primary resistance and shorter remission durations.32 Pim-mediated phosphorylation and inactivation of FoxO3a suppresses the transcription of cyclin-dependent kinase inhibitor p27.28 Pim kinases also negatively regulate p27 at the posttranslational level via phosphorylation at Thr157 and Thr198, which induces binding with 14-3-3 protein and subsequent nuclear export and proteasomal degradation. Our results are in agreement with these reports as treatment of MV-4-11 cells with low doses of SGI-1776 (0.1-0.3μM) resulted in a 3- to 4-fold increase in p27 (Figure 2C-D). This removal of p27 suppression may affect cell cycle progression and inhibit Pim-mediated cell survival and proliferation.
Pim kinase involvement in translational control may also be a mechanism for SGI-1776–mediated cytotoxicity in AML cells. Pim kinase-target 4E-BP1 is a repressor of protein translation that binds to eIF4E to inhibit the translation initiation complex assembly. 4E-BP1, when phosphorylated at multiple sites, releases eIF4E resulting in protein synthesis initiation. Of note, in quiescent cells 4E-BP1 is hypophosphorylated and can interact strongly with eIF4E, whereas in contrast, 4E-BP1 is hyperphosphorylated on multiple sites after extracellular stimulation by growth factors, cytokines and hormones.33 4E-BP1 phosphorylation is hierarchical, and phosphorylation at Thr37/Thr46 are considered priming events for subsequent phosphorylation on other sites.34 Treatment with SGI-1776 resulted in decline in the level of phosphorylated 4E-BP1(Thr37/Thr46) protein levels in both AML cell lines (Figure 2A-B, supplemental Figure 3) and primary cells (Figure 6B-C), irrespective of FLT3 mutation status. Thus, inhibition of 4E-BP1 phosphorylation by SGI-1776 may be an attractive strategy to tackle multiple signaling pathways that up-regulate 4E-BP1 activation. There has been growing interest in identifying and targeting molecules that are downstream of multiple oncogenic signaling pathways that converge to drive proliferation, and 4E-BP1 has been indentified as one such protein.35 In addition, the effects of inhibiting 4E-BP1 phosphorylation may not be limited to protein translational regulation. Abrogation of 4E-BP1 phosphorylation and release of eIF4E may also affect c-Myc cell signaling; eIF4E has been shown to cooperate with c-Myc in B-cell lymphomagenesis36 through 2 combined events. eIF4E can suppress c-Myc–induced apoptosis and in-tandem c-Myc can antagonize eIF4E-induced cellular senescence. Interestingly, there also appears to be a regulation feedback loop between eIF4E and c-Myc at the transcriptional and translational level; eIF4E promotes the expression of c-Myc,37,38 which can in turn drive eIF4E expression.39 The utility of Pim kinase inhibition may be through the reduction of 4E-BP1 phosphorylation as well as altered eIF4E expression through inhibition of c-Myc (Ser62) phosphorylation.
For cancer cells, the essential readout of transcription and translation inhibition is oncoprotein synthesis, especially on those with short transcript and protein half-lives such as Mcl-1. Mcl-1 plays an oncogenic prosurvival role40 and is overexpressed in AML blasts after relapse.41 Recently, Mcl-1 was reported to facilitate Myc-induced AML development in mice and deletion of one allele of Mcl-1 from Myc-induced AML cells prolonged the survival of transplanted mice from 20 days to almost 50 days.42 In AML cell lines, ectopic expression of Mcl-1 has been shown to provide protection from cytotoxic agents.43 In AML, high levels of Mcl-1 protein are correlated with FLT3-ITD activating mutations,44 and Mcl-1-mediated cell survival in AML stem cells was identified to be via FLT3-ITD–driven activation of STAT5.These data highlight the significance of Mcl-1 as a therapeutic target in AML and our investigations described here provide a mechanism-based rationale for Pim kinase inhibition. In the present work in AML cell lines and primary cells, we observe a decline in Mcl-1 transcripts (Figure 3B) and in Mcl-1 protein levels after SGI-1776 treatment. SGI-1776 mechanism in AML may be via a blockade at multiple levels; through reduction in Mcl-1 protein translation by inhibiting 4E-BP1 phosphorylation, as well as through the inhibition of Mcl-1 transcription (Figure 7D).
In AML cells with FLT3 activating mutations, inhibition of Mcl-1 transcription (Figure 3B) may also arise through FLT3-STAT signaling. STAT-activated transcription may be inhibited via alteration in upstream FLT3 signaling by Pim kinase inhibition. We demonstrate that FLT3 protein is modulated after SGI-1776 treatment in FLT3-ITD cell line MV-4-11 (Figure 2C). FLT3-WT exists largely as the mature 150 kDa form, whereas as FLT3-ITD is partially retained as the 130 kDa form and is associated with endoplasmic reticulum (ER) transmembrane chaperone calnexin.45 A recent report describes Pim-1 stabilization of the 130 kDa isoform of FLT3.25 MV-4-11 cell line is homozygous for FLT3-ITD mutation, and SGI-1776 treatment shifts the equilibrium from the 130 kDa isoform to the 150 kDa isoform of FLT3 (Figure 2C). This may arise because FLT3 is also a client protein of heat shock protein 90 (HSP90),46 and Pim-1 has been reported to enhance FLT3 binding with calnexin and HSP90, which increases the half-life of the 130 kDa FLT3 isoform and decreases the half-life of the 150 kDa isoform. FLT3-ITD retention in the ER is dependent on its phosphorylation, and unlike wild-type FLT3, FLT3-ITD can induce transformation from an intracellular location. Using engineered ER-anchored FLT3 mutants, Böhmer and coworkers demonstrate that ER-retained FLT3-ITD resulted in sustained STAT5 activation, which is lost with ER-retained wild-type FLT3.47 There was also robust activation of STAT1/STAT3 in ER-retained FLT3-ITD.47 Mcl-1, Pim-1 and c-Myc are all downstream of STAT3,48-50 and thus activation of this pathway would up-regulate these proteins and result in inhibited apoptotic signaling. Given the identification of FLT3-ITD mutation as a negative prognostic indicator, Pim kinase inhibition may be a strategy to elicit therapeutic response in this subset of AML through modulation of FLT3-ITD signaling and down-regulation of Mcl-1. In addition to Pim kinase pathway, direct inhibitory action of SGI-1776 on FLT3 can play a biologic role in AML cells with FLT3 activating mutations. Our data suggest that SGI-1776 was effective in all 3 cell lines with varied FLT3 status, however potency was maximal in cells with FLT3-ITD. Overall a dual inhibition of FLT3 and Pim kinase by SGI-1776 may be an optimal approach for hard to treat AML with FLT3 activating mutation.
Cell death induction by SGI-1776 may be via targeting several key oncogenic pathways simultaneously, and the dual inhibition of both Mcl-1 transcription (Figure 3B) and protein synthesis (Figure 3C) may be a key factor in the utility of SGI-1776 in leukemia. Decrease in Bad(Ser112) phosphorylation and total protein (Figures 2A-B; 6A) may also play a role in SGI-1776 effect in AML, because binding partner 14-3-3 scaffold protein is then free to sequester antiapoptotic Bcl-2. An important feature of SGI-1776 is its selective induction of apoptosis in leukemia cells with only limited cytotoxicity in normal lymphocytes.10 However, biologic consequence of SGI-1776 in normal myeloid cells needs to be evaluated.
Pim kinase inhibitors are emerging as a new class of cancer therapeutics and we demonstrate here that during in vitro investigation in AML cell line and primary cells that SGI-1776 is effective in inhibiting Pim kinase pathways and inducing apoptosis. Furthermore, efficacy was observed in vivo in AML xenograft models; SGI-1776 treatment led to tumor volume reduction without significant changes in body weight (Figure 5A-B). These results are consistent with our previous investigations, which revealed that SGI-1776 was cytotoxic to CLL cells but not normal lymphocytes.10 Because SGI-1776 is the first Pim kinase inhibitor that is currently in clinical trial for patients with prostate cancer, our data provide rationale for clinical investigations in hematologic malignancies. Furthermore, our previous and current studies identify biomarkers that could be tested during clinical trial for indolent disease such as CLL and aggressive leukemias such as AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Yuling Chen for obtaining blood samples and to Mary Beth Rios for providing information on patient characteristics. The authors would like to acknowledge Dr Zhun Wang (CrownBio International) for his help with the xenograft studies.
This work was supported by the Translational Research Program of the Leukemia & Lymphoma Society of America (#LLS 0123-10; to V.G.) and a Leukemia SPORE (P50 CA100632) Career Development Award (to L.S.C.).
National Institutes of Health
Authorship
Contribution: L.S.C. designed research, performed experiments, analyzed results, and wrote the manuscript; V.G. supervised research, analyzed data, and wrote the manuscript; J.E.C. identified patients for this study; and S.R. and P.T. provided input on the xenograft animal model and reviewed the manuscript.
Conflict-of-interest disclosure: S.R. and P.T. are employees of SuperGen Inc. The remaining authors declare no competing financial interests.
Correspondence: Dr Varsha Gandhi, Dept of Experimental Therapeutics, University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Blvd, Unit 71, Houston, TX 77030-4009; e-mail: vgandhi@mdanderson.org.

![Figure 1. Pim kinases in cancer. (A) Pim kinase pathways (adapted from Chen et al26). (B) Chemical structure of imidazo[1,2-b]pyridazine compound SGI-1776.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/3/10.1182_blood-2010-12-323022/4/m_zh89991174840001.jpeg?Expires=1767709938&Signature=rw4kDKmHitkIAqx7pQwpDDKf6w5SiRczLjuV54O-6wW8HyMbCy-KWzXVsf58T9I220laTe3IOt0s1yERuQXq3vlZV2cghmrxq4Qmo5jB-Bn9~jS~fsMqhNV7owAr4ohzZzJNushAFVxZ4iU31UzauY5QRzkIfGf8YOxauBMD2W4pTizmhqcYD0YdoxkbtGxqGTjn8mtz0CsjbsakNW1Wlt-ovGzaKelki~6TGWaHlhJ7XwO36fo~0yhpVFmSnHe6aZiG8IxogivqUnT~pczhp9NkFlKTck2Aa9Se4Xnk2bT3VT2u49-hTgHRsnlGm9kPSKpV7grYohY3o2pGY64UPQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Effect of SGI-1776 in AML cell lines. (A) Inhibition of RNA synthesis in AML cell lines treated with SGI-1776. MV-4-11 (squares), MOLM-13 (triangles), and OCI-AML-3 (circles) lines were incubated with 0.1% DMSO alone, or 0.1, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours, then 1 hour before harvesting the cells, [3H]uridine was added to the cell culture as described in “RNA protein synthesis assay.” The results represent an average of triplicate experiments ± SEM. (B) Dose-dependent reduction of MCL-1 transcript levels in AML cell line treated with SGI-1776. MV-4-11 cells were treated with 0.1% vehicle DMSO alone, 0.1, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours, and the RNA was isolated. MCL-1 transcript levels were measured using real-time RT-PCR and normalized using 18S transcripts. Each RNA sample was assayed in triplicate and the results are expressed as a percentage of the MCL-1 expression level in cells treated with vehicle DMSO alone. The results represent an average of triplicate experiments ± SEM (C) Inhibition of protein synthesis in AML cell line treated with SGI-1776. MOLM-13 cells were incubated with 0.1% DMSO alone, or 0.1, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours, then 1 hour before harvesting the cells, [3H]leucine was added to the cell culture as described in “RNA protein synthesis assay.” The results represent an average of triplicate experiments ± SEM.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/3/10.1182_blood-2010-12-323022/4/m_zh89991174840003.jpeg?Expires=1767709938&Signature=Zf9A29HmtGlL1xjwAwrHZvjrQa~vMm20M9m1vo9ageGhuxvbLGV~4RP4H~FVR2JAdbKSATceXCin2YLNMJGunXFRxKg3dm9WC3dQaGbJP4BujjlNgJmUd~s14lua5RsPH14q-pYg2Ue7cYXFfbaP~FU57Mi0Cugeu6-8ptiMyClU8N~sTxgC4GOVHLrp4-Yp3sepn3ZUSOQwDaqohaASY5oST0s3BO-g8QpwyVKjg1391xZzqhf8fuSSzx6lAoTLWPyuOos5OpIFuyzINfMf6IwFUrev-IhtS2kjZzYb10ZmGYss6wMO-xVKXBS3MLBzlzeO-W0ECKvnotA51MZX8Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 6. Molecular mechanism of action of SGI-1776 in primary AML cells. (A) Immunoblot analysis of Pim kinase targets in AML primary cells treated with SGI-1776. AML blasts were treated with 0.1% vehicle DMSO alone, 0.3, 1, 3, or 10μM SGI-1776 for 24 hours, then harvested and lysed. Immunoblot analysis of the total and phospho-protein levels of (A) histone H3 (Ser10), c-Myc(Ser62), and Bad(Ser112) from lysates (patient 1). (B) Immunoblot analysis of the total and phospho-protein levels of 4E-BP1 (Thr37/46) from AML primary cells (patients 4 and 5). (C) Quantitation of phospho-4E–BP1 protein levels normalized to total 4E-BP1 levels in AML primary cells (patients 1, 2, 3, 4, and 5). (D) Immunoblot analysis of Mcl-1 protein using GAPDH as a loading control in AML primary cells (patients 4, 5). (E) Quantification of Mcl-1 protein levels normalized to GAPDH levels in 5 AML primary samples (patients 1, 2, 3, 4, and 5) treated with SGI-1776. (F) Inhibition of RNA synthesis in AML primary cells treated with SGI-1776. AML primary cells (patients 1, 2, 3, 4, and 6) were incubated with 0.1% DMSO alone, 1, 3, or 10μM SGI-1776 for 24 hours, then 1 hour before harvesting the cells, [3H]uridine was added to the cell culture as described in “RNA/protein synthesis assay.” (G) Immunoblot of anti- and proapoptotic proteins Bcl-xL, Bcl-2, Bak, and Bax in AML primary cells (patients 4 and 5).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/118/3/10.1182_blood-2010-12-323022/4/m_zh89991174840006.jpeg?Expires=1767709938&Signature=HmDwYafriNE4hEYtTx8BnBp92YkugSVoxd8R5p0-IoRWrD7ERdlX8vgwIH3B-xRWu56hFB9j04LmZ7zcqnwzPg8~lutH865AkNRAjSJRrgbZD7axNMhXfev349Er5S9AuGtz3KzL~UsitM3CYFzmMcO5~vBsfA0ojZFQWdwDFNclyyLEexpfO1N3qDC0y3ZsO0BDgkhp~DxzWalN1yPllQKY397NVHXcIIgq-HAbzhP5nGver7mWsT0So3zk45rVTUtLyvDkFEWHGgVTveRsVx6lIeUaAn7crEC7iu5aLuQog35QW6QQgeu8zCtxclWNS657Bk5rNS35iaJFYdPp8Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)