Abstract
Polycomb-group (PcG) proteins are essential regulators of hematopoietic stem cells (HSCs). In contrast to Bmi1, a component of Polycomb repressive complex 1 (PRC1), the role of PRC2 and its components in hematopoiesis remains elusive. Here we show that Ezh2, a core component of PRC2, is essential for fetal, but not adult, HSCs. Ezh2-deficient embryos died of anemia because of insufficient expansion of HSCs/progenitor cells and defective erythropoiesis in fetal liver. Deletion of Ezh2 in adult BM, however, did not significantly compromise hematopoiesis, except for lymphopoiesis. Of note, Ezh2-deficient fetal liver cells showed a drastic reduction in trimethylation of histone H3 at lysine 27 (H3K27me3) accompanied by derepression of a large cohort of genes, whereas on homing to BM, they acquired a high level of H3K27me3 and long-term repopulating capacity. Quantitative RT-PCR revealed that Ezh1, the gene encoding a backup enzyme, is highly expressed in HSCs/progenitor cells in BM compared with those in fetal liver, whereas Ezh2 is ubiquitously expressed. These findings suggest that Ezh1 complements Ezh2 in the BM, but not in the fetal liver, and reveal that the reinforcement of PcG-mediated gene silencing occurs during the transition from proliferative fetal HSCs to quiescent adult HSCs.
Introduction
Polycomb-group (PcG) proteins are involved in the maintenance of gene silencing through chromatin modifications and have been implicated in stem cell self-renewal. They reside in 2 main complexes, termed Polycomb repressive complex 1 (PRC1) and PRC2.1-3 Human PRC2 consists of 4 subunits: EZH2, EED, SUZ12, and RBAP48. PRC2 possesses catalytic activity specific for trimethylation of histone H3 at lysine 27 (H3K27me3).3-5 In contrast, PRC1 contains 4 core components, RING2 (Ring1B in mice), BMI1, HPH, and CBX, and possesses E3 ubiquitin ligase activity for the mono-ubiquitylation of histone H2A at lysine 119 (H2AK119ub1).5,6 These 2 types of complexes cooperate in the maintenance of gene silencing.
Among PcG proteins, Bmi1, a component of PRC1, and its role in the inheritance of the stemness of adult stem cells have been well characterized.7-9 Bmi1 maintains the self-renewal capacity of adult stem cells, at least partially, by repressing the Ink4a/Arf locus, which encodes a cyclin-dependent kinase inhibitor, p16Ink4a, and a tumor suppressor, p19Arf.10,11 Deletion of both Ink4a and Arf in Bmi1-deficient mice substantially restored the defective self-renewal capacity of hematopoietic stem cells (HSCs) and neural stem cells.11,12 On the other hand, comprehensive genome-wide analyses demonstrated that PRC1, PRC2, and trithorax-group complexes mark developmental regulator gene promoters with bivalent domains consisting of overlapping repressive (H3K27me3 and H2AK119ub1) and activating (H3K4me3) histone modifications to keep developmental regulators “poised” for activation in embryonic stem (ES) cells.13-16 Promoters with bivalent domains are resolved into a monovalent state, either active or repressive, on differentiation. Thus, transcriptional regulation of a large cohort of developmental regulator genes via bivalent domain is essential to maintain ES cells in an undifferentiated pluripotent state.17,18 Bivalent domains are partially shared by adult stem cells and have been detected in murine CD150+KSL HSCs and human CD34+CD133+ HSCs.19,20 Corresponding to these findings, we demonstrated that Bmi1 keeps differentiation programs poised for activation in HSCs to maintain their multipotency by repressing hematopoietic developmental regulator genes via bivalent domains as they do in ES cells.21
Contrary to studies of PRC1, there have been relatively few reports detailing the role of PRC2 in HSCs. Heterozygosity for an Eed null allele causes myeloproliferative and lymphoproliferative disease in mice.22 A hypomorphic mutation of Suz12 and heterozygosity for an Ezh2 null allele mildly but significantly ameliorate the HSC defects and reduced platelet numbers in mice that lacked the thrombopoietin receptor.23,24 These findings evoke the possibility that PRC2 restricts HSC/progenitor activity. In contrast, however, overexpression of Ezh2 in HSCs efficiently prevents exhaustion of the long-term repopulating potential of HSCs during serial transplantation,25 and mice with conditional knockout of Ezh2 in the hematopoietic system demonstrate a block in early B-cell differentiation with impaired rearrangement of the immunoglobulin heavy chain gene and a differentiation block of T cells at the early CD4−CD8− (DN) stage in thymus.26,27
In this study, to understand the contradictory and complicated findings on the PRC2 function based on the analyses of hypomorphic or haploinsufficient PRC2 mutant mice, we performed detailed analyses of mice in which Ezh2 was completely deleted in fetal and adult hematopoietic cells. We found that Ezh2 is essential for fetal hematopoiesis, but its function is largely dispensable in BM. Thus, dependency on Ezh2 changes during the transition from fetal to adult HSCs, suggesting that the epigenetic status of HSCs is regulated in a developmental stage and/or environment-specific manner.
Methods
Mice
For conditional deletion of Ezh2, the Ezh2fl/fl mice28 were crossed with either Tie2-Cre29 or Rosa28;Cre-ERT mice (TaconicArtemis GmbH). To induce Cre-ERT activity, mice were injected with 100 μL of tamoxifen dissolved in corn oil at a concentration of 10 mg/mL intraperitoneally for 5 consecutive days. C57BL/6 (B6-CD45.2) mice were purchased from Japan SLC. C57BL/6 mice congenic for the Ly5 locus (B6-CD45.1) were purchased from Sankyo-Lab Service. Mice were bred and maintained in the Animal Research Facility of the Graduate School of Medicine, Chiba University in accordance with institutional guidelines.
Flow cytometry and antibodies
Monoclonal antibodies (mAbs) recognizing the following antigens were used in flow cytometry and cell sorting: CD45.2 (104) CD45.1 (A20), Gr-1 (RB6-8C5), CD11b/Mac-1 (M1/70), Ter-119, CD127/IL-7Rα (A7R34), B220 (RA3-6B2), CD4 (L3T4), CD8α (53-6.7), CD117/c-Kit (2B8), Sca-1 (D7), and CD16/32/FcγRII-III (93). The mAbs were purchased from BD Biosciences, eBioscience, or BioLegend. Dead cells were eliminated by staining with propidium iodide (1 μg/mL; Sigma-Aldrich). All flow cytometric analyses and cell sorting were performed on a JSAN (Bay Bioscience), FACSAria II, or FACSCanto II (BD Biosciences).
Purification of hematopoietic cells
Hematopoietic cells harvested from embryonic day 12.5 (E12.5) fetal liver or BM were triturated and passed through 70-μm nylon mesh to obtain a single-cell suspension. Mononuclear cells were isolated on Ficoll-Paque PLUS (GE Healthcare) and incubated with a mixture of biotin-conjugated mAbs against lineage markers, including Gr-1, Mac-1, Ter-119, B220, IL-7Rα, CD4, and CD8α (Mac-1 was removed from the mixture for the fetal liver). The cells were further stained with allophycocyanin (APC)–Cy7-conjugated streptavidin and a combination of mAbs, including FITC-conjugated anti–CD34, PE- or PE-Cy7-Sca-1, PE-FcγR, and APC-c-Kit. Pacific Blue-CD45.2 mAb was used as an additional marker for donor-derived cells in the BM of B6-CD45.1 recipient mice. HSCs (Lin−CD34−/lowc-Kit+Sca-1+), multipotent progenitors (MPPs; Lin−CD34+c-Kit+Sca-1+), common myeloid progenitors (CMPs; Lin−Sca-1−c-Kit+CD34+FcγRlow), granulocyte-macrophage progenitors (GMPs; Lin−Sca-1−c-Kit+CD34+FcγRhi), and megakaryocyte-erythroid progenitors (MEPs; Lin−Sca-1−c-Kit+CD34−FcγRlow) fractions were defined as described previously.
BM transplantation
E12.5 fetal liver and BM cells from B6-CD45.2 mutant mice (test cells) were transplanted intravenously into 8-week-old B6-CD45.1 recipients irradiated at a dose of 9.5 Gy with or without BM cells from 8-week-old B6-CD45.1 congenic mice (competitor cells). At 3 months after transplantation, serial transplants were carried out by transferring 2 × 106 BM cells to secondary recipients. The chimerism of donor-derived hematopoiesis was monitored monthly by flow cytometry. Peripheral blood (PB) cells were stained with a mixture of mAbs that included PE-anti–Gr-1, PE-anti–Mac-1, APC-anti–B220, APC-Cy7-anti–CD4, APC-Cy7-anti–CD8α, FITC-CD45.2, and PE-Cy7-anti–CD45.1. The proportion of donor cells was evaluated by dividing the number of CD45.2 single-positive cells by the total number of CD45-positive cells (CD45.1 + CD45.2).
Quantitative RT-PCR
Total RNA was isolated using TRIZOL LS solution (Invitrogen) and reverse-transcribed by the ThermoScript RT-PCR system (Invitrogen) with an oligo-dT primer. Real-time quantitative PCR was performed with an ABI Prism 7300 Thermal Cycler (Applied Biosystems) using FastStart Universal Probe Master (Roche Applied Science) and the indicated combinations of Universal Probe Library (Roche Applied Science) and primers listed in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
ChIP assay
ChIP assays were performed as described previously.30 Briefly, cells were fixed with 1% formaldehyde for 15 minutes and then incubated in the presence of 0.125M glycine for 10 minutes. The cells were lysed with lysis buffer (50mM Tris-HCl, pH 8.0, 10mM EDTA, 0.1% SDS, and a proteinase inhibitor cocktail, Complete Midi) on ice for 20 minutes and sonicated. The lysate was separated into 4 samples diluted with an equal volume of dilution buffer (50mM Tris-HCl, pH 8.0, 150mM NaCl, 1% Triton X-100, 0.1% SDS, and a proteinase inhibitor cocktail, Complete Midi). Before the immunoprecipitation, 25 μL of Dynabeads Protein G (Invitrogen) was incubated with an anti–H3K27me3 antibody (07-449; Millipore) or an anti–monoubiquitinated H2A (H2Aub1; E6C5, Millipore) together with an anti–mouse IgMμ (12-488; Millipore) for 2 hours at 4°C. Chromatin was immunoprecipitated overnight at 4°C with antibody-conjugated Dynabeads. The immunoprecipitates were washed extensively and subjected to a quantitative PCR analysis with SYBR Premix Ex Taq II (Takara) using the primers listed in supplemental Methods.
Western blot analysis for histone tail methylation
Cells were lysed in 100mM Tris (pH 7.5), 150mM NaCl, 1.5mM MgCl2, 0.65% NP-40, and protease inhibitor cocktail (PIC; Roche Applied Science). Nuclei pelleted from the lysis buffer were extracted with 0.2 N HCl, and acid-soluble histones were precipitated with 20% trichloroacetic acid. Histones were separated by SDS-PAGE, transferred to a PVDF membrane (Immobilon; Millipore), and detected by Western blotting using the following antibodies: anti-H3 (ab1791, Abcam) and anti-H3K27me3 (07-449, Millipore).
Microarray analysis
A 1-color microarray-based gene expression analysis system (Agilent Technologies) containing 39 429 clones was used (Agilent Whole Mouse Genome 4 × 44k array Version 2, Design ID 026655), according to the manufacturer's instructions. Total RNA was extracted from MEPs. A total of 250 ng of total RNA was mixed with spike-in controls using an Agilent One Color Spike Mix Kit (Agilent Technologies), amplified and labeled with Cyanine 3 using a Quick Amp Labeling Kit (Agilent Technologies) according to the manufacturer's instructions, which generated single-color labeled cRNA. A total of 1650 ng of the labeled cRNA was used for each hybridization. The process of hybridization and washing was performed using a Hi-RPM Gene Expression Hybridization Kit (Large; Agilent Technologies) and a Gene Expression Wash Pack (Agilent Technologies), respectively. A DNA microarray scanner (Agilent Technologies) was used for array scanning. All data are MIAME compliant, and the raw data were deposited in Gene Expression Omnibus (accession number GSE32929).
Results
Tie2-Cre;Ezh2fl/fl embryos die at mid-gestation because of anemia
Ezh2−/− mice show early developmental defects and die by E7.5 or earlier.31 To delineate Ezh2 function in hematopoiesis, we conditionally deleted Ezh2 by crossing Ezh2fl/fl mice with Tie2-Cre mice, which specifically express Cre in hematopoietic and endothelial cells (Tie2-Cre;Ezh2fl/fl). We confirmed the efficient deletion of Ezh2 in fetal liver hematopoietic cells from Tie2-Cre;Ezh2fl/fl mice by genomic PCR (supplemental Figure 1). Tie2-Cre;Ezh2fl/fl embryos were recovered at nearly the expected Mendelian ratio at E12.5, but approximately 25% of them were already dead (Table 1). Tie2-Cre;Ezh2fl/fl embryos were pale, and their fetal livers were significantly smaller than those of the littermate controls (Figure 1A-B), and no mutant embryo was born alive (Table 1). In contrast, no significant difference was detected between Tie2-Cre;Ezh2+/+ and Tie2-Cre;Ezh2fl/fl yolk sacs in numbers of total yolk sac cells, c-Kit+ progenitor cells, and Ter119+ erythroblasts (supplemental Figure 2A). In addition, the vascular networks were normal in Tie2-Cre;Ezh2fl/fl embryos (supplemental Figure 2B).
Analysis of Ezh2 heterozygous intercross progenies
| . | Ezh2+/+ . | Ezh2fl/+ . | Ezh2fl/fl . | Tie2-Cre . | Tie2-Cre;Ezh2fl/+ . | Tie2-Cre;Ezh2fl/fl . | Total . |
|---|---|---|---|---|---|---|---|
| E12.5 | 31 | 68 | 36 | 32 | 80 | 37 (9 dead) | 284 |
| E13.5 | 9 | 13 | 10 | 8 | 13 | 3 (3 dead) | 56 |
| E14.5 | 12 | 6 | 5 | 6 | 5 | 4 (2 dead) | 38 |
| P0 | 8 | 12 | 6 | 6 | 16 | 0 | 48 |
| . | Ezh2+/+ . | Ezh2fl/+ . | Ezh2fl/fl . | Tie2-Cre . | Tie2-Cre;Ezh2fl/+ . | Tie2-Cre;Ezh2fl/fl . | Total . |
|---|---|---|---|---|---|---|---|
| E12.5 | 31 | 68 | 36 | 32 | 80 | 37 (9 dead) | 284 |
| E13.5 | 9 | 13 | 10 | 8 | 13 | 3 (3 dead) | 56 |
| E14.5 | 12 | 6 | 5 | 6 | 5 | 4 (2 dead) | 38 |
| P0 | 8 | 12 | 6 | 6 | 16 | 0 | 48 |
Tie2-Cre;Ezh2fl/fl embryos die at mid-gestation. (A) Appearance of Tie2-Cre and Tie2-Cre;Ezh2fl/fl embryos at E12.5. (B-D) Absolute cell numbers of whole fetal liver at E12.5 (Tie2-Cre;Ezh2, n = 27; Tie2-Cre;Ezh2fl/fl, n = 32) (B), KSL cells per fetal liver at E12.5 (Tie2-Cre;Ezh2, n = 11; Tie2-Cre;Ezh2fl/fl, n = 15) (C), and myeloid progenitors per fetal liver at E12.5 (Tie2-Cre, n = 10; Tie2-Cre;Ezh2fl/fl, n = 6) (D). Data are mean ± SD. (E) FACS profiles of erythroid differentiation in Tie2-Cre;Ezh2fl/fl fetal livers at E12.5. Representative FACS profiles are shown. (F) Absolute numbers of erythroblasts in the fractions defined by expression of CD71 and Ter119. Data are mean ± SD (Tie2-Cre, n = 3; Tie2-Cre;Ezh2fl/fl, n = 7). **P < .005.
Tie2-Cre;Ezh2fl/fl embryos die at mid-gestation. (A) Appearance of Tie2-Cre and Tie2-Cre;Ezh2fl/fl embryos at E12.5. (B-D) Absolute cell numbers of whole fetal liver at E12.5 (Tie2-Cre;Ezh2, n = 27; Tie2-Cre;Ezh2fl/fl, n = 32) (B), KSL cells per fetal liver at E12.5 (Tie2-Cre;Ezh2, n = 11; Tie2-Cre;Ezh2fl/fl, n = 15) (C), and myeloid progenitors per fetal liver at E12.5 (Tie2-Cre, n = 10; Tie2-Cre;Ezh2fl/fl, n = 6) (D). Data are mean ± SD. (E) FACS profiles of erythroid differentiation in Tie2-Cre;Ezh2fl/fl fetal livers at E12.5. Representative FACS profiles are shown. (F) Absolute numbers of erythroblasts in the fractions defined by expression of CD71 and Ter119. Data are mean ± SD (Tie2-Cre, n = 3; Tie2-Cre;Ezh2fl/fl, n = 7). **P < .005.
Flow cytometric analysis revealed that both Tie2-Cre;Ezh2+/+ and Tie2-Cre;Ezh2fl/fl fetal livers contained comparable numbers of c-Kit+Sca-1+ lineage marker− (KSL) cells, which includes HSCs and MPPs, at E11.5 (supplemental Figure 3A), indicating that the number of HSCs seeding the Tie2-Cre;Ezh2fl/fl liver is not significantly different from the control fetal liver. However, KSL cells expanded 40-fold in Tie2-Cre fetal livers, but only 12-fold in Tie2-Cre;Ezh2fl/fl fetal livers by E12.5, resulting in 4-fold reduction in KSL numbers in Tie2-Cre;Ezh2fl/fl liver at E12.5 (Figure 1C). Thus, expansion of HSCs and progenitor cells is impaired in Tie2-Cre;Ezh2fl/fl fetal livers. Furthermore, annexin V+ cells were increased in Tie2-Cre;Ezh2fl/fl KSL cells (supplemental Figure 3B), indicating that increased apoptosis also contributes to the reduction in HSCs and progenitor cells in Ezh2-deficient fetal liver.
Among myeloid progenitors, the number of MEPs was reduced 10-fold in Tie2-Cre;Ezh2fl/fl embryos compared with the controls, whereas no significant reduction was detected in numbers of CMPs or GMPs (Figure 1D). Detailed flow cytometric analyses demonstrated a maturation delay of erythroblasts from CD71+Ter119− cells (proerythroblasts and basophilic erythroblasts) to CD71+Ter119+ cells (basophilic and early polychromatic erythroblasts) in Tie2-Cre;Ezh2fl/fl embryos compared with the controls (Figure 1E), and revealed a significant reduction in numbers of CD71+Ter119+ erythroblasts in Tie2-Cre;Ezh2fl/fl fetal livers (Figure 1F). Ezh2Δ/Δ fetal liver CFU-E (CD45−c-Kit+CD71+Ter119−) were also reduced in number, whereas BFU-E (CD45+c-Kit+CD71−/lowTer119−), though significantly increased in frequency, were present at an absolute number similar to the control fetal livers (supplemental Figure 3C). These findings demonstrate profound defects in MEPs and CFU-E, as well as in the transition from CD71+Ter119− to CD71+Ter119+ erythroblasts during erythroid differentiation. We propose that this impaired erythroid differentiation, as well as the inefficient expansion of HSCs and progenitor cells, contributes to the fatal anemia in Tie2-Cre;Ezh2fl/fl embryos.
Ezh2Δ/Δ fetal liver cells reconstitute hematopoiesis in recipient BM
We next performed competitive reconstitution assays to examine whether Ezh2 is essential for maintaining the self-renewal capacity of HSCs. Surprisingly, we saw long-term reconstitution by Ezh2Δ/Δ fetal liver cells in 5 of 10 recipients, despite the insufficient expansion of Ezh2Δ/Δ HSCs/progenitors in the fetal liver (Figure 2A-B). As reported previously,26,27 the reconstitution of lymphoid lineage cells by Ezh2Δ/Δ fetal liver cells was severely compromised. Nevertheless, Ezh2Δ/Δ cells efficiently reconstituted myeloid lineage cells (Figure 2A-B). Ezh2Δ/Δ fetal liver cells retained a long-term reconstitution capacity in secondary transplantation and efficiently reconstituted the KSL population in BM (Figure 2C-D). To further evaluate the capacity of Ezh2Δ/Δ fetal liver HSCs to reconstitute hematopoiesis in detail, we next transplanted 200 and 1000 purified KSL cells along with BM competitor cells. With a limiting dose at 200 cells, Ezh2Δ/Δ KSL cells exhibited successful engraftment at the frequency similar to the control KSL cells (Ezh2Δ/Δ 1 of 4 vs control 2 of 6), suggesting a similar frequency of functional HSCs. Ezh2Δ/Δ cells reconstituted the myeloid lineage cells in PB at levels comparable with the controls, but barely the lymphoid lineage cells (supplemental Figure 4). Transplantation of 1000 Ezh2Δ/Δ KSL cells successfully reconstituted myeloid lineage cells in PB of all recipients and reconstituted the KSL population in BM to the levels comparable with the controls (supplemental Figure 4). These findings indicate that Ezh2Δ/Δ fetal liver HSCs retain long-term reconstituting capacity and suggest that the lower engraftment efficiency of Ezh2Δ/Δ fetal liver cells in primary transplantation observed in Figure 2A-B could be attributed to the low numbers of functional HSCs in Ezh2Δ/Δ fetal livers compared with the controls.
Ezh2-deficient fetal liver cells reconstitute hematopoiesis in BM. (A) Donor chimerism in competitive reconstitution assays using the same number of E12.5 fetal liver test cells and BM competitor cells (ranging from 2 × 105 to 1 × 106 cells, n = 10). The chimerism of CD45.2+ donor-derived cells in PB of each recipient mouse is shown. Lineage contribution of donor cells to myeloid (Gr-1+ and/or Mac-1+), B (B220+), or T (CD4+ and/or CD8+) cells is also shown. Five of 10 recipients infused with Tie2-Cre;Ezh2fl/fl fetal liver cells showed no engraftment of donor cells. The recipients indicated by asterisks were subjected to secondary transplantation at 12 weeks after transplantation. (B) Donor chimerism in PB of recipients with successful engraftment in panel A at 12 weeks after transplantation. Recipient mice with donor cell chimerism > 1.0% for myeloid and for B- and T-lymphoid lineages were considered successfully reconstituted. The data were summarized in bar graphs and presented as the mean ± SD (Tie2-Cre, n = 10; Tie2-Cre;Ezh2fl/fl, n = 5). (C) Donor chimerism in secondary transplantation. A total of 5 × 106 BM cells from the primary recipients indicated by asterisks in panel A were infused into lethally irradiated secondary recipients at 12 weeks after transplantation. Primary recipients *1 and *2 were used for experiments 1 (Exp. 1) and 2 (Exp. 2), respectively. The chimerism in PB is shown. Data are mean ± SD (n = 6). (D) Donor chimerism in BM KSL cells in Exp. 2 in secondary recipients. Data are mean ± SD (n = 6). *P < .05. **P < .005.
Ezh2-deficient fetal liver cells reconstitute hematopoiesis in BM. (A) Donor chimerism in competitive reconstitution assays using the same number of E12.5 fetal liver test cells and BM competitor cells (ranging from 2 × 105 to 1 × 106 cells, n = 10). The chimerism of CD45.2+ donor-derived cells in PB of each recipient mouse is shown. Lineage contribution of donor cells to myeloid (Gr-1+ and/or Mac-1+), B (B220+), or T (CD4+ and/or CD8+) cells is also shown. Five of 10 recipients infused with Tie2-Cre;Ezh2fl/fl fetal liver cells showed no engraftment of donor cells. The recipients indicated by asterisks were subjected to secondary transplantation at 12 weeks after transplantation. (B) Donor chimerism in PB of recipients with successful engraftment in panel A at 12 weeks after transplantation. Recipient mice with donor cell chimerism > 1.0% for myeloid and for B- and T-lymphoid lineages were considered successfully reconstituted. The data were summarized in bar graphs and presented as the mean ± SD (Tie2-Cre, n = 10; Tie2-Cre;Ezh2fl/fl, n = 5). (C) Donor chimerism in secondary transplantation. A total of 5 × 106 BM cells from the primary recipients indicated by asterisks in panel A were infused into lethally irradiated secondary recipients at 12 weeks after transplantation. Primary recipients *1 and *2 were used for experiments 1 (Exp. 1) and 2 (Exp. 2), respectively. The chimerism in PB is shown. Data are mean ± SD (n = 6). (D) Donor chimerism in BM KSL cells in Exp. 2 in secondary recipients. Data are mean ± SD (n = 6). *P < .05. **P < .005.
Deletion of Ezh2 in adult mice affects the differentiation, but not self-renewal, of HSCs
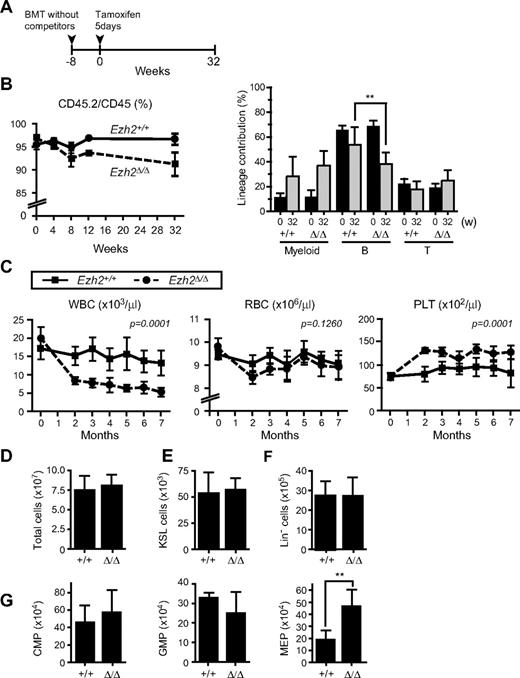
To evaluate the role of Ezh2 in adult BM hematopoiesis, we next prepared Cre-ERT;Ezh2fl/fl mice. To exclude any influences from the BM microenvironment on hematopoiesis, we transplanted BM cells from Cre-ERT control and Cre-ERT;Ezh2fl/fl mice without competitor cells into lethally irradiated wild-type recipient mice and deleted Ezh2 by inducing nuclear translocation of Cre by intraperitoneal injection of tamoxifen at 8 weeks after transplantation (Figure 3A). The deletion of Ezh2 was quite efficient and little residual floxed allele was detected by genomic PCR of CD45.2 donor cells even at 32 weeks after the injection of tamoxifen (supplemental Figure 1). The chimerism of Ezh2Δ/Δ cells in PB dropped slightly after the deletion of Ezh2 because of a reduced contribution of Ezh2Δ/Δ cells to the lymphoid lineage, particularly B lymphoid lineage (Figure 3B). Correspondingly, the white blood cell count in PB significantly decreased in recipient mice repopulated with Ezh2Δ/Δ cells, while the platelet count significantly increased (Figure 3C). At 32 weeks after the deletion of Ezh2, impaired differentiation of lymphoid cells was clearly observed as reported previously (supplemental Figure 5).26,27 The cell number in thymus was reduced 10-fold and a differentiation block was obvious at the CD4−CD8− DN stage. B cell differentiation was compromised at the transition from the pro-B to pre-B cell stage. In contrast, no significant change was detected in numbers of KSL cells or myeloid progenitors except for a 2-fold increase in MEPs (Figure 3D-G).
Depletion of Ezh2 in hematopoietic cells in adult mice does not greatly compromise hematopoiesis. (A) Schema of conditional deletion of Ezh2 in adult mice. BM cells from 8-week-old Cre-ERT and Cre-ERT;Ezh2fl/fl mice were transplanted into lethally irradiated recipient mice without competitor cells. At 8 weeks after transplantation, recipient mice were injected with tamoxifen for 5 consecutive days, and their PB was monitored for 32 weeks. (B) Donor chimerism and lineage contribution in PB. The chimerism of CD45.2+ donor-derived cells in PB of recipient mice was monitored for 32 weeks after injection of tamoxifen (left panel). Lineage contribution of donor cells to myeloid (Gr-1+ and/or Mac-1+), B (B220+), or T (CD4+ and or CD8+) cells is also shown in the right panel. Data are mean ± SD (n = 10). (C) PB cell counts of recipients repopulated with Cre-ERT and Cre-ERT;Ezh2fl/fl BM cells after deletion of Ezh2 by tamoxifen injection. Data are mean ± SD (Tie2-Cre, n = 8; Tie2-Cre;Ezh2fl/fl, n = 9). Statistical analysis was performed using 2-way repeated-measures ANOVA. (D-G) Absolute numbers of BM cells (D), KSL HSC/MPPs (E), Lin− cells (F), and CMPs, GMPs, and MEPs (G) in BM of the recipient mice at 32 weeks after tamoxifen injection. Data are mean ± SD (n = 6). **P < .005.
Depletion of Ezh2 in hematopoietic cells in adult mice does not greatly compromise hematopoiesis. (A) Schema of conditional deletion of Ezh2 in adult mice. BM cells from 8-week-old Cre-ERT and Cre-ERT;Ezh2fl/fl mice were transplanted into lethally irradiated recipient mice without competitor cells. At 8 weeks after transplantation, recipient mice were injected with tamoxifen for 5 consecutive days, and their PB was monitored for 32 weeks. (B) Donor chimerism and lineage contribution in PB. The chimerism of CD45.2+ donor-derived cells in PB of recipient mice was monitored for 32 weeks after injection of tamoxifen (left panel). Lineage contribution of donor cells to myeloid (Gr-1+ and/or Mac-1+), B (B220+), or T (CD4+ and or CD8+) cells is also shown in the right panel. Data are mean ± SD (n = 10). (C) PB cell counts of recipients repopulated with Cre-ERT and Cre-ERT;Ezh2fl/fl BM cells after deletion of Ezh2 by tamoxifen injection. Data are mean ± SD (Tie2-Cre, n = 8; Tie2-Cre;Ezh2fl/fl, n = 9). Statistical analysis was performed using 2-way repeated-measures ANOVA. (D-G) Absolute numbers of BM cells (D), KSL HSC/MPPs (E), Lin− cells (F), and CMPs, GMPs, and MEPs (G) in BM of the recipient mice at 32 weeks after tamoxifen injection. Data are mean ± SD (n = 6). **P < .005.
We next examined the long-term reconstitution capacity of Ezh2Δ/Δ cells from adult BM. To this end, Cre-ERT;Ezh2+/+ control and Cre-ERT;Ezh2fl/fl BM cells were transplanted along with the same number of BM competitor cells into lethally irradiated wild-type recipients. At 8 weeks after transplantation, Ezh2 was deleted by intraperitoneal injection of tamoxifen (Figure 4A). Ezh2Δ/Δ BM cells again efficiently reconstituted myeloid lineage cells as well as BM KSL cells at a level comparable to the control (Figure 4B-C). Secondary transplantation analyses also confirmed the long-term reconstitution capacity of Ezh2Δ/Δ BM cells to be intact (Figure 4B).
Reconstitution capacity of Ezh2-deficient BM cells is almost intact. (A) Schema of competitive reconstitution assays to evaluate Ezh2-deficient BM cells. BM cells from 8-week-old Cre-ERT and Cre-ERT;Ezh2fl/fl mice were transplanted into lethally irradiated recipient mice with the same number of competitor BM cells (1 × 106 cells). At 8 weeks after transplantation, recipient mice were injected with tamoxifen for 5 consecutive days; and after a further 12 weeks, secondary transplantation was performed. (B) Donor chimerism in PB at 12 weeks after primary and secondary transplantations. Donor chimerism in PB at the point just before tamoxifen injection was arbitrarily set to 100%. Data are mean ± SD (n = 6): ■ represents wild-type BM cells; and  , Ezh2Δ/Δ BM cells. (C) Donor chimerism in BM KSL cells at 12 weeks after primary transplantation. Data are mean ± SD (n = 4).
, Ezh2Δ/Δ BM cells. (C) Donor chimerism in BM KSL cells at 12 weeks after primary transplantation. Data are mean ± SD (n = 4).
Reconstitution capacity of Ezh2-deficient BM cells is almost intact. (A) Schema of competitive reconstitution assays to evaluate Ezh2-deficient BM cells. BM cells from 8-week-old Cre-ERT and Cre-ERT;Ezh2fl/fl mice were transplanted into lethally irradiated recipient mice with the same number of competitor BM cells (1 × 106 cells). At 8 weeks after transplantation, recipient mice were injected with tamoxifen for 5 consecutive days; and after a further 12 weeks, secondary transplantation was performed. (B) Donor chimerism in PB at 12 weeks after primary and secondary transplantations. Donor chimerism in PB at the point just before tamoxifen injection was arbitrarily set to 100%. Data are mean ± SD (n = 6): ■ represents wild-type BM cells; and  , Ezh2Δ/Δ BM cells. (C) Donor chimerism in BM KSL cells at 12 weeks after primary transplantation. Data are mean ± SD (n = 4).
, Ezh2Δ/Δ BM cells. (C) Donor chimerism in BM KSL cells at 12 weeks after primary transplantation. Data are mean ± SD (n = 4).
Levels of trimethylation of H3K27 in the absence of Ezh2
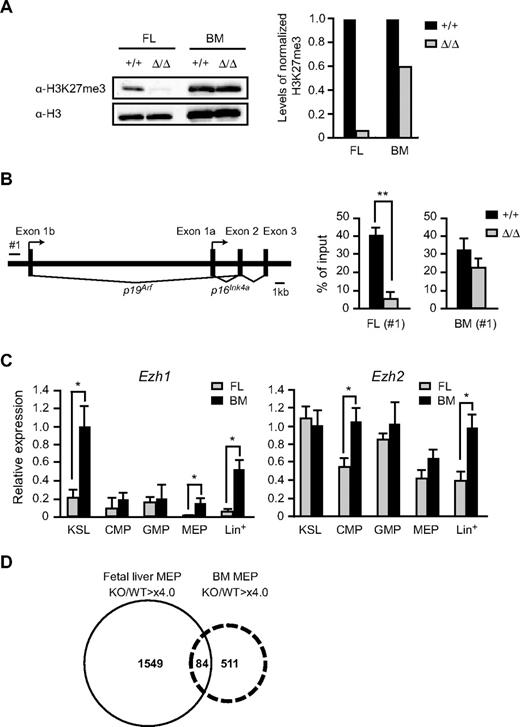
To understand the different effects of the deletion of Ezh2 on fetal liver and BM hematopoiesis, we checked the levels of H3K27me3 in Ezh2Δ/Δ CD45+Lin− immature hematopoietic cells by Western blot analysis. As expected, the amount of H3K27me3 in Ezh2Δ/Δ fetal liver cells was significantly reduced to less than one-tenth of the control level (Figure 5A). Of note, however, the Ezh2Δ/Δ fetal liver cells that reconstituted the recipient BM gained a high level of H3K27me3, approximately two-thirds of the control value (Figure 5A). We next analyzed the PRC2-mediated histone modification at the Ink4a/Arf locus, one of the PcG targets, in purified immature hematopoietic cells. ChIP assays showed a significant reduction in H3K27me3 levels at the Ink4a/Arf promoter. The level of H3K27me3 was reduced 8-fold in Ezh2Δ/Δ fetal liver cells but was maintained at two-thirds of the control value in Ezh2Δ/Δ BM (Figure 5B). The ChIP data corresponded well to the results of Western blotting of global H3K27me3. Immunofluorescence analysis of the levels of H3K27me3 further supported the results of Western blot and ChIP analyses in both CD45+Lin− and KSL cells, although the reduction in H3K27me3 levels in the absence of Ezh2 was relatively moderate compared with those detected by Western blot and ChIP analyses. This could be because of higher background signals in immunostaining analyses (supplemental Figure 6).
Levels of trimethylation of H3K27 in the absence of Ezh2. (A) Levels of trimethylation at H3K27 in Ezh2Δ/Δ fetal livers. Histones were extracted from CD45+Lin− immature hematopoietic cells purified from E12.5 Tie2-Cre (Ezh2+/+) or Tie2-Cre;Ezh2fl/fl (Ezh2Δ/Δ) fetal liver cells (left 2 lanes) and from recipients' BM repopulated by E12.5 Tie2-Cre (Ezh2+/+) or Tie2-Cre;Ezh2fl/fl (Ezh2Δ/Δ) fetal liver cells (right 2 lanes), and were analyzed by Western blotting using an anti–H3K27me3 antibody. Levels of trimethylated H3K27 were normalized to the amount of H3 and are indicated relative to Tie2-Cre control values. (B) A schematic representation of the Ink4a/Arf locus (left panel). ChIP assays were performed using an anti–H3K27me3 antibody. The regions amplified from the precipitated DNA by site-specific quantitative PCR are indicated by a bar (#1). Quantitative ChIP analysis of the Ink4a/Arf locus in CD45+Lin− cells from E12.5 Tie2-Cre (Ezh2+/+) and Tie2-Cre;Ezh2fl/fl (Ezh2Δ/Δ) fetal livers and CD45+Lin−c-Kit+ cells from recipients' BM repopulated with Cre-ERT (Ezh2+/+) or Cre-ERT;Ezh2fl/fl (Ezh2Δ/Δ) BM cells at 32 weeks after tamoxifen treatment. Percentages of input DNA are shown as the mean ± SD (right panel). (C) Quantitative RT-PCR analysis of Ezh1 and Ezh2 expression in E14.5 fetal liver and BM from 8-week-old mice. mRNA levels in indicated hematopoietic fractions were normalized to Hprt1 expression. Expression levels relative to those in the BM KSL cells are shown as the mean ± SD for triplicate analyses. Lin+ indicates lineage marker-positive differentiated hematopoietic cells. (D) A Venn diagram depicting derepressed genes (> 4-fold) in Ezh2-deficient fetal liver MEPs and BM MEPs.
Levels of trimethylation of H3K27 in the absence of Ezh2. (A) Levels of trimethylation at H3K27 in Ezh2Δ/Δ fetal livers. Histones were extracted from CD45+Lin− immature hematopoietic cells purified from E12.5 Tie2-Cre (Ezh2+/+) or Tie2-Cre;Ezh2fl/fl (Ezh2Δ/Δ) fetal liver cells (left 2 lanes) and from recipients' BM repopulated by E12.5 Tie2-Cre (Ezh2+/+) or Tie2-Cre;Ezh2fl/fl (Ezh2Δ/Δ) fetal liver cells (right 2 lanes), and were analyzed by Western blotting using an anti–H3K27me3 antibody. Levels of trimethylated H3K27 were normalized to the amount of H3 and are indicated relative to Tie2-Cre control values. (B) A schematic representation of the Ink4a/Arf locus (left panel). ChIP assays were performed using an anti–H3K27me3 antibody. The regions amplified from the precipitated DNA by site-specific quantitative PCR are indicated by a bar (#1). Quantitative ChIP analysis of the Ink4a/Arf locus in CD45+Lin− cells from E12.5 Tie2-Cre (Ezh2+/+) and Tie2-Cre;Ezh2fl/fl (Ezh2Δ/Δ) fetal livers and CD45+Lin−c-Kit+ cells from recipients' BM repopulated with Cre-ERT (Ezh2+/+) or Cre-ERT;Ezh2fl/fl (Ezh2Δ/Δ) BM cells at 32 weeks after tamoxifen treatment. Percentages of input DNA are shown as the mean ± SD (right panel). (C) Quantitative RT-PCR analysis of Ezh1 and Ezh2 expression in E14.5 fetal liver and BM from 8-week-old mice. mRNA levels in indicated hematopoietic fractions were normalized to Hprt1 expression. Expression levels relative to those in the BM KSL cells are shown as the mean ± SD for triplicate analyses. Lin+ indicates lineage marker-positive differentiated hematopoietic cells. (D) A Venn diagram depicting derepressed genes (> 4-fold) in Ezh2-deficient fetal liver MEPs and BM MEPs.
These findings indicate that back-up enzymes for trimethylation at H3K27 might function in adult BM, but not in fetal liver HSCs/progenitor cells. We then analyzed the expression of Ezh1, the gene encoding another enzyme for trimethylation at H3K27. Quantitative RT-PCR revealed that Ezh1 was highly expressed in KSL cells in BM compared with fetal liver cells and was barely expressed in fetal liver MEPs while Ezh2 was ubiquitously expressed (Figure 5C). These expression profiles of Ezh1 might partly account for the difference in function of back-up enzymes between fetal and adult hematopoiesis.
The early stages of fetal liver hematopoiesis mostly generate RBCs to supply sufficient oxygen to developing embryos. Deletion of Ezh2 in fetal liver caused lethal anemia but did not significantly affect erythropoiesis in adult BM as evident from the normal RBC numbers and hemoglobin content in PB (Figure 3C; and data not shown). To understand the physiologic significance of reduced levels of H3K27me3 in Ezh2Δ/Δ cells, we performed gene expression profiling of fetal and BM Ezh2Δ/Δ MEPs compared with the control. Microarray analysis identified a large number of genes derepressed in Ezh2Δ/Δ fetal liver MEPs. This was true even in Ezh2Δ/Δ BM MEPs, although the erythropoiesis was considered almost normal in BM in the absence of Ezh2. The extent of derepression was well correlated with the levels of reduction in H3K27me3, and 3-fold more genes were derepressed in fetal liver MEPs compared with BM MEPs (Figure 5D; detailed data are available in supplemental Microarray Dataset). Of interest, expression of the transcription factor genes essential for erythroid differentiation was not greatly affected by the absence of Ezh2 except for Gata2 (supplemental Table 1). Up-regulation of Gata2, an essential regulator for HSCs and early progenitors, in Ezh2Δ/Δ fetal liver MEPs might reflect the compensatory mechanisms operating for impaired expansion of HSCs and progenitors. The majority of derepressed genes did not overlap between Ezh2Δ/Δ fetal liver and BM MEPs, indicating that the PcG complexes regulate quite different sets of genes in fetal liver and BM MEPs (Figure 5D).
Discussion
In this study, we found that deletion of Ezh2 profoundly compromises fetal liver hematopoiesis but not the self-renewal capacity of HSCs in BM. Notably, loss of Ezh2 in fetal liver resulted in a drastic reduction in H3K27me3 levels, but Ezh2-deficient fetal liver HSCs successfully homed to adult BM and acquired a high level of H3K27me3 and long-term reconstitution capacity. These findings suggest that the regulation of epigenetic status by the PcG complexes is dramatically altered during the transition from fetal to adult HSCs. Ezh1, closely related to Ezh2, forms similar PRC2 complexes to Ezh2 and complements Ezh2 in maintaining stem cell identity and executing pluripotency in Ezh2−/− ES cells.32,33 Depletion of Ezh1 in Ezh2−/− ES cells abolishes residual methylation at H3K27 and derepresses H3K27me3 target genes.33 Given that Ezh1 and Ezh2 are the only histone methyltransferases for trimethylation at H3K27, our findings suggest that Ezh1 may function as a compensatory mechanism for Ezh2 in BM, but not so sufficient in fetal liver. Although the function of PRC2 containing Ezh1 as a histone methyltransferase is reportedly weaker than that containing Ezh2,32 we detected unexpectedly high levels of residual H3K27me3 in Ezh2-deficient cells. Thus, the activity of Ezh1 as a histone methyltransferase could be strong enough to rescue Ezh2 deficiency in BM.
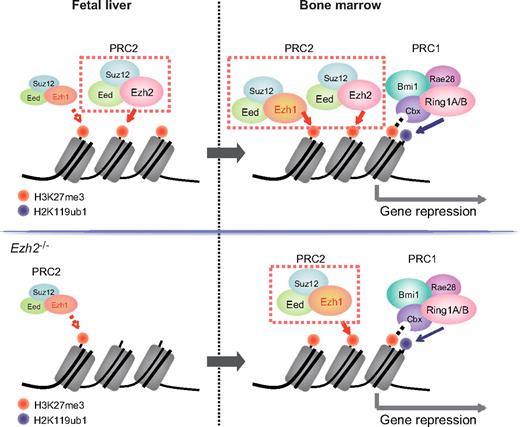
It is generally accepted that HSCs arise in both yolk sac and aorta-gonad-mesonephros and seed the fetal liver, where they expand and acquire the capacity to engraft and to reconstitute the BM.34 HSCs extensively undergo cell division in the fetal liver while they are mostly in a quiescent state in the BM. HSCs receive highly distinctive signals from niche cells in fetal liver and BM. These signals are thought to regulate the cell cycle status of HSCs and promote maturation of HSCs. Bmi1 has been demonstrated to be dispensable for fetal liver hematopoiesis.8,9 Likewise, Ezh1 was expressed at a low level in fetal liver compared with BM and did not appear to complement Ezh2 function in fetal liver in this study. These findings indicate that PcG-mediated gene silencing is highly dependent on Ezh2 in proliferative fetal liver HSCs but reinforced in quiescent BM HSCs by the collaboration of Ezh2 with Ezh1 and Bmi1-containing PRC1. Thus, the stringency of PcG-mediated histone modifications might define the cell cycle status of HSCs by modulating gene expression profiles (Figure 6). Indeed, fetal and adult HSCs display marked differences in their self-renewal, differentiated cell output, and gene expression profiles.35 These functional changes may be partially attributable to differential activities of the PcG proteins.
PcG proteins behave differentially in fetal liver and adult BM HSCs. In this study, Ezh1 contributed little to fetal liver HSCs similar to a PRC1 protein, Bmi1, even in the absence of Ezh2 (top left panel, wild-type HSCs; bottom left panel, Ezh2−/− HSCs). In BM, Ezh2-mediated gene silencing is reinforced by Ezh1 and mono-ubiquitylation at H2AK119 by Bmi1-containing PRC2 (top right panel, wild-type HSCs; bottom right panel, Ezh2−/− HSCs). Differential epigenetic regulation by PcG proteins might have a strong impact on the cell cycle status of proliferative fetal and quiescent adult HSCs.
PcG proteins behave differentially in fetal liver and adult BM HSCs. In this study, Ezh1 contributed little to fetal liver HSCs similar to a PRC1 protein, Bmi1, even in the absence of Ezh2 (top left panel, wild-type HSCs; bottom left panel, Ezh2−/− HSCs). In BM, Ezh2-mediated gene silencing is reinforced by Ezh1 and mono-ubiquitylation at H2AK119 by Bmi1-containing PRC2 (top right panel, wild-type HSCs; bottom right panel, Ezh2−/− HSCs). Differential epigenetic regulation by PcG proteins might have a strong impact on the cell cycle status of proliferative fetal and quiescent adult HSCs.
To test this hypothesis, we have to await a comparative genome-wide analysis of PcG histone modifications between fetal liver and BM HSCs. It is also possible that niche signals directly regulate the expression of PcG genes and/or post-transcriptional modifications of PcG proteins critical to their function. Elucidating how environmental factors regulate PcG function is important to understanding the precise role of PcG proteins in HSCs.
PcG proteins are also known to regulate developmental regulator genes involved in differentiation. We previously demonstrated that Bmi1 negatively regulates B lineage master regulator genes, Ebf1 and Pax5, in HSCs and MPPs to keep the B lineage differentiation program poised for activation.21 Ezh2 has been reported to repress premature activation of differentiation-related genes in epidermal progenitor cells.36 In both cases, PcG proteins negatively regulate differentiation. In contrast, deletion of Ezh2 causes blocks in T- and B-cell differentiation.26,27 In this study, we showed that deletion of Ezh2 impairs erythroid differentiation in fetal livers but not in BM. Conversely, deletion of Ezh2 enhanced platelet production in adult mice. A hypomorphic mutation of Suz12 in mice has been reported to complement reductions of platelet production in mice lacking thrombopoietin.23 Taken together, PRC2 plays an essential role in fetal liver erythropoiesis but negatively regulates megakaryocytic differentiation in BM. These findings highlight important roles of PcG proteins in fine-tuning cell differentiation.
The human EZH2 gene is located at 7q, a region often deleted in hematologic disorders, such as myelodysplastic syndrome and myeloproliferative neoplasm. Recently, loss-of-function mutations of EZH2 were identified in myelodysplastic syndrome and myeloproliferative neoplasm.37 These findings indicate that EZH2 could function, not only as an oncogene4 but also as a tumor suppressor gene. Although we did not observe any hematologic neoplasms developing in Ezh2-deficient mice, reduced levels of H3K27me3 could lead to derepression of oncogenes. Actually, in contrast to Bmi1-deficient mice with a severe self-renewal defect of HSCs, mice with hypomorphic mutations of Eed and Suz12 unexpectedly showed enhanced hematopoiesis.22-24 These contradictory phenotypes are in line with our observation in this study and are suggestive of a broad range of target genes of the PcG complexes. Thus, the PcG complexes fine-tune the growth of hematopoietic cells in both a positive and a negative manner. In this regard, our findings also provide an important insight into the tumor suppressive function of EZH2 in human myeloid malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Yoko Koseki, Makiko Yui, and Satomi Tanaka for technical support, George Wendt for critical reading of the manuscript, and Mieko Tanemura for laboratory assistance.
This work was supported in part by MEXT KAKENHI (20052009, 21790906, and 22118004) and the Global COE Program (Global Center for Education and Research in Immune System Regulation and Treatment), MEXT, Japan, the Japan Science and Technology Corporation (Grant-in-aid for Core Research for Evolutional Science and Technology), the Takeda Science Foundation, and Astellas Foundation for Research on Metabolic Disorders (research grant).
Authorship
Contribution: M.M.-K. performed the experiments, analyzed results, made the figures, and actively wrote the manuscript; Y.M., S.M., M.N., A.S., and T.K. assisted with the experiments, including the hematopoietic analyses, ChIP analyses, and immunostaining; J.S. and H.K. provided conditional Ezh2 knockout mice; and A.I. conceived of and directed the project, secured funding, and actively wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Atsushi Iwama, 1-8-1 Inohana, Chuo-ku, Chiba, 260-8670 Japan; e-mail: aiwama@faculty.chiba-u.jp.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal