Abstract
Despite an increase in survival for children with acute lymphoblastic leukemia (ALL), the outcome after relapse is poor. To understand the genetic events that contribute to relapse and chemoresistance and identify novel targets of therapy, 3 high-throughput assays were used to identify genetic and epigenetic changes at relapse. Using matched diagnosis/relapse bone marrow samples from children with relapsed B-precursor ALL, we evaluated gene expression, copy number abnormalities (CNAs), and DNA methylation. Gene expression analysis revealed a signature of differentially expressed genes from diagnosis to relapse that is different for early (< 36 months) and late (≥ 36 months) relapse. CNA analysis discovered CNAs that were shared at diagnosis and relapse and others that were new lesions acquired at relapse. DNA methylation analysis found increased promoter methylation at relapse. There were many genetic alterations that evolved from diagnosis to relapse, and in some cases these genes had previously been associated with chemoresistance. Integration of the results from all 3 platforms identified genes of potential interest, including CDKN2A, COL6A2, PTPRO, and CSMD1. Although our results indicate that a diversity of genetic changes are seen at relapse, integration of gene expression, CNA, and methylation data suggest a possible convergence on the WNT and mitogen-activated protein kinase pathways.
Introduction
The cure rate for childhood acute lymphoblastic leukemia (ALL) has improved dramatically over the past 4 decades, yet up to 20% of patients experience disease recurrence. Although children who relapse late (≥ 3 years from initial diagnosis) fare better than those who relapse early in treatment, the prognosis for these children remains poor, even with aggressive treatment.1,2 Thus, new approaches to prevent and treat relapsed ALL are urgently needed. To discover the underlying biologic pathways that may play a role in drug resistance and relapse, we have previously determined differences in gene expression and copy number in matched diagnosis/relapse paired samples. We identified a distinct signature of differentially expressed genes from diagnosis to relapse associated with early relapse, but not for late relapse, perhaps because of the smaller cohort of patients.3 Likewise, we and others have surveyed genome-wide copy number profiles in paired and associated germline samples using Affymetrix 500K SNP arrays.4,5 Because of the known biologic heterogeneity of childhood ALL, we have now extended our analysis to a larger group of matched diagnosis/relapse samples using newer platforms that greatly expand the amount of genetic information acquired. In addition, we explore the role of epigenetic dysregulation in the development of relapsed ALL through the use of a genome-wide methylation array. This integrated analysis, of 3 high throughput platforms, suggests that although multiple defects evolve from diagnosis to relapse, some may converge on distinct pathways, such as the WNT and mitogen-activated protein kinase signaling pathways.
Methods
Patient samples
Ficoll-enriched, cryopreserved bone marrow samples from diagnosis, remission, and relapse were obtained from the Children's Oncology Group (COG) from pediatric patients with relapsed B-precursor ALL. All patients had a bone marrow or bone marrow/central nervous system combined relapse and were initially treated on COG protocols for newly diagnosed ALL. Patient characteristics are detailed in supplemental Table 1 (available on the Blood Web site; see the Supplemental Materials link at the top of the online article). All patients (or parents) had provided informed consent for use of these samples for research studies in accordance with the Declaration of Helsinki. This study was approved by the institutional review board of all participating institutions.
Gene expression analysis
RNA was isolated from 49 diagnostic and relapse bone marrow samples using QIAGEN RNEasy Mini Kits, and quality was verified by an Agilent 2100 Bioanalyzer. A total of 1 μg of total RNA was used as a template in a double amplification protocol using RiboAmp RNA amplification kits (Arcturus). In vitro transcription was completed with biotinylated UTP and CTP for labeling using the ENZO BioArray HighYield RNA Transcript Labeling kit (Enzo Diagnostics). A total of 13 μg of labeled cRNA was fragmented and hybridized to Affymetrix U133Plus2 microarrays according to Affymetrix protocol. Raw Affymetrix CEL files were processed with the standard Affymetrix probe modeling algorithm RMA. Background correction and quantile normalization were applied during the process. The processed data were stored in a matrix containing one intensity value per probe set in the GCT format file. Analyses were based on the GCT format files with a class label file created in the CLS format. Different versions of the CLS file were created when comparing different subsets of samples. Gene expression comparison between other platforms can be found in supplemental Methods. Expression data discussed in this publication have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession number GSE28460.
Copy number analysis
DNA was isolated using QIAGEN Gentra Puregene Core Kit A. Copy number analyses were performed following the procedures described previously5 with slight modifications (supplemental Methods). DNA from patient samples was applied to Affymetrix Genome-Wide Human SNP Array Version 6.0 as per the manufacturer's recommendation. Raw signal intensities were first summarized by CNAT Version 4.0 using an independent set of germline DNA arrays (250 ALL patients enrolled on COG 9906) as reference. Signal intensities were then normalized to a median intensity value consistent across chips. Segmented DNA copy numbers were inferred using DNAcopy, a circular binary segmentation algorithm developed on the R platform, and normalized to a median of 2 copies. Somatic copy number abnormalities (CNAs) in the diagnosis and relapse sample were determined by comparison with the matched germline sample as previously described.5
Methylation analysis
Genomic DNA isolated from patient samples (as described in “Copy number analysis”) was bisulfite converted with the MethylEasy Xceed Bisulphite Conversion kit (Human Genetic Signatures) according to the manufacturer's protocol. Bisulfite-converted DNA was processed for hybridization to the Infinium HumanMethylation27 BeadChip following the manufacturer's directions (Illumina). Array chemistry and processing were performed by ServiceXS (www.serviceXS.com). Imaging was performed using the Illumina BeadArray Reader, and images were processed with the DNA methylation module within BeadStudio Version 3.0 software. Additional methods describing methylation classification are described in supplemental Methods. Methylation data discussed in this publication have been deposited in the National Center for Biotechnology Information GEO (http://www.ncbi.nlm.nih.gov/geo) and are accessible through GEO Series accession number GSE28461.
Integration and validation
For correlation between methylation and gene expression, a methylation status transition matrix was coded with 0, 1, and 2 to indicate no methylation status change, hypermethylation, and hypomethylation, respectively. Gene expression data were reformatted to reflect the pair-wise change between diagnosis and relapse. To identify those genes with the highest likelihood of playing a role in chemoresistance across multiple biologic subtypes, we prioritized those genes with the greatest correlation between methylation and gene expression. The correlation between methylation and gene expression was normally distributed, with the mean concordance of 37.4% ± 11.5% (supplemental Figure 1), and we focused on genes that were at least 1 SD above the mean with respect to methylation/expression concordance. Genes were considered to be concordant for methylation and copy number status if they were hypomethylated or hypermethylated and had a focal gain or loss of copy number in at least 2 patients. Correlation between copy number and gene expression was calculated using CNAs that occurred in 2 or more patients compared with gene expression changes occurring within those same patients.
Validation of gene expression of several of the top target genes was performed by quantitative RT-PCR. Genes that were differentially expressed between diagnosis and relapse were validated in an independent cohort of patients. Those genes that were differentially expressed between diagnosis and relapse and distinct between late or early relapse were validated using a combination of independent and previously analyzed patient samples. Genes that were differentially expressed, with concordant methylation changes, were validated in the individual patients where the methylation changes were seen. Quantitative RT-PCR was performed as previously described.3 Quantitative PCR primers are listed in supplemental Methods.
Validation of methylation status was performed using the SEQUENOM MassArray Epityper platform. The locus surrounding the exact CpG sites from the Illumina array that passed the matrix threshold were assayed in all the patient samples. See supplemental Methods for primer sequences.
An integrated pathway analysis was performed using Ingenuity Pathway Analysis (Version 8.8) to evaluate pathways that were altered at relapse. Analysis was performed using genes that were differentially expressed or methylated at relapse, as well as those with a focal CNA at relapse. For gene expression, genes with a P value < .002 and family-wise error rate (FWER) < 0.1 from the new 49 pairs, as well as those in the expanded early and late relapse cohort were used. The 984 genes that were hypomethylated or hypermethylated were included. Lastly, genes with CNA were included if there were 2 or more focal deletions at relapse and 4 or more focal gains at relapse. A cut-off of 4 or more focal gains was used to be more discriminatory because there were a much larger number of genes with 2 focal gains compared with 2 focal losses.
Statistical analysis
Significance values for gene expression data were computed using false discovery rate (Benjamini and Hochberg procedure), FWER, feature-specific P value, and Bonferroni correction. Pair-wise t test was used to compare methylation status at diagnosis and relapse. Pair-wise t test was used for quantitative RT-PCR validation of gene expression changes from diagnosis to relapse in experiments with > 9 samples. Wilcoxon paired signed-rank test was used for quantitative RT-PCR validation in experiments with 9 or less samples.
Results
Multiplatform analysis of relapsed pediatric ALL
Frozen bone marrow aspirates taken at the time of original diagnosis and subsequent relapse were obtained for 61 pediatric ALL patients (122 samples) from the COG tissue bank. From a single aliquot of Ficoll-enriched blast cells, total RNA and genomic DNA were extracted. Gene expression profiling completed on Affymetrix U133Plus2 array, copy number alterations on Affymetrix SNP Version 6.0, and promoter methylation analysis on the Infinium Human Methylation27 BeadArray platform was performed in 49, 56, and 33 pairs, respectively (supplemental Table 1). Fifty-two of the patients were studied on more than one platform, with 25 patients studied on all 3 platforms. In addition, there were 21 patients who were studied by gene expression and CNA analysis and 6 who were studied by CNA and promoter methylation analysis.
Gene expression analysis reveals a distinct profile for pediatric relapsed ALL patients at both early and late time points
Gene expression analysis was performed in 49 patients: 27 patients had an early marrow relapse and 22 patients had a late marrow relapse (< 36 months and ≥ 36 months, respectively). Unsupervised analysis of the pairs did not reveal clustering of the early and late pairs (supplemental Figure 2). Twenty-one pairs showed diagnosis and relapse samples clustering adjacently, indicating quite similar diagnosis/relapse gene expression profiles. We examined the correlation coefficient of the gene expression pattern at diagnosis and relapse for each individual patient. In contrast to our previous data,3 the correlation between diagnosis and relapse samples did not vary as a function of time to relapse (supplemental Figure 3).
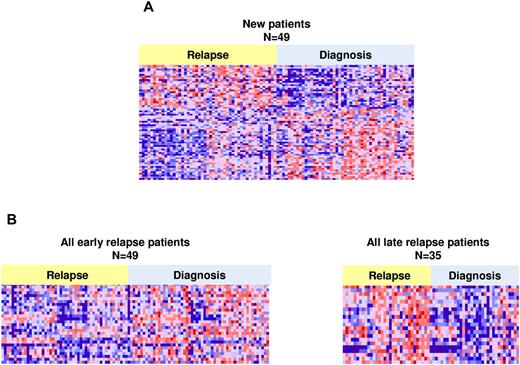
Using a false discovery rate < 10% and a P value < .002, we defined a signature of 3609 probe sets that were differentially expressed between diagnosis and relapse. Of these, 2222 were down-regulated and 1387 were up-regulated. In an effort to identify those genes that were probably of highest statistical and potentially biologic significance, a shorter, more stringent list of differentially expressed genes was identified using an FWER < 0.1 and P value < .002. FWER was chosen as it is a more stringent statistic and better for multiple, pair-wise comparisons. There were 64 probes representing 60 genes: 38 genes down-regulated and 22 genes up-regulated (Figure 1A; supplemental Table 2). Genes that were up-regulated included FANCD2, FOXM1, CENPM, and OBSL1 and down-regulation was seen in ALOX5 and ITPR1.
Heat maps of differentially expressed genes at diagnosis and relapse. Significant differentially expressed probes between diagnosis and relapse (A) in 49 relapse pairs (64 probes, 60 genes). (B) Heat maps for early relapse (n = 49, 166 probes, 148 genes) and late relapse (n = 35, 177 probes, 166 genes), respectively. Probes shown have a FWER less than < 0.1 and P value < .002.
Heat maps of differentially expressed genes at diagnosis and relapse. Significant differentially expressed probes between diagnosis and relapse (A) in 49 relapse pairs (64 probes, 60 genes). (B) Heat maps for early relapse (n = 49, 166 probes, 148 genes) and late relapse (n = 35, 177 probes, 166 genes), respectively. Probes shown have a FWER less than < 0.1 and P value < .002.
Given differences in outcome between patients who relapse early in therapy compared with those who relapse late, we aimed to determine whether distinct gene expression signatures characterized these 2 groups. To achieve optimal statistical power for detecting expression differences between early and late relapse cases, we combined data from the 49 patients reported here with our previously published patients,3 giving a total of 49 early and 35 late pairs (84 pairs total, 168 samples). Whereas the current patients were analyzed on the Affymetric U133Plus2 microarrays, the previous patients had been analyzed using the U133A microarrays. We analyzed pair-wise (diagnosis to relapse) gene expression differences for those probes present on both platforms (supplemental Methods). The most significantly differentially expressed genes were identified using an FWER < 0.1 and P value < .002. Early and late relapse was characterized by expression changes in different sets of genes (Figure 1B). The early relapse signature consisted of 148 genes (166 probes, supplemental Table 3), and the late relapse signature consisted of 166 genes (177 probes, supplemental Table 4) with 12 genes in common. FOXM1, exonuclease NEF-sp, BIRC5, NCAPH, GTSE1, CENPM, KIAA0101, C10orf56, BUB1B, UBE2V1, POLQ, and TMEM97 were up-regulated in both early and late relapse. Interestingly, a set of genes, including PAICS, TYMS, IMPA2, CAD, ATIC, and GART, involved in nucleotide biosynthesis and folate metabolism, were specifically up-regulated at late relapse. Changes in RNA expression at relapse, in early, late, or the entire cohort were confirmed in the majority of genes tested by quantitative RT-PCR (P < .05; Table 1). Failure to validate the results obtained by expression arrays may be the result of relatively small sample sets for PCR validation in some cases.
Validation of gene expression changes by quantitative RT-PCR
| Gene . | Analysis list* . | Test . |
|---|---|---|
| t test | (E,L) | |
| FANCD2 | Diagnosis vs relapse | 0.011 |
| FOXM1 | Diagnosis vs relapse | 0.002 |
| HRK | Diagnosis vs relapse | 0.590 |
| CENPM | Diagnosis vs relapse | 0.017 |
| SMEK2 | Diagnosis vs relapse | 0.140 |
| IFIT5 | Diagnosis vs relapse | 0.110 |
| LAPTM4B | Diagnosis vs relapse | 0.280 |
| OBSL1 | Diagnosis vs relapse | 0.002 |
| CCNB2 | Early relapse | 0.055, 0.089 |
| KCNH2 | Early relapse | 0.013, 0.065 |
| TYMS | Late relapse | 0.140, 0.028 |
| ATIC | Late relapse | 0.028, 0.009 |
| CAD | Late relapse | 0.051, 0.035 |
| GART | Late relapse | 0.062, 0.034 |
| PAICS | Late relapse | 0.037, 0.013 |
| IMPA2 | Late relapse | 0.030, 0.001 |
| RAD51AP1 | Late relapse | 0.100, 0.002 |
| BIRC5 | Early and late relapse | 0.005 |
| BUB1B | Early and late relapse | 0.018 |
| Wilcoxon test† | ||
| WT1 | M/E | 0.05 |
| GATA5 | M/E | 0.05 |
| CSMD1 | M/E | 0.05 |
| COL6A | M/E | 0.05 |
| CDKN2A | M/E | 0.05 |
| TOP1MT | M/E | > 0.05 |
| NOTCH4 | M/E | > 0.05 |
| Gene . | Analysis list* . | Test . |
|---|---|---|
| t test | (E,L) | |
| FANCD2 | Diagnosis vs relapse | 0.011 |
| FOXM1 | Diagnosis vs relapse | 0.002 |
| HRK | Diagnosis vs relapse | 0.590 |
| CENPM | Diagnosis vs relapse | 0.017 |
| SMEK2 | Diagnosis vs relapse | 0.140 |
| IFIT5 | Diagnosis vs relapse | 0.110 |
| LAPTM4B | Diagnosis vs relapse | 0.280 |
| OBSL1 | Diagnosis vs relapse | 0.002 |
| CCNB2 | Early relapse | 0.055, 0.089 |
| KCNH2 | Early relapse | 0.013, 0.065 |
| TYMS | Late relapse | 0.140, 0.028 |
| ATIC | Late relapse | 0.028, 0.009 |
| CAD | Late relapse | 0.051, 0.035 |
| GART | Late relapse | 0.062, 0.034 |
| PAICS | Late relapse | 0.037, 0.013 |
| IMPA2 | Late relapse | 0.030, 0.001 |
| RAD51AP1 | Late relapse | 0.100, 0.002 |
| BIRC5 | Early and late relapse | 0.005 |
| BUB1B | Early and late relapse | 0.018 |
| Wilcoxon test† | ||
| WT1 | M/E | 0.05 |
| GATA5 | M/E | 0.05 |
| CSMD1 | M/E | 0.05 |
| COL6A | M/E | 0.05 |
| CDKN2A | M/E | 0.05 |
| TOP1MT | M/E | > 0.05 |
| NOTCH4 | M/E | > 0.05 |
E,L indicates diagnosis vs early relapse, diagnosis vs late relapse; and M/E, correlation of methylation and gene expression.
All comparisons are from diagnosis to relapse.
Wilcoxon paired signed-rank test.
DNA copy number analysis identifies relapse specific alterations
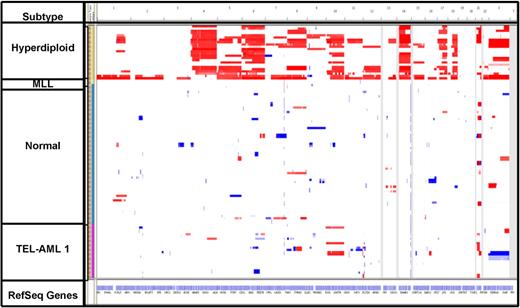
Genome-wide copy number analysis of 56 patients revealed various numbers of genetic lesions, ranging from 4 to 118 CNAs per sample. These CNAs included gross copy number changes consistent with conventional cytogenetic analysis; however, the majority of the lesions were cryptic with a median size of 133 kb (45.8% < 100 kb, and 32.4% < 1 Mb). Overall, the majority of CNAs observed were shared between diagnosis and relapse in the same patient: 82.3% of CNAs at diagnosis were present at relapse, and 87.2% of CNAs at relapse were also seen at diagnosis (Figure 2). All 56 cases exhibited genetic lesions persisting from diagnosis to relapse, whereas 52 of 56 patients (92.8%) also either lost or acquired a genetic lesion.
Heat map of total copy number data. Diagnosis samples (yellow) and relapse samples (brown) appear next to each other. Samples are grouped according to cytogenetic subtype.
Heat map of total copy number data. Diagnosis samples (yellow) and relapse samples (brown) appear next to each other. Samples are grouped according to cytogenetic subtype.
CNAs involving several previously reported transcription regulators, essential in early lymphoid specification and B lineage commitment, and cell cycle genes were also seen in this relapsed ALL cohort (Table 2). One of the most frequent CNAs involved deletions of CDKN2A/B at 9p21.3, occurring in 20 patients (35.7%), with deletions in 17 patients persisting from diagnosis to relapse. In addition, occurring on chromosome 9p, deletions of PAX5 were observed in 14 (25%) patients, with 7 patients exhibiting PAX5 deletion at both diagnosis and relapse. Copy number losses of IKZF1 (7p12.2) were present in 22 patients (39.3%), 16 (28.6%) of which occurred at both diagnosis and relapse. Two patients developed IKZF1 deletions at relapse that were not present at diagnosis. Similarly, focal deletion of EBF1 (5q33.3) was shared at diagnosis and relapse in 5 cases (8.9%) and present only at relapse in 1 additional case. When the presence of these commonly occurring lesions was compared with CNAs profiled in a cohort of B-precursor diagnosis samples,6 similar incidences were reported for CDKN2A/B (35.7% vs 33.9%) and PAX5 (25% vs 29.7%), respectively. However, there was an increased prevalence noted for IKZF1 (39.3% vs 8.9%), EBF1 (12.5% vs 4.2%), and BTG1 (14.3% vs 6.7%) CNAs in this relapse cohort compared with patients at diagnosis6 (supplemental Table 5).
Common CNA events at diagnosis or relapse ranked by overall observance
| Common CNAs . | No. of total CNAs (focal) (n = 56) . | Incidence, % . | No. of total CNAs* (focal) (n = 76) . | Overall observed, % . | No. of relapse specific events* (focal) . |
|---|---|---|---|---|---|
| Deletions | |||||
| CDKN2A/B | 20 (15) | 35.7 | 32 (23) | 42.1 | 2 (1) |
| IKZF1 | 22 (19) | 39.3 | 29 (23) | 38.2 | 4 (1) |
| VPREB1 | 21 (18) | 37.5 | 27 (23) | 35.5 | 6 (5) |
| PAX5 | 14 (9) | 25 | 22 (14) | 28.9 | 3 (2) |
| EBF1 | 7 (5) | 12.5 | 12 (10) | 15.8 | 3 (2) |
| BTG1 | 8 (8) | 14.3 | 12 (12) | 15.8 | 2 (2) |
| Common CNAs . | No. of total CNAs (focal) (n = 56) . | Incidence, % . | No. of total CNAs* (focal) (n = 76) . | Overall observed, % . | No. of relapse specific events* (focal) . |
|---|---|---|---|---|---|
| Deletions | |||||
| CDKN2A/B | 20 (15) | 35.7 | 32 (23) | 42.1 | 2 (1) |
| IKZF1 | 22 (19) | 39.3 | 29 (23) | 38.2 | 4 (1) |
| VPREB1 | 21 (18) | 37.5 | 27 (23) | 35.5 | 6 (5) |
| PAX5 | 14 (9) | 25 | 22 (14) | 28.9 | 3 (2) |
| EBF1 | 7 (5) | 12.5 | 12 (10) | 15.8 | 3 (2) |
| BTG1 | 8 (8) | 14.3 | 12 (12) | 15.8 | 2 (2) |
Selected CNAs were listed and ranked by overall observance.
Inclusion of 20 patients previously published in Yang et al.5
In line with our previous findings, we again observed recurrent relapse specific CNAs involving mismatch repair genes (eg, MSH6 gene at chromosome 2p16.3 in 2 patients).5 In both cases, a hemizygous deletion was seen at relapse involving MSH6, FBXO11, and KCNK12 (supplemental Figure 4). In one case, the deletion was restricted to MSH6, FBXO11, and KCNK12 exclusively, and in the other, the deletion was more extensive, involving several additional genes within the region. Similarly, we also observed focal deletions at relapse in a small number of other genes (supplemental Table 6), including BTG1. BTG1 was found to be deleted in a total of 8 patients, 2 of which harbored relapse specific deletions. Deletions were also found in NR3C1 and were specific to relapse in 2 patients (one focal, one nonfocal).
DNA methylation profiling distinguishes diagnosis from relapse samples
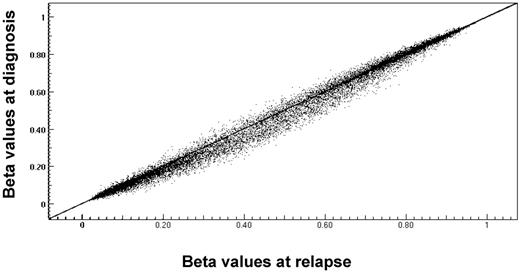
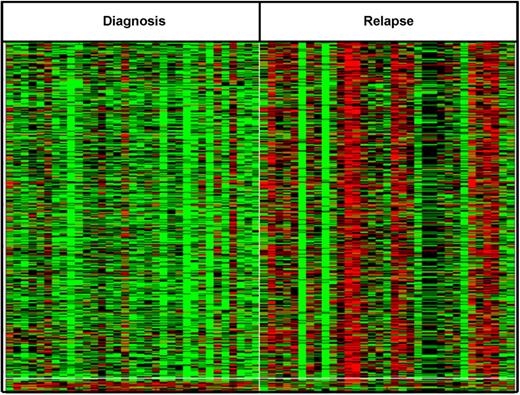
Genome-wide DNA methylation profiling was performed on 33 pairs. To explore whether these patients showed a relapse specific change in global promoter methylation, the DNA methylation patterns seen at relapse were compared with those from diagnosis. Overall, the relapse cohort had a distinctly higher CpG methylation level compared with diagnosis, P value = .000002 (Figure 3). The change in methylation status of individual CpG sites, from diagnosis to relapse, was evaluated using a methylation state transition matrix (supplemental Methods; supplemental Figure 5). Using this matrix, 11 336 CpG sites (41%) showed no change in methylation status. However, to identify genes whose changes in methylation status might be correlated with relapse across biologic subtypes, we looked for changes that were shared among a significant number of the pairs analyzed. We identified 1228 CpG sites (top 5%) that included CDKN2A, a benchmark gene already known to be hypermethylated at relapse (supplemental Figure 6). Of these, 1147 CpG sites (corresponding to 905 genes) were preferentially hypermethylated and 81 CpG loci (corresponding to 79 genes) were preferentially hypomethylated on relapse (Figure 4). No genes appeared to undergo a change involving both hypermethylation and hypomethylation. Supplemental Table 7 lists genes that are hypermethylated or hypomethylated on relapse.
Comparison of average methylation β values of genes at diagnosis and relapse. CpG loci β values were graphed per patient to compare overall change from diagnosis to relapse. Central diagonal line through graph indicates no change between diagnosis and relapse sample. Points above line indicate a change to hypomethylation at relapse; and points below line indicate change to hypermethylation at relapse.
Comparison of average methylation β values of genes at diagnosis and relapse. CpG loci β values were graphed per patient to compare overall change from diagnosis to relapse. Central diagonal line through graph indicates no change between diagnosis and relapse sample. Points above line indicate a change to hypomethylation at relapse; and points below line indicate change to hypermethylation at relapse.
Differentially methylated probes between diagnosis and relapse show unique signature. Top differentially methylated genes at relapse. A total of 867 CpG sites were altered between diagnosis and relapse in more than 30% of patients with P < .01. Red represents hypermethylation; and green, hypomethylation.
Differentially methylated probes between diagnosis and relapse show unique signature. Top differentially methylated genes at relapse. A total of 867 CpG sites were altered between diagnosis and relapse in more than 30% of patients with P < .01. Red represents hypermethylation; and green, hypomethylation.
Cross platform integration of genomic profiles identifies relapse-specific pathways
Each of the genome-wide profiling methods identified individual genes that may be involved in the development of chemotherapy-resistant leukemia. To create a hierarchy of genes involved in relapse and to strengthen the association with relapse specific events, we hypothesized that genes identified through more than one method might be of higher biologic significance. It is possible that identification of genes hypermethylated/hypomethylated or deleted/amplified with concordantly altered expression at relapse may indicate a relapse-driving event. A particular pathway may be altered by different mechanisms in distinct patients with the same end biologic effect.
First, to correlate changes in DNA methylation with downstream effects on gene expression, we integrated the methylation status of the 984 differentially methylated genes with gene expression changes in the 25 patients that were assayed on both platforms using a maximum likelihood procedure (supplemental Methods; supplemental Figure 5). There were 251 genes that exhibited promoter hypermethylation with concordant down-regulation and 29 genes that showed hypomethylation with concordant up-regulation of expression on relapse, suggesting that DNA methylation is a mechanism regulating expression of these genes in relapsed ALL (supplemental Table 8). Concordant changes in RNA expression were confirmed in individual patients for a subset of genes (Table 1).
We also correlated the 356 genes with CNA that were shared in 2 or more patients with gene expression changes in the 46 patients that were assayed on both platforms. Of the 612 individual copy number events in these patients, 236 (39%) had concordant gene expression changes (supplemental Table 9), a value in line with similar analyses in other tumor types.7
Next, we compared the differentially methylated genes with those showing CNAs. Ten genes were hypermethylated and focally lost in 2 or more patients (RERG, LRRC3, PTPRT, FUCA2, OGDHL, CDKN2A/2B, COL6A2, PTPRO, and CSMD1), whereas 5 genes were hypomethylated and gained (SLC7A7, TMEM128, NRM, NOTCH4, and TOP1MT). Further integration with gene expression yielded 4 of these genes (CDKN2A, COL6A2, PTPRO, and CSMD1) that were hypermethylated and down-regulated at relapse, and also focally deleted. Two genes (NOTCH4 and TOP1MT) were hypomethylated and up-regulated at relapse, with focal amplification. These CNAs, although focal, were seen at diagnosis and relapse, except for CDKN2A, which had some relapse specific deletions. Most of these genes have been previously implicated in different types of cancer; furthermore, epigenetic regulation has previously been observed for some of them.8 The copy number alterations occurred in the same patients as the methylation changes (PTPRO and CDKN2A) or in different patients (CDKN2A, COL6A2, CSMD1, NOTCH4, and TOP1MT), suggesting that some patients do indeed have > 1 dysregulating event in a single gene and in other patients only one event is seen. The methylation status of the promoter regions for all 6 genes was independently validated using SEQUENOM. The change in methylation status from diagnosis to relapse of CDKN2A, COL6A2, PTPRO, and NOTCH4 was validated in a majority of patients (supplemental Figure 7). Moreover, significant methylation changes for these 4 genes were detected in additional patients not originally detected by the Illumina array. An alteration in methylation state was not validated in CSMD1 and TOP1MT because the SEQUENOM assay was not optimal for these genes.
Finally, to identify biologic pathways that were differentially regulated at relapse, Ingenuity Pathway Analysis software was used on a combined list of genes that were altered on any of the 3 platforms and an integrated list of genes that appeared on at least 2. One of the top pathways identified was the WNT/β-catenin pathway (P = .009 and P = .002 for all or shared genes, respectively). Many known negative regulators of the WNT pathway were deleted and/or hypermethylated and/or down-regulated at relapse, including the direct WNT inhibitors9,10 : WIF1, PTPRO, sFRP2, sFRP4, sFRP5, FZD10, DKK2, and DKK3. Inhibitors of the β-catentin/TCF/LEF complex, including APC, WT1, and several of the cadherin (CDH1, CDH11, and CD13) and SOX genes (SOX2, SOX3, SOX8, SOX9, SOX11, SOX14 and SOX21) were also all negatively regulated. In accordance, we observed up-regulation of BIRC5, a downstream target of the WNT pathway, by gene expression array and quantitative RT-PCR. PTPRO is also normally induced by the WNT/β-catenin pathway and functions as a negative feedback inhibitor by binding to WNT and blocking its association with other receptors.11 In our cohort, this negative feedback loop may be interrupted because of the hypermethylation of PTPRO. Indeed, rather than induction of expression, PTPRO is down-regulated. Several of the other top networks identified by Ingenuity Pathway Analysis centered around genes in the MAPK cascade, which included up-regulation of BRAF, MAP2K1 (MEK1), and MAPK1 (ERK2). Interestingly, crosstalk has been observed between these 2 pathways.12,13
Discussion
Genome-wide expression analysis of paired diagnosis/relapse samples provides an opportunity to discover the biologic pathways that may contribute to drug resistance and has been performed previously by several groups, including ours.3,14-16 Our current results examining an expanded cohort confirm many of our earlier findings but also offer new insights into the underlying mechanisms of relapse.
We validated 56 of the 126 differentially expressed probes that were previously identified in our smaller cohort of B-precursor cases (false discovery rate < 0.1, P < .019).3 Many of the differentially expressed genes play a role in cell cycle, DNA replication and repair, and cell death, as has been previously described.3,14-16 BIRC5 is particularly interesting, as it is associated with a wide range of cancers and has been shown to be a marker of poor prognosis. We have demonstrated that down-regulation of BIRC5 in vitro leads to increased chemosensitivity,17 and we have established a pediatric clinical trial targeting BIRC5 with a locked nucleic acid oligonucleotide for relapsed ALL (www.clinicaltrials.gov: #NCT01186328). In addition to confirming many of our previous findings, this current analysis has identified additional genes of interest, including up-regulation of FOXM1, an oncogenic transcription factor, which is expressed in all embryonic tissues and proliferating cells. Similar to BIRC5, FOXM1 is also often up-regulated in many types of human cancers, including glioblastoma, breast cancer, and colorectal cancer.18,19
Another new finding of this study is the identification of distinct differential gene expression signatures associated with the timing of relapse. Although the absolute gene expression values at diagnosis and relapse vary considerably among individual patients, the change in expression during that time is consistent across patients. Of particular interest is the up-regulation of genes involved in nucleotide biosynthesis and folate metabolism in late relapse, but not early relapse. PAICS, TYMS, CAD, ATIC, and GART are involved in purine and/or pyrimidine synthesis, and it is interesting that these genes are up-regulated in late relapse, given that methotrexate and mercaptopurine are integral components of maintenance therapy.
The larger number of samples analyzed here also allows us to better compare the frequency of CNAs contributing to relapse versus newly diagnosed ALL.6 The prevalence of CNA events in CDKN2A/B and PAX5 in our relapse cohort were similar to that that seen in samples from newly diagnosed patients indicating a role in transformation, but not necessarily in resistant disease. In contrast, deletions involving IKZF1, VPREB1, and EBF1 were all enriched in our cohort of patients who relapsed. The IKZF1 deletion was found in 39.3% of relapse patients, with 6 patients harboring the lesion in exons 3 to 6, which leads to expression of a dominant negative isoform.20,21 Both IKZF1 and VPREB1 were recently associated with a poor prognosis when detected at diagnosis,22,23 which is consistent with our data.
Somatic deletions in genes that have previously been linked to chemoresistance were also seen at relapse, and these may contribute to disease recurrence. Decreased expressions of MSH6, BTG1, and NR3C1 have been linked to resistance to thiopurines and alkylators (MSH6) and glucocorticoids (BTG1 and NR3C1).5,24 We previously reported deletions of MSH6 at relapse associated with ex vivo resistance to thiopurines and others have observed deletions in patients with brain tumors who fail temozolomide therapy.5,25 We also observed 2 relapse specific deletions of NR3C1, the glucocorticoid (GC) receptor. Functional NR3C1 is required for response to GC; therefore, somatic deletion at relapse is likely to lead to resistance to this class of drugs. Indeed, it has been shown that the LC50 values for prednisone and dexamethasone are higher in relapsed blasts than diagnostic blasts.26 In addition to the antiproliferative and apoptotic activity of BTG1, it was also recently shown that BTG1 expression also correlates with GC resistance.24 Loss of BTG1 expression causes GC resistance both by reducing GC receptor expression and by controlling GC receptor-mediated transcription, as BTG1 is an essential binding partner of PRMT1, which is required to promote GC-receptor expression. Overall, 14.3% of patients harbored deletions in BTG1 (compared with ∼ 7% in diagnosis and 10% in high-risk diagnosis cases6,27 ).
Genome-wide methylation studies in both acute myelogenous leukemia and ALL have been used to identify distinct biologic subtypes and to create outcome prediction models.28-31 We discovered that the chemotherapy-resistant relapse samples are hypermethylated compared with matched diagnosis samples. This is consistent with a report that chemotherapy-resistant infant MLL rearranged ALL is globally hypermethylated compared with MLL-wild type ALL and normal controls.31,32 Another study identifying a list of methylation-targeted genes in ALL found an inverse correlation between the number of methylated genes and overall survival.33
Genome-wide and gene-targeted methylation assays have identified many cancer-related genes as substrates for methylation and altered gene expression. In this study, 6 genes were significantly altered from diagnosis to relapse when assayed on all 3 platforms: CDKN2A, PTPRO, CSMD1, COL6A, NOTCH4, and TOP1MT. We were able to validate gene expression changes in CSMD1, COL6A, and CDKN2A by quantitative RT-PCR in the exact patient samples that were predicted to have altered methylation with concordant expression. For PTPRO, we were unable to detect a PCR product at diagnosis to validate down-regulation of expression. This may have been because of an already low level of expression at diagnosis. Quantitative RT-PCR results for NOTCH4 and TOP1MT did not meet statistical significance. It is possible that the small sample size (5 and 4 patients, respectively) was a factor. CDKN2A, PTPRO, and CSMD1 have previously been identified as tumor suppressors in multiple cancers34-37 and are epigenetically silenced by DNA hypermethylation in ALL,38 chronic lymphocytic leukemia,8 hepatocellular carcinoma,36 lung cancer,37 and squamous cell carcinoma.39 For these genes, promoter hypermethylation correlated with down-regulation of mRNA expression, and expression of these genes could be reactivated by treatment with 5-azacytidine.39-41 Although a direct role of COL6A, encoding the collagen protein VI, has not been identified in cancer initiation/progression, the gene has been identified by gene expression analysis as part of a 10-gene classifier for lung squamous cell carcinoma42 and by genome-wide methylation arrays as being hypermethylated in pediatric ALL.30
Overall, our results indicate that multiple genetic lesions, unique to individual patients, occur in ALL relapse and differ between early and late relapse. Some lesions offer an obvious explanation for drug resistance, such as deletions of BTG1 and NR3C1, which account for the well-known resistance of leukemic blasts to glucocorticoids. Therefore, therapeutic efforts to restore glucocorticoid receptor expression, which has been accomplished with histone deacetylase inhibitors,43 should be a therapeutic priority. Moreover, overexpression of TYMS has been linked to methotrexate resistance, whereas in vitro resistance to methotrexate has been linked with amplification of CAD.44 Similarly, both PAICS and GART are part of the purine metabolic pathway. Down-regulation of these genes correlates with the decreased de novo purine synthesis in EVT6/RUNX1 ALL and has been postulated to account for the chemosensitivity of this genetic subtype.45 Overexpression in the blasts of patients who have a late relapse after prolonged maintenance therapy, offers an explanation for resistance to antimetabolites. Thus, increasing the dose and/or schedule of antimetabolite therapy may prevent and/or better treat late relapse. However, these results are not straightforward, as overexpression of IMPA2 correlates with an increase in methotrexate polyglutamation46-48 and we see up-regulation of IMPA2.
In conclusion, we were able to harness the power of 3 different, yet complementary, platforms to identify differentially regulated genes and, consequently, biologic pathways that could have relevance to relapsed pediatric ALL. One of the main pathways identified by this approach was the WNT/β-catenin pathway. The WNT pathway has previously been shown to have a role in controlling proliferation, survival, and differentiation of hematopoietic stem cells.47 It is frequently mutated in cancers and more recently was linked to hematologic malignancies, including acute myelogenous leukemia and ALL. Dysregulation of the WNT pathway in cancer usually occurs through one of 2 mechanisms: down-regulation of negative regulators or activation and stabilization of β-catenin through mutation. Recently, several other papers have identified epigenetic regulation of the WNT inhibitors, DKK3, sFRP2,4,5, and WIF1 in acute myelogenous leukemia and ALL.48,49 Notably, we identified the WNT pathway through an integrated analysis of 3 genomic platforms and, while we observe hypermethylation of these same genes and others, we also report concordant gene expression changes and in some instances, complete loss of an allele. In addition to WNT signaling, we also observed constitutive activation of the MAPK cascade, which is a common event in cancer, including pediatric ALL.50 Crosstalk and correlation between up-regulation of the WNT pathway and MAPK signaling have previously been documented in bladder cancer12 and colorectal cancer,13 but not pediatric ALL. Activation of the WNT pathway and genes in the MAPK cascade appears to be a theme shared by many cases of relapsed ALL and requires further preclinical investigation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Children's Oncology Group for patient specimens.
This work was supported by the National Institutes of Health (R01 CA140729; W.L.C.) and New York University Cancer Center (support grant 5 P30 CA16087-30). L.E.H. was supported by St Baldrick's Foundation and the American Society of Hematology. J.A.M. was supported by (National Institutes of Health T32CA009161). S.P.H. is the Ergen Family Chair in Pediatric Cancer. This work was supported by National Institutes of Health grants to the Children's Oncology Group (U10 CA98543, U10 CA98413, and U24 CA114766).
National Institutes of Health
Authorship
Contribution: L.E.H., J.A.M., J.Y., N.W., D.B., and D.J.M. designed and performed research, collected, analyzed, and interpreted data, performed statistical analysis, and wrote the manuscript; J.W. analyzed and interpreted data, performed statistical analysis, and wrote the manuscript; W.Y. analyzed and interpreted data; G.C. collected data; S.P.H., E.R., R.S., and M.V.R. designed the research and wrote the manuscript; and W.L.C. designed and directed the research, analyzed and interpreted data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for L.E.H. is Stony Brook University, Stony Brook, NY.
Correspondence: William L. Carroll, New York University Cancer Institute, Smilow 1201, 522 First Ave, New York, NY 10016; e-mail: william.carroll@nyumc.org.
References
Author notes
L.E.H., J.A.M., and J.Y. contributed equally to this study.